Abstract
Nucleus accumbens associated 1 (NACC1) is a cancer-associated BTB/POZ (pox virus and zinc finger/bric-a-brac tramtrack broad complex) gene, and is involved in several cellular functions in neurons, cancer and stem cells. Some of the BTB/POZ proteins associated with cancer biology are SUMOylated, which appears to play an important role in transcription regulation. We show that NACC1 is SUMOylated on a phylogenetically conserved lysine (K167) out of three consensus SUMOylation motif sites. Amino acid substitution in the SIM sequence (SIM/M) within the BTB/POZ domain partially reduced K167 SUMOylation activity of NACC1. Overexpression of GFP-NACC1 fusion protein leads to formation of discrete nuclear foci similar to promyelocytic leukemia nuclear bodies (PML-NB), which colocalized with SUMO paralogues (SUMO1/2/3). Both NACC1 nuclear body formation and colocalization with SUMO paralogues were completely suppressed in the GFP-NACC1-SIM/M mutant, whereas they were partially maintained in the NACC1 K167R mutant. Confocal immunofluorescence analysis showed that endogenous and exogenous NACC1 proteins colocalized with endogenous PML protein. A pull-down assay revealed that the consensus motifs of the SUMO acceptor site at K167 and the SIM within the BTB/POZ domain were both necessary for efficient binding to PML protein. Our study demonstrates that NACC1 can be modified by SUMO paralogues, and cooperates with PML protein.
Keywords: Migration, nucleus accumbens associated 1, promyelocytic leukemia nuclear body, SUMO-interacting motif, SUMOylations
SUMOylation, covalent modification of a protein by a small ubiquitin-related modifier (SUMO) peptide on a lysine residue, has emerged as an important posttranslational modification in regulating cellular processes.1 Similar to ubiquitination, the process of SUMOylation requires the E1-activating and the E2-conjugating (UBC9) enzymes.2 UBC9 catalyzes the formation of an isopeptide bond between the C-terminus of SUMO and the amino group of the target lysine. In general, SUMOylation occurs on the lysine residue within a consensus motif ΨKXE (where Ψ is a large hydrophobic residue and X is any residue).3 However, in a few cases, SUMOylation can still occur even without this consensus sequence flanking lysine.4 An emerging theme, however, is that SUMOylation often promotes interactions between modified proteins and downstream factors containing SUMO-interacting motifs (SIM).5 The known SIMs have a hydrophobic core [(V/I) X (V/I) (V/I)] followed or preceded by a negatively charged cluster of amino acids.6,7 The increasing number of SUMOylated factors being identified has revealed that SUMOylation largely occurs in the nuclear compartment and that most identified SUMO substrates are known transcription factors and coregulators.8,9
Among them, attention has focused on the roles of BTB/POZ (bric-a-brac tramtrack broad complex/pox virus and Zn finger) family proteins, which are recognized as transcription repressors. There are over 300 candidate members of the BTB/POZ family in the human genome,10 and several of these genes have been shown to be involved in cancer development, such as BCL6,11 promyelocytic leukemia zinc finger (PLZF),12 leukemia/lymphoma-related factor (LRF)/Pokemon,13,14 Bach2 (BTB and CNC homology 1, basic leucine zipper transcription factor 2)15 and hypermethylated in cancer-1 (HIC1).16 Some of the BTB/POZ proteins, such as HIC1, PLZF and Bach2, are covalently SUMOylated, which appears to play an important role in transcription repression.16–18 For example, Bach2 forms nuclear foci associated with promyelocytic leukemia nuclear bodies (PML-NB), and shuttles between the cytoplasm and nucleus.19 A candidate mediator of such regulation of shuttling is the BTB/POZ domain of Bach2, which is involved in protein–protein interactions.20 Among the proteins containing BTB/POZ domains and known to form nuclear foci are PLZF and nucleus accumbens associated 1 (NACC1; also known as NAC1 or BTBD14B).21,22 NACC1 is found in both the cytoplasm and the nucleus.23
Nucleus accumbens associated 1 is a cancer-associated BTB/POZ gene that is significantly overexpressed in several types of human malignancies, its intense immunoreactivity being significantly correlated with tumor recurrence in ovarian cancer,22 uterine carcinoma24 and malignant melanoma.23 Evidence for a role of NACC1 in tumor cell biology, particularly cell proliferation activity25,26 and drug resistance,27,28 has been increasing. We have demonstrated that NACC1 is involved in the deacetylation activity of histone deacetylase 6 (HDAC6), and regulates tumor cell migration through deacetylation of an actin cross-linking protein, cortactin, and α-tubulin of microtubules.23
In the present study, we show that a conserved lysine (K167) of NACC1 is an important SUMO acceptor. Furthermore, the NACC1 SIM sequence within the BTB/POZ domain, which is also highly conserved in several species, is necessary for NACC1 SUMOylation, because of the requirement for binding to UBC9, which is a SUMO E2 ligase. Finally, this SUMO modification is also required for the recruitment of NACC1 protein to the PML-NB.
