Abstract
Hepatocellular carcinoma (HCC) is among the leading causes of cancer-related death in China. Deregulation of microRNA (miRNA) contributes to HCC development by influencing cell growth, apoptosis, migration or invasion. It has been proved that miR-940 plays important roles in various cancers. Here we investigated the role of miR-940 in HCC. We found that miR-940 was remarkably decreased in HCC tissues and cell lines. Importantly, lower miR-940 expression in HCC tissues significantly correlated with the reduced patient’s survival rate. Overexpression of miR-940 inhibited HCC cell line growth and induced cell apoptosis, and vice versa. Estrogen-related receptor gamma (ESRRG) was targeted by miR-940, and suppression of ESRRG inhibited HCC cell lines growth and induced cell apoptosis. In conclusion, we found that a lower level of miR-940 in HCC promoted cellular proliferation via ESRRG, which may lead to the short survival period of HCC patients.
Keywords: Cell proliferation, estrogen-related receptor gamma, hepatocellular carcinoma, miR-940, patients’ survival
Hepatocellular carcinoma (HCC) is among the leading causes of cancer-related death in Asia, especially in China.1,2 Poor prognosis makes HCC the third leading cause of cancer-related mortality, responsible for 600 000 deaths annually cross the globe.3 Surgery is the most effective treatment with curative potential, but only 30–40% of HCC patients are diagnosed in the early stages and are candidates for potentially curative surgical therapy. Patients who have undergone radical tumor resection have a high rate of relapse, and the 5-year survival rate has been found to be only around 60% in well-selected patients.4,5 MicroRNA (miRNA) are a class of endogenous non-coding, single-stranded small regulatory RNA molecules, which are approximately 22 nucleotides in length.6 Besides multiple genetic and epigenetic changes of protein coding genes in HCC,2 growing evidence indicates that deregulation of miRNA can also contribute to HCC development by influencing cell growth, apoptosis, migration or invasion.7–23
Recently, several studies have revealed the role of miR-940 in various types of cancer. Ma et al.24 show that lower miR-940 promotes tumorigenesis in nasopharyngeal carcinoma cells via nestin. Rajendiran et al.25 reveal that miR-940 inhibits the migratory and invasive potential of cells, attenuates their anchorage-independent growth ability, and increases E-cadherin expression in prostate cancer.
In the present study, we investigated the role of miR-940 in HCC. We found that miR-940 was remarkably decreased in HCC tissues and cell lines. Importantly, lower miR-940 expression in HCC significantly correlated with shorter survival of HCC patients. In addition, the potential mechanism and targets of miR-940 were investigated. Our results demonstrate that miR-940 may be a useful diagnostic marker as well as a therapeutic agent for HCC.
Materials and Methods
Patients
Surgical specimens from 23 HCC patients and matched tumor-adjacent normal tissues were obtained postoperatively in 2011 from the Department of General Surgery, West China Hospital, Sichuan University (Chengdu, China). All patients gave signed, informed consent for their tissues to be used for scientific research. Ethical approval for the study was obtained from Sichuan University (Chengdu, China). All diagnoses were based on pathological evidence. The histological features of the specimens were evaluated by senior pathologists of West China Hospital according to the World Health Organization classification criteria. Tissues were obtained before chemotherapy and radiotherapy and were immediately frozen and stored at −80°C prior to qRT-PCR assay. 23 patients had been followed up for 3 years and complete clinical data were electronically recorded. The clinical information for the 23 patients is listed in Table S1.
Cell lines and reagents
All cell lines were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China). HEK293, Huh7 cell line and normal human hepatic cell line L-02 were maintained in RPMI1640 with 10% FBS (PAA Laboratories, Pasching, Austria). Human HCC cell lines HepG2, Hep3B, SMMC-7721, Huh7 and normal human hepatic cell line Chang were maintained in DMEM with 10% FBS (PAA Laboratories, Pasching, Austria). Antibodies specific to estrogen-related receptor gamma and β-actin were obtained from Sigma (Beijing, China).
RNA quantification
Expression of miR-940 was assessed using a mirVana qRT-PCR miRNA Detection Kit (Ambion, Austin, TX, USA). The primers were designed and synthesized by the Shengong Company (Shanghai, China). Small nucleolar RNA-U6 was used as an internal control.