Materials and Methods
Plasmids and mutagenesis
Plasmids expressing NACC1, SUMO1-3, PML (isoform I), and UBC9, SENP1 and SENP2 were made (Data S1). Mutations were generated with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). For the expression of GFP/Cherry fusion proteins, pcDNA-DEST53 Gateway Vector (Life Technologies, Carlsbad, CA, USA) and pmCherry-C1 Vector (CLONTECH Laboratories, Palo Alto, CA, USA) were used. For the synthesis of recombinant proteins by wheat germ extract systems, pF3KWG (BYDV) FlexiR Vector (Promega, Madison, WI, USA) was used.
Reagents and antibodies
All antibodies were commercially purchased (Data S1).
Cell culture and transfection
HeLa and MCF-7 were obtained from Health Science Research Resources Bank (HSRRB, Osaka, Japan). They were maintained under appropriate conditions. Cells were transfected with expression vectors within OptiMEM using Lipofectamine 2000 according to the manufacturer’s protocol (Life Technologies).
Immunoprecipitation and immunoblotting
Immunoprecipitation (IP) and immunoblotting (IB) protocols are shown in Data S1.
In vitro and in vivo SUMOylation assays
In vitro transcription and translation reactions were performed with the TNT-coupled wheatgerm extract system (Data S1).
Confocal microscopy
Cells were fixed with 4% paraformaldehyde, and imaged with a Zeiss LSM510 Meta confocal microscope (Carl Zeiss Jena GmbH, Jena, Germany) (Data S1).
Results
SUMO modification of nucleus accumbens associated 1 protein
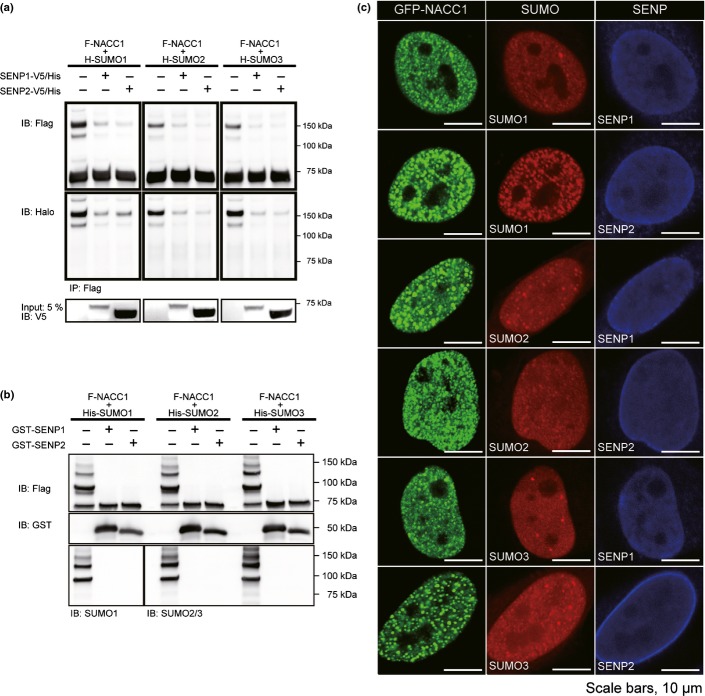
To demonstrate that endogenous NACC1 proteins are modified by SUMO, we first examined HeLa and MCF-7 by IP with anti-NACC1 antibody, but no migrating band was observed (data not shown). We examined whether NACC1 is modified by SUMO family in HeLa transiently coexpressing Flag (F)-NACC1 and Halo (H)-SUMO1-3 (Fig.1a). IB analysis using anti-Flag antibody revealed that additional migrating bands were observed when cells were coexpressed with H-SUMO vectors. The additional bands were confirmed to be SUMOylated bands of NACC1 (Fig.1a). The major band migrated at a position corresponding to a molecular mass. The difference in molecular weights between the two proteins was consistent with the addition of one molecule (50 KDa) of H-SUMO protein. The faint bands above F-NACC1 seemed to be endogenous SUMO1 and SUMO2/3 modified F-NACC1. We conclude that NACC1 protein is modified by SUMO proteins in vivo.
Fig 1.
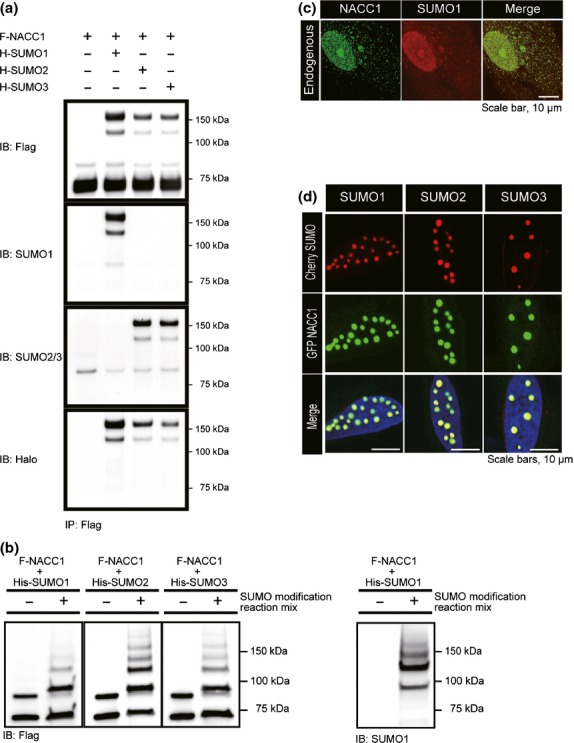
In vivo and in vitro SUMO modification of NACC1. (a) In vivo SUMOylation of nucleus accumbens associated 1 (NACC1) in HeLa cells. (b) Analysis of SUMO modification by reconstituted in vitro SUMOylation reactions. Slow-migrating bands due to addition of SUMO paralogues were observed. (c) Confocal immunofluorescence of endogenous NACC1 and SUMO1 proteins in HeLa cells (NACC1, green; SUMO1, red). Both NACC1 and SUMO1 were observed in nucleus and cytoplasm with a granular pattern. Bar, 10 μm. (d) Colocalization of GFP-NACC1 and Cherry-SUMO paralogues (NACC1, green; SUMO1, red). Bar, 10 μm.
To investigate whether NACC1 is SUMOylated in vitro, we next performed in vitro SUMOylation assays (Fig.1b). In vitro-translated, full-length F-NACC1 generated with the wheat germ extract system and recombinant His-SUMO proteins were subjected to an in vitro modification reaction by incubation with a mix containing recombinant Aos1/uba2, recombinant Ubc9, and ATP in either the presence (+) or the absence (−) of recombinant SUMO. Slower-migrating bands were obtained in the same way as for the in vivo SUMOylation assays (Fig.1b). Using SUMO antibodies, we confirmed that these lower migrating bands were a result of SUMO addition (right side panel of Fig.1b). Thus, NACC1 is modified by all SUMO paralogues in vitro.
To investigate colocalization of both NACC1 and SUMO1/2/3, we examined endogenous NACC1 and SUMO1 proteins. Both proteins were localized predominantly in the nucleus, and their staining revealed a granular pattern. They were also observed in the cytoplasm, again with a granular pattern. The majority of both proteins colocalized (Fig.1c). We also generated GFP-NACC1 and Cherry-SUMO fusion proteins, and cotransfected them into HeLa cells. Colocalization of ectopic NACC1 and SUMO proteins was observed with a discrete nuclear body pattern (Fig.1d). To investigate whether the colocalization of SUMO paralogues with NACC1 occurs through direct conjugation of proteins, we first examined the capability of SENP1 and SENP2 to deconjugate all of the SUMO paralogues from NACC1 in vivo. SENP1 and SENP2 were capable of deconjugating SUMO from NACC1 (Fig.2a). In in vitro deconjugation assays, SENP1 and SENP2 were also potent in removing all of the SUMO paralogues from NACC1 (Fig.2b). Furthermore, when SENP1 or SENP2 was used to transfect HeLa cells, the nuclear speckles of NACC1 and SUMO paralogues were abolished (Fig.2c). These observations suggest that NACC1-SUMO modification occurs through direct conjugation of SUMO paralogues.
Fig 2.
Deconjugation of nucleus accumbens associated 1 (NACC1)-SUMO by SENP1 and SENP2. (a) HeLa cells were cotransfected with F-NACC1 and H-SUMO1–3. Conjugation of SUMO1-3 to F-NACC1 was reversed by transfection with SENP1 or SENP2. (b) Analysis of deSUMOylation by reconstituted in vitro SUMOylation reactions. (c) Confocal immunofluorescence of GFP-NACC1 and Cherry-SUMO1/2/3 protein overexpression with SENP1 or SENP2 (NACC1, green; SUMO1/SUMO2/SUMO3, red; SENP1/SENP2, blue). When expressed in the presence of SENP1 or SENP2, GFP-NACC1 and Cherry-SUMO1/2/3 failed to form large nuclear foci. Bar, 10 μm.