MiRNA mimics, miRNA antisense, oligonucleotides, estrogen-related receptor gamma siRNA
MiRNA-940 mimics (Cat. no.MSY0004983), miR-940 ASO (Cat. no. MIN0004983), its negative control (Cat. no. MSY0002505), ESRRG siRNA-1 (Cat. no. SI00025977), ESRRG siRNA-2 (Cat. no. SI00025991), ESRRG siRNA-3 (Cat. no. SI00025998) and its negative control (Cat. no.1022076) were purchased from Qiagen (Venlo, Limburg, the Netherlands). MiRNA ASO, miRNA mimics, ESRRG siRNA and respective negative controls were transfected into cells at a concentration of 50 nM using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were collected for further experiments 48 h later.
Cell proliferation assay
Cell proliferation was assayed by MTT assay as described previously.26–28 After the treatment, cells (1 × 103 cells per well) were seeded into 96-well plates. The absorbance of the samples was measured with a spectrophotometer reader at 490 nm. Each assay was performed in triplicate and repeated three times independently.
Apoptosis analysis
For apoptosis analysis, the treated cells were harvested after 48 h, and then stained with PI-labeled and FITC-labeled anti-annexin-V antibody, and analyzed by FACS.
Western blot
After transfection, cells were subjected to western blot analysis with anti-ESRRG (1:100; Merck) or β-actin (1:1500; Santa Cruz Biotechnology, Dallas, TX, USA) antibodies. Cells were harvested and lysed in 0.5 mL of lysis buffer (10 mM Tris–HCl, pH 7.6, 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF, 0.1 mM Na3VO4, 1% Triton X-100, 1 mM phenylmethyl sulfonyl fluoride and protease inhibitor cocktail (Roche Applied Science, Branford, CT, USA). Protein (20 μg) was separated by SDS-PAGE on 10% gels. The proteins were electrophoretically transferred to PVDF membranes (Millipore, Billerica, MA, USA) and incubated with primary antibody, followed by incubation with an HRP-conjugated secondary antibody (Santa Cruz Biotechnology). After washing with TBS, the bound antibodies were visualized by enhanced chemiluminescence (Pierce, Rockford, IL, USA).
MicroRNA targets prediction
TargetScanHuman (www.targetscan.org/vert_61) was applied to identify the potential target of miR-940.29–32
Estrogen-related receptor gamma 3′UTR reporter analysis
Reporter gene analysis in this study was performed by Shengong Company (Shanghai, China). Briefly, after the construction of estrogen-related receptor gamma (ESRRG) 3′UTR reporter plasmids (pRL-ESRRG), mutation in the miR-940 seed regions of the ESRRG 3′UTR was generated using the QuikChang Multi site-directed mutagenesis kit (Promega, Fitchburg, WI, USA). RL reporter plasmids (3.6 fmol) and pGL3-control (500 ng for normalization) (Promega, Madison, WI, USA) were transfected with Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) into HEK293 (6 × 104 cells per well). Cells were collected after 48 h for assay using the Dual Luciferase reporter assay system (Promega, Madison, WI, USA).33
Statistical analysis
Data was presented as the mean ± SD from at least three independent experiments. The difference between groups was analyzed using the two-tailed Student’s t-test when only two groups were compared. The difference between groups was analyzed using anova when three or more than three groups were compared. The Wilcoxon matched-pairs signed rank test was used to determine whether there was a statistically significant difference in the expression of miR-940 between matched pairs. Correlation analysis was performed using Pearson’s two-tailed correlation coefficient analysis. Overall survival of patients was determined using the Kaplan–Meier method. Statistical analysis was performed using SPSS software (version 16.0). P < 0.05 was considered significantly different.
Results
Lower level of miR-940 in hepatocellular carcinoma tissues correlated with reduced patient survival rate
Initially, the miR-940 levels in 23 HCC tissues were assayed by qRT-PCR, and the matched tumor adjacent normal tissues were used as controls. We found that there were 17 pairs with lower miR-940 levels in HCC tissues than in the matched tumor adjacent normal tissues (Fig.1a). The mean level of miR-940 in HCC tissues and the tumor adjacent normal tissues was calculated, and we found that the mean level of miR-940 in HCC tissues was lower than in tumor adjacent normal tissues (Fig.1b). To evaluate the significance of miR-940 in HCC, the prognoses of the 23 HCC patients were investigated, and statistical analysis with the Kaplan–Meier method revealed that lower miR-940 levels in HCC tissues significantly correlated with reduced patient survival rates (Fig.1c). The median value of all 23 cases was chosen as the cutoff point for separating miR-940 high-expression cases from miR-940 low-expression cases.