Target lysine of nucleus accumbens associated 1 for SUMO modification
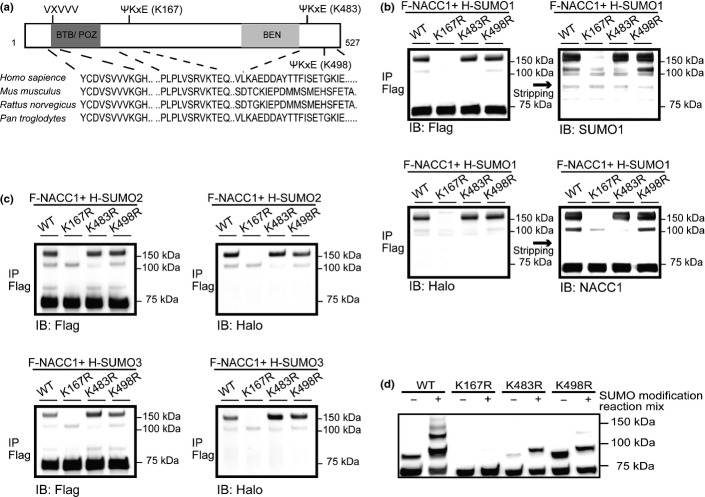
Analysis of NACC1 using the SUMOplot algorithm (http://www.abgent.com.cn/doc/sumoplot/login.asp) for the presence of consensus SUMOylation motifs revealed that three lysine residues (K167, K483 and K498) had a high probability of being sites for SUMOylation (Fig.3a). K167 is highly conserved in several species, but the other two lysine sites are not (Fig.3a). A SIM sequence is found within the N-terminus of the BTB/POZ domain and is also phylogenetically conserved (Fig.3a).
Fig 3.
Covalent SUMO modifications at the consensus SUMOylation motif sites. (a) Schematic representation showing that human nucleus accumbens associated 1 (NACC1) protein contains two conserved domains, three SUMO consensus motifs (Ψ-Lys-X-Glu/Asp) and a SUMO-interaction motif (SIM, Val-X-Val-Val). (b) HeLa cells were transfected with F-NACC1-WT or F-NACC1 mutants with a replacement of Lys by Arg together with H-SUMO-1. (c) HeLa cells were transfected with a plasmid encoding each F-NACC1, together with a plasmid encoding either H-SUMO-2 or H-SUMO-3. (d) In vitro-translated, F-NACC1, F-NACC1 K167R, K483R or K498R were analyzed by SDS-PAGE directly or subjected to an in vitro modification reaction mix by incubation with recombinant E1, E2 and His6-tagged SUMO1. Slow-migrating bands were observed in WT and in mutants with K483R and K498R, but not with K167R.
To identify the lysine residue of NACC1 modified by SUMO, mutants were generated (with lysine to arginine substitutions) and analyzed for SUMOylation status. Lysates from cells expressing either F-NACC1 WT or F-NACC1 (K483R and K498R) mutants displayed migrating bands, but these bands were not visible in lysates from K167R mutant (Fig.3b). These results suggest that the K167 of NACC1 is a target for SUMO1 modification. IP followed by IB analyses also revealed that mutation of K167 in the SUMOylation consensus sequence prevented conjugation of F-NACC1 with H-SUMO2 and SUMO3 (Fig.3c). In the in vitro SUMOylation assay, slower migrating bands were also obtained, except for the case of the F-NACC1-K167R mutant (Fig.3d).
Effects of nucleus accumbens associated 1 mutant at K167 for cellular sublocalization
Because SUMOylation of several proteins has been shown to change their subcellular localization,29 we analyzed the localization of NACC1. Wild type GFP-NACC1 and Cherry-SUMO1/2/3 fusion proteins colocalized in the nucleus and formed a large nuclear body. Notably, only a portion of the K167R mutant of GFP-NACC1 did not merge with Cherry-SUMO1 protein, although formation of NACC1 nuclear bodies was observed (Fig.4). Two other lysine mutants did not affect colocalization of the two fusion proteins (Fig.4). Experiments using Cherry-SUMO2/3 led to similar results (Fig. S1).
Fig 4.
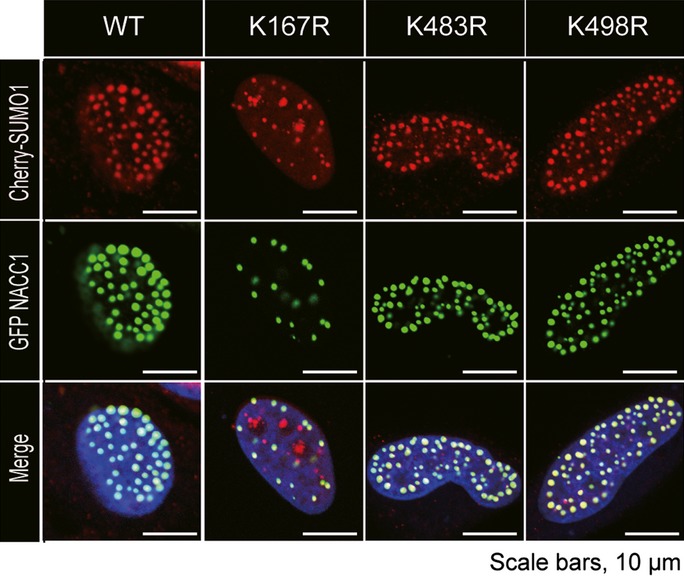
Confocal immunofluorescence observations of GFP-nucleus accumbens associated 1 (NACC1) and Cherry-SUMO1 fusion proteins. HeLa cells were cotransfected with fusion protein expression vectors (NACC1, green; SUMO1, red; DAPI, blue). Colocalization was observed with WT, K438 and K498R, but colocalization with K167R did not always occur. Bar, 10 μm.
SIM within nucleus accumbens associated 1 affects SUMOylation and nucleus accumbens associated 1 body formation
Several proteins containing SIM sequences show enhanced binding affinity to SUMOlyated proteins.4 Such non-covalent binding to SUMOylated proteins mediated by SIM is observed between UBC9 and SUMO-targeted proteins.30 The SIM amino acid sequence may contribute to SUMO modification of target proteins, including NACC1. Furthermore, both the SIM sequence and covalent SUMOylation at lysine residues mediate PML-NB formation.31,32 In the BTB/POZ domain of NACC1, we found a SIM (VSVV, residues 32 to 35 of NACC1, Fig.5a). Therefore, we hypothesized that the SIM in NACC1 affects SUMOylation of NACC1 itself as well as nuclear body formation.
Fig 5.
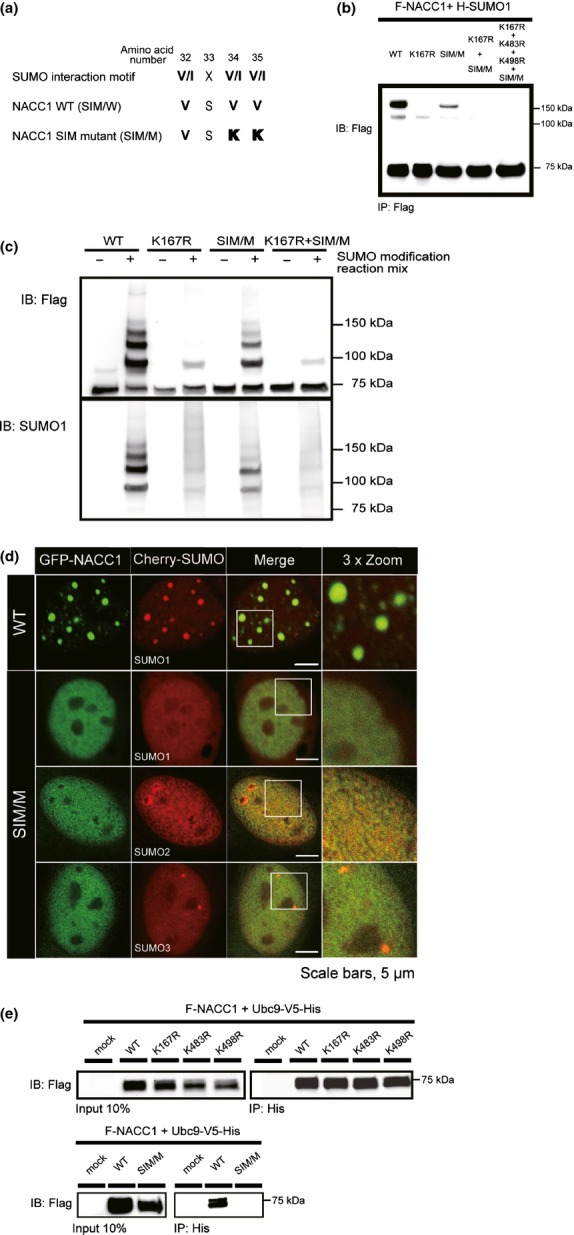
Disruption of the SIM sequence of nucleus accumbens associated 1 (NACC1) and the interaction with NACC1 and UBC9. (a) SIM consensus motif aligned with the NACC1-WT and NACC1-SIM mutant sequence (SIM/M). (b) HeLa cells were transfected with a plasmid encoding either F-NACC1-WT or F-NACC1-SIM/M, together with H-SUMO1. Slow-migrating bands were observed with SIM/M, but their density was reduced in comparison with those of the WT. A weak slow-migrating band was found in K167R but not in K167R + SIM/M or K167R + K483R + K498R + SIM/M. (c) Analysis of SUMO modification by reconstituted in vitro SUMOylation reactions. Slow-migrating bands due to SUMO1 were observed in WT and SIM/M, although the density of the migrating bands was weaker in SIM/M than in WT and was further reduced in K167R and K167R + SIM/M. (d) Confocal immunofluorescence of GFP-NACC1-WT or -SIM/M mutants and Cherry-SUMO1/2/3 fusion proteins (NACC1, green; SUMO1/SUMO2/SUMO3, red). Both fusion proteins colocalized in WT with a discrete nuclear body pattern. Complete disruption of the nuclear body pattern was observed when the SIM/M construct was used. Bar, 5 μm. (e) Cotransfection and immunoprecipitation analyses for the binding between UBC9 and NACC1. HeLa cells were cotransfected with plasmids encoding various F-NACC1-WT, -K167R, -K483R, -K498 or -SIM/M and UBC9-V5/His vectors. The interaction between NACC1 and UBC9 was suppressed only in the case of Flag-NACC1 SIM/M.
To test this hypothesis, we mutated two hydrophobic amino acids of this SIM to lysines (V34K and V35K) (Fig.5a).7,32 Plasmids expressing NACC1 or NACC1-SIM mutant were cotransfected with H-SUMO1. NACC1-SIM/M displayed a reduced SUMO conjugation activity when compared to NACC1-WT (Fig.5b,c). Furthermore, the migrating bands almost disappeared in the K167R mutant (Fig.5b,c), indicating that SUMOylation activity is markedly reduced in double mutants at K167 and SIM. These experiments suggest that the SIM sequence is also necessary for NACC1 SUMO modification.