Fig 1.
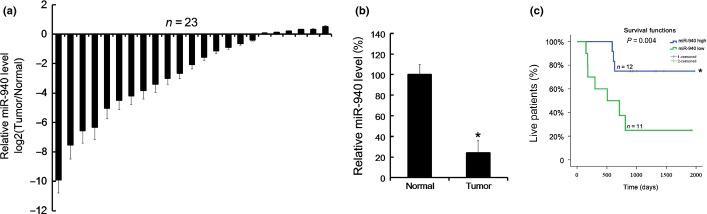
Low miR-940 level in hepatocellular carcinoma (HCC) tissues was correlated with a low survival rate of HCC patients. A total of 23 pairs of HCC tissues and the matched tumor-adjacent normal tissues were collected from West China Hospital, and were analyzed by qRT-PCR for miR-940 levels. The differences between tumor and matched normal tissues were compared using the formula log2(Tumor/Normal) (a). The mean miR-940 expression in the 23 pairs of HCC and matched tumor-adjacent normal tissues (b). Kaplan–Meier plot of overall survival in HCC patients post-operation according to the expression of miR-940. +, censored data (c). Data are mean ± SD of three separate experiments. $P < 0.05.
Overexpression of miR-940 inhibited hepatocellular carcinoma cell lines growth and induce cells apoptosis
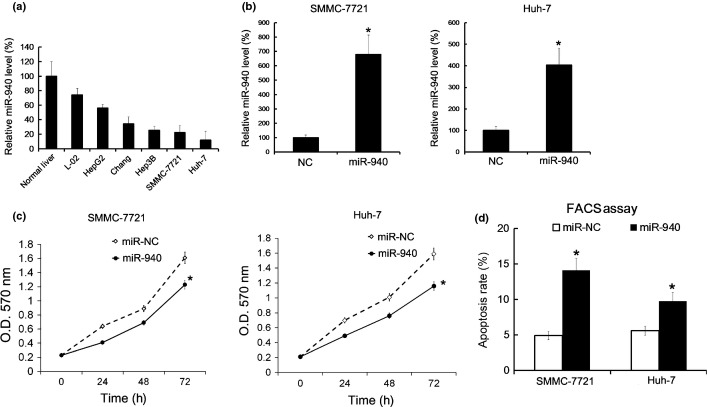
To investigate the role of miR-940 in HCC, we studied the effect of miR-940 on cell proliferation and apoptosis of HCC cell lines in vitro. The levels of miR-940 in normal liver tissue and HCC cell lines were assayed by qRT-PCR. SMMC-7721 and Huh-7 cell lines showed lower miR-940 levels than other cell lines and normal liver tissues (Fig.2a). Thus, we transfected SMMC-7721 and Huh-7 with miR-940 mimics to overexpress the miR-940 levels in vitro (Fig.2b). Then the cell proliferation was evaluated by MTT analysis, and we found that overexpression of miR-940 inhibited the proliferation of SMMC-7721 and Huh-7 cells (Fig.2c). Next, cell apoptosis were evaluated by FACS analysis, and we found that overexpression of miR-940 upregulated the apoptosis rate in both cell lines (Fig.2d).
Fig 2.
Transfection with miR-940 mimics inhibited proliferation of SMMC-7721 and Huh-7 and induced cell apoptosis. Normal liver tissues, L-02, HepG2, Chang, Hep3B, SMMC-7721 and Huh-7 were prepared for the miR-940 level by qRT-PCR analysis; the level of miR-940 in normal liver was arbitrarily defined as 100% (a). The level of miR-940 in SMMC-7721 and Huh-7 were analyzed by qRT-PCR 48 h after transfection of miR-940 mimics. The level of miR-940 in the miR-NC-treated group was arbitrarily defined as 100% (b). The Optical Density (OD) value was measured at the indicated time point in MTT analysis experiment, the cellular proliferation curves were shown (c). 48 h after transfection of miR-940 mimics, cell apoptosis was measured by Annexin V-FITC/PI double staining FACS analysis (d). All data are mean ± SD of three separate experiments. $P < 0.05.
Suppression of miR-940 promoted hepatocellular carcinoma cell line growth and inhibited cells apoptosis
As L-02 and HepG2 showed a relatively higher level of miR-940, we downregulated the miR-940 level in L-02 and HepG2 cells by miR-940 ASO transfection (Fig.3a). Similarly, the cell proliferation was evaluated by MTT analysis, and we found that suppression of miR-940 promoted the proliferation of L-02 and HepG2 cells (Fig.3b). Next, 48 h after miR-940 ASO transfection, the cell apoptosis was revealed by FACS analysis, and we found that miR-940 ASO transfection led to a reduced cell apoptosis rate (Fig.3c).
Fig 3.
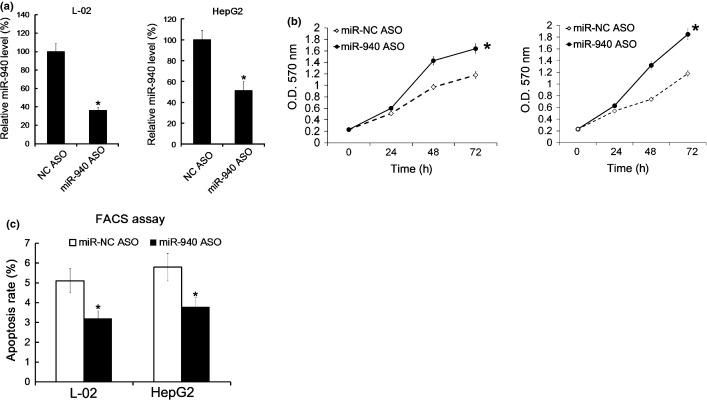
Transfection with miR-940 ASO promoted proliferation of SMMC-7721 and Huh-7 and inhibited cell apoptosis. 48 h after miR-940 ASO transfection, the levels of miR-940 in SMMC-7721 and Huh-7 were analyzed by qRT-PCR. The level of miR-940 in miR-NC the ASO treated group was arbitrarily defined as 100% (a). The OD value was measured at the indicated time point in the MTT analysis experiment; the cellular proliferation curves were shown (b). 48 h after miR-940 ASO transfection, cell apoptosis was measured by Annexin V-FITC/PI double staining FACS analysis (d). All data are mean ± SD of three separate experiments. $P < 0.05.
The estrogen-related receptor gamma gene was targeted by miR-940
To investigate the potential target genes of miR-940 involved in HCC, bioinformatics algorithms were applied to predict the target genes. Many genes were predicted (data not shown), including ESRRG. A recent study showed that the miR-545/374a cluster was partially responsible for the poor prognosis for HCC patients, and ESRRG had a reverse correlation with the expression of miR-545.34 Thus, ESRRG was chosen for further study. The binding sites and mutated sites in ESRRG are shown in Figure 4(a). The intact 3′UTR of ESRRG and its mutated version were cloned into luciferase reporter plasmids, which were used for co-transfection with miR-940 into HEK293 cells. We found that miR-940 reduced the luciferase activity of wild type 3′ UTR reporter, while the difference between miR-NC and miR-940 in the luciferase activity of reporter with mutated version was not significantly different (Fig.4b). Next, 48 h after transfection of miR-940 mimics, the ESRGG protein level in HEK293 cells was assayed by western blot analysis, and we found that miR-940 mimics significantly inhibited the ESRGG protein level (Fig.4c). Furthermore, the ESRGG mRNA expression in the 10 HCC tissues were examined by qRT-PCR, and we found that the ESRGG mRNA level and the miR-940 level were negatively correlated (Fig.4d).
Fig 4.