We next tested whether the NACC1 SIM is required for NACC1 body formation, as for PML-NB. Plasmids expressing GFP-NACC1-WT or GFP-NACC1 mutants and Cherry-SUMO1 were transfected into HeLa cells, and confocal analysis was performed (Fig.5d). As demonstrated in a preceding section, cells transfected with GFP-NACC1-WT contained 10–20 NACC1-NB of various sizes per nucleus. Almost all of them colocalized with Cherry-SUMO1 protein. Surprisingly, although in vivo and in vitro SUMOylation assays suggested that SIM/M constructs (with intact K167) are partially SUMOylated and the loss of SUMOylation signal density was inferior to that of the K167R mutant (Fig.5b,c), colocalization with Cherry-SUMO1 and NACC1-NB formation itself were completely disrupted in this mutant (Fig.5d). NACC1-SIM/M displayed a diffused nuclear staining pattern (Fig.5d). Similar results were obtained with cotransfection of GFP-NACC1-SIM/M and Cherry-SUMO2 or 3. Thus, the NACC1 SIM sequence is required for NACC1 body formation. Moreover, almost all of the WT, K167R and SIM/M fusion proteins were observed only in the nucleus, and, therefore, we think that SUMO modification of NACC1 is not associated with the nuclear-cytoplasm trafficking of NACC1.
Nucleus accumbens associated 1 interacts with UBC9
To investigate the interaction of NACC1 with UBC9 mediated by SIM and SUMOylated lysine sites, we performed IP analysis in HeLa cells transiently expressing F-NACC1 or F-NACC1 mutants together with UBC9-V5/His. IB analysis showed that none of the lysine mutants significantly affected binding to UBC9, but SIM mutations disrupted the binding between NACC1 and UBC9 (Fig.5e). Consequently, the SIM sequence is very important for the UBC9 recognition of NACC1 as a substrate for E2/E3 substrate ligase.
Nucleus accumbens associated 1 recruitment within promyelocytic leukemia nuclear bodies
Because overexpression of WT-NACC1 caused formation of discrete round and oval NACC1 nuclear bodies similar to PML-NB, we tested whether NACC1 proteins participate in PML-NB. To analyze the interplay between PML, DAXX, SP100 and NACC1, cells were exposed to arsenite as previously described.33–36 Immunofluorescence study showed that endogenous PML and NACC1 colocalized in nuclei of HeLa (Fig.6a). We next overexpressed WT-GFP-NACC1 fusion protein in HeLa cells and stained them with antibodies against endogenous PML, DAXX and SP-100 proteins. All of the proteins were well colocalized. To investigate physical interactions between NACC1 and PML protein, we performed pull-down assays using H-PML and F-NACC1 plasmids (WT, K167R and SIM/M). A dense signal was observed for the interaction of F-WT-NACC1 and H-PML, but signals for K167R or SIM/M were markedly reduced (Fig.6b). These data indicate that NACC1 protein is recruited within the PML body by covalent and non-covalent binding at K167 and SIM, respectively.