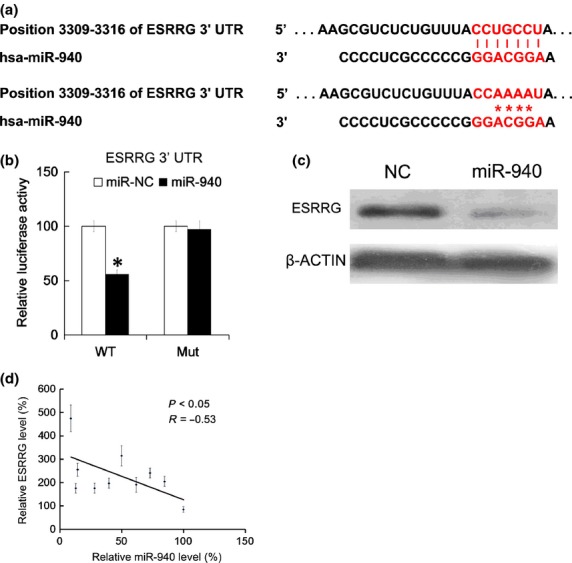
The estrogen-related receptor gamma (ESRRG) gene was targeted by miR-940. Putative targeted genes were predicted by TargetScanHuman; the binding site of the putative targeted gene and the mutated site of miR-940 are shown (a). The RL reporter plasmids (RL-control, RL-ESRRG and RL-mutated ESRRG) and miR-940 or miR-NC were co-transfected into HEK293 cells, along with a firefly luciferase reporter (pGL control) for normalization. Luciferase activities were measured after 48 h. Then the ratio of RL activity of firefly luciferase activity in the miR-940 treated group were calculated and compared with the ratio in the miR-NC group, which was arbitrarily defined as 100% (b). Following 48 h transfection of miR-940 mimics, the HEK293 cells were collected for ESRRG protein level assay by Western blot (c). ESRRG mRNA levels and miR-940 levels in 10 HCC tissues were analyzed by qRT-PCR, the correlation between ESRRG mRNA and miR-940 was analyzed by two-tailed Person’s correlation coefficient analysis (d). All data are mean ± SD of three separate experiments. $P < 0.05.
Suppression of estrogen-related receptor gamma inhibited hepatocellular carcinoma cell lines growth and induce cells apoptosis
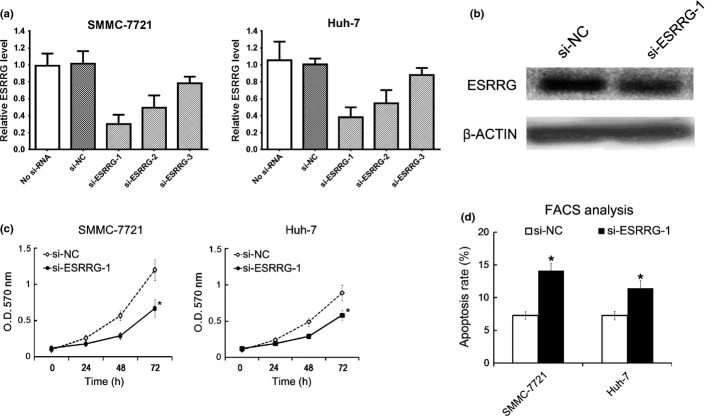
To further investigate the role of ESRRG in HCC, we downregulated the ESRRG level in SMMC-7721 and Huh-7 cells by ESRRG siRNA (si-ESRRG) transfection, and found that si-ESRRG-1 showed the most effectiveness (Fig.5a). In addition, the western blot analysis showed that si-ESRRG-1 inhibited the ESRRG protein level significantly (Fig.5b). Thus, si-ESRRG-1 was used in the next experiments. The cell proliferation was then evaluated by MTT analysis, and we found that downregulation of ESRRG inhibited the proliferation of SMMC-7721 and Huh-7 cells (Fig.5c). Next, 48 h after si-ESRRG-1 transfection, the cell apoptosis was revealed by FACS analysis, and we found that si-ESRRG-1 transfection caused the increased cell apoptosis rate (Fig.5d).
Fig 5.
Si-ESRRG-1 transfection inhibited HCC cell lines growth and induce cells apoptosis. 48 h after si-ESRRG (1–3) transfection, the levels of ESRRG in SMMC-7721 and Huh-7 were analyzed by qRT-PCR. The level of ESRRG in si-NC treated group was arbitrarily defined as 100% (a). Following 48 h si-ESRRG-1 transfection, the SMMC-7721 cells were collected for ESRRG protein level assay by Western blot (b). The Optical Density (OD) value were measured at the indicated time point in MTT analysis experiment, the cellular proliferation curves were shown (c). 48 h after si-ESRRG-1 transfection, cell apoptosis was measured by Annexin V-FITC/PI double staining FACS analysis (d). All data are mean ± SD of three separate experiments. $P < 0.05.