Fig 6.

Interaction with nucleus accumbens associated 1 (NACC1) and promyelocytic leukemia (PML) proteins. (a) Confocal immunofluorescence observations of endogenous and ectopic NACC1 and endogenous PML, DAXX and SP-100 proteins (NACC1, green; PML/DAXX/SP-100, red; DAPI, blue). Endogenous NACC1 and PML proteins, detected by immunofluorescence with anti-NACC1 and anti-PML antibodies, were expressed as a nuclear speckle pattern (first row). GFP-NACC1-WT was clearly localized in discrete round and oval nuclear bodies in association with endogenous PML (second row), DAXX (third row) and SP100 proteins (fourth row). Bars, 5 μm. (b) Pull-down assay for interaction between in vitro-translated NACC1 and PML proteins with SUMO conjugation.
To confirm the occurrence of in vivo NACC1 SUMOylation in other cell types, we also performed some experiments using MCF-7. The results we obtained were similar to those for HeLa (Fig. S2).
Discussion
Nucleus accumbens associated 1 has been highlighted not only as a key player in neurogenesis and cancer biology,22,23,25,27,28 but also as a transcription factor for the maintenance of pluripotency of ES and other types of stem cells.37 Our study is the first to report that NACC1 is modified by SUMO paralogues and recruited to PML-NB. The biological function of NACC1 in PML-NB remains unclear, but these results could provide clues for additional study.
Many proteins including SP100 and DAXX have been found to localize to PML-NB. It has been implicated in the regulation of numerous biological functions such as cell cycle regulation, transcriptional repression, DNA damage response, apoptosis and cellular senescence.38 A small binding interface between the SUMO acceptor site (ΨKXE) and the SIM is thought to be essential for binding between the PML protein and its partner proteins (such as SP100 and DAXX),38,39 such that the machinery has been referred to as “two beads on a string.”40,41 However, NACC1 has never been shown to be an aggregated protein within PML-NB, even though it has both SUMO acceptor and SIM signatures. The PML protein itself has three consensus SUMO acceptor sites and a SIM site.31 Pandolfi and his colleagues provide a novel model of PML-NB formation and degradation in relation to the cell cycle, as follows.31 In interphase, PML protein is highly SUMOylated. Subsequently, non-covalent interaction between PML molecules mediated by both the SIM and the RBCC motif (which contains a RING domain and is essential for PML dimerization) promotes the growth of concentrated PML networks. Many of the proteins that are constitutive components of PML-NB are themselves SUMOylated and some of them, such as DAXX, contain a SIM sequence that allows them to interact with SUMOylated PML proteins. In mitotic cells, PML protein is deSUMOylated and the binding partners dissociate from PML-NB. Hence, their aggregation model of PML and its partner proteins is concordant with our result that NACC1 mutants at the SIM and the phylogenetically-conserved lysine (K167) disrupt NACC1 recruitment within PML-NB. The PML-NB have been intensively studied over the past few decades, and the variety of the nuclear body (NB) components suggests a wide range of possible biological roles. Interaction between NACC1 and PML might provide a chance to clarify the role of PML-NB.