Discussion
In our study, we investigated the role of miR-940 in HCC, and found that a lower level of miR-940 in HCC promoted cellular proliferation in HCC. The roles of miRNA in HCC have been investigated in previous studies. Hou et al.35 studied the miRNomes in HCC and reveal that miR-199a/b-3p is a potential therapeutic target for HCC. Li et al.36 show that miR-99a was remarkably decreased in HCC tissues and cell lines, and lower miR-99a expression in HCC tissues was significantly correlated with shorter survival of HCC patients. Our data may add new miRNA makers in the miRNomes of HCC.
Recent studies prove that miR-940 is intensively involved in HCC. MiR-940 exhibits tumor suppression function in nasopharyngeal carcinoma cells by downregulating nestin24 and inhibits the migratory and invasive potential of cells.25 Our data also indicate the protection role of miR-940 in HCC. However, the targeted gene in our study was ESRRG.
The estrogen receptor-related receptor (ESRR) can bind to the estrogen response element. Previous researchers found that ESRRA, a member of ESRR, can promote p53 gene expression and induce HCC progression in female patients.37 Another study shows that miRNA 545 is higher in male HCC patients compared to female HCC patients, and the ESRRG gene had a reverse correlation with the expression of miR-545.34 Here our data indicated that miR-940 could target ESRRG and the reverse correlation between miR-940 and ESRRG. These data reveal that miR-940 may play a role in the gender disparity of HCC. We will investigate this in a future study.
Hepatitis B virus infection is a major risk factor for HCC.38 Hou et al.35 applied massively parallel signature sequencing to carry out an in-depth analysis of the miRNomes in normal human liver, hepatitis liver and HCC, and found that the three types of tissue showed different miRNA profiles. We suppose that miR-940 may play an important role in the carcinogenesis of HBV-related HCC, which requires further investigation.
In conclusion, we studied the role of miR-940 in HCC, and found that a lower level of miR-940 in HCC tissues correlated with a reduced patient survival rate. Overexpression of miR-940 inhibited HCC cell line growth and induced cell apoptosis, and vice versa. The ESRRG gene was targeted by miR-940. Suppression of ESRRG inhibited HCC cell line growth and induced cells apoptosis. We hope that our study will provide some hints for further HCC research.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1 The clinical information of the 23 patients.
References
- El-Serag HB. Hepatocellular carcinoma. N Eng J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Hou J, Zhou Y, Zheng Y, et al. Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell. 2014;25:49–63. doi: 10.1016/j.ccr.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–21. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–9. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Tamori A, Itami S, et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer. 2013;13:99. doi: 10.1186/1471-2407-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Majumder S, Nuovo G, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–61. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–36. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Eng J Med. 2009;361:1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Yamashita T, Budhu A, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–80. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- Song G, Sharma AD, Roll GR, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51:1735–43. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura S, Honda M, Yamashita T, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–92. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- Ma J, Sun F, Li C, et al. Depletion of intermediate filament protein Nestin, a target of microRNA-940, suppresses tumorigenesis by inducing spontaneous DNA damage accumulation in human nasopharyngeal carcinoma. Cell Deat Dis. 2014;5:e1377. doi: 10.1038/cddis.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendiran S, Parwani AV, Hare RJ, Dasgupta S, Roby RK, Vishwanatha JK. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. 2014;13:250. doi: 10.1186/1476-4598-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol. 2011;731:237–45. doi: 10.1007/978-1-61779-080-5_20. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou C, Gao F, et al. MiR-34a in age and tissue related radio-sensitivity and serum miR-34a as a novel indicator of radiation injury. Int J Biol Sci. 2011;7:221–33. doi: 10.7150/ijbs.7.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZB, Yang Z, Chi Y, et al. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151. Cell Physiol Biochem. 2013;31:823–32. doi: 10.1159/000350100. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–46. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–86. [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS ONE. 2014;9:e109782. doi: 10.1371/journal.pone.0109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–43. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Li D, Liu X, Lin L, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–85. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Yeh KH, Liu WH, et al. Elevated p53 promotes the processing of miR-18a to decrease estrogen receptor-alpha in female hepatocellular carcinoma. Int J Cancer. 2015;136:761–70. doi: 10.1002/ijc.29052. [DOI] [PubMed] [Google Scholar]
- Brechot C, Gozuacik D, Murakami Y, Paterlini-Brechot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) Semin Cancer Biol. 2000;10:211–31. doi: 10.1006/scbi.2000.0321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The clinical information of the 23 patients.