Nucleus accumbens associated 1 controls cell growth and survival by repressing the transcription factor of GADD45GIP1/CRIF1/CKBBP2, which is the growth arrest and DNA-damage-inducible 45-γ interacting protein.25 Nakayama et al. report novel results that a dominant-negative form of NACC1 truncated protein containing only BTB/POZ (N130, amino acids 1–129) conferred a growth inhibitory effect that can be partially rescued by GADD45GIP1 siRNA knockdown.22 Moreover, NACC1 knockdown decreased FOXQ1 expression and promoter activity, and was involved in cell migration.42 The BTB/POZ domain is well conserved, and SIM sequences are common in the domains of this family of proteins. In the oncogenic BTB/POZ family members, namely ZBTB27 (BCL6), ZBTB16 (PLZF), ZBTB7A (Pokemon), Bach2 and ZBTB29 (HIC1), the SIM sequence is found in all except Bach2, and two or more consensus SUMO acceptor sites are found in the center portion or the C-terminus. Our experimental results could be explained by a previously proposed mechanism, as follows.4 A SIM-containing protein binds to SUMO covalently linked to the active site cysteine of UBC9. Binding enhances the local concentration of UBC9 in the vicinity of the substrate, allowing UBC9 to recognize lysine residues not necessarily contained within optimal consensus SUMOylation motifs. However, all the abovementioned BTB/POZ members are not highly SUMOylated. In fact, we have confirmed that BCL6 is not SUMOylated in vitro or in vivo (data not shown). It is well known that SUMO conjugation may be affected by the unique conformation of each SUMO acceptor site as determined by the flanking amino acid sequences.43
A unique SUMO acceptor site, the phosphorylation-dependent SUMO consensus motif, has been identified in several proteins.42,44 Phosphorylation of the serine residue within the motif leads to increased levels of SUMO conjugation by UBC9. Although NACC1 does not have this motif, the K167 site matches another extended consensus SUMO acceptor motif; namely, the negatively charged amino acid-dependent sumoylation motif (NDSM).45 The signature of this site consists of the core SUMOylation motif (ΨKXE) followed by at least two further acidic residues in the C-terminal tail, one of which is located between amino acids 3 and 6 within the 10-amino-acid region located immediately downstream.44 The matching sequence at K167 of NACC1 is VKTEQQESDSVQCM. The downstream acidic patch can bind a corresponding basic patch on UBC9 and enhance substrate binding. Our data showed substantial SUMO conjugation activity at K167, whereas activity at K483 and K498 was not evident. However, the amino acid sequences flanking K498 are compatible with the signature called acetylation/SUMOylation switch motif (ΨKXEP), which has been identified in the center region of the BTB/POZ family member HIC1.16 HIC1 is a tumor suppressor gene essential for mammalian development; it is epigenetically silenced in many human tumors and is involved in complex pathway regulating TP53 tumor suppression activity. A phylogenetically conserved lysine (K314) of HIC1 is a target for both SUMOylation and acetylation. Furthermore, HIC1 transcriptional repression activity is positively controlled by two types of deacetylase, SIRT1 and HDAC4, which increase its deacetylation and SUMOylation, respectively.16 The acetylation/SUMOylation switch at K498 of NACC1 requires further study.
Disclosure Statement
The authors have no conflict of interest to declare.
Supporting Information
Additional supporting information may be found in the online version of this article:
Data S1. Protocols for experiments.
Fig. S1. Confocal immunofluorescence observations of GFP-nucleus accumbens associated 1 (NACC1) and Cherry-SUMO2/3 fusion proteins.
Fig. S2. In vivo nucleus accumbens associated 1 (NACC1) SUMOylation in MCF-7. The reproducibility of NACC1 SUMOylation in vivo was confirmed in MCF-7 cells.
References
- Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–82. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–9. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhu S, Guzzo CM, et al. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–15. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–27. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–8. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–41. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–9. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. BioEssays. 2006;28:1194–202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Polo JM, Dell’Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–35. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- Yeyati PL, Shaknovich R, Boterashvili S, et al. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–34. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Pandolfi PP. The transcription factor Pokemon: a new key player in cancer pathogenesis. Cancer Res. 2005;65:8575–8. doi: 10.1158/0008-5472.CAN-05-1055. [DOI] [PubMed] [Google Scholar]
- Motamed-Khorasani A, Jurisica I, Letarte M, et al. Differentially androgen-modulated genes in ovarian epithelial cells from BRCA mutation carriers and control patients predict ovarian cancer survival and disease progression. Oncogene. 2007;26:198–214. doi: 10.1038/sj.onc.1209773. [DOI] [PubMed] [Google Scholar]
- Stankovic-Valentin N, Deltour S, Seeler J, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–75. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao TT, Chang CC, Shih HM. SUMO modification modulates the transrepression activity of PLZF. Biochem Biophys Res Commun. 2007;358:475–82. doi: 10.1016/j.bbrc.2007.04.157. [DOI] [PubMed] [Google Scholar]
- Tashiro S, Muto A, Tanimoto K, et al. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol Cell Biol. 2004;24:3473–84. doi: 10.1128/MCB.24.8.3473-3484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A, Tashiro S, Tsuchiya H, et al. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J Biol Chem. 2002;277:20724–33. doi: 10.1074/jbc.M112003200. [DOI] [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–8. [PubMed] [Google Scholar]
- Dong S, Zhu J, Reid A, et al. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-alpha fusion protein. Proc Natl Acad Sci USA. 1996;93:3624–9. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nakayama N, Davidson B, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci USA. 2006;103:18739–44. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda K, Oikawa H, Tada H, et al. Nucleus accumbens-associated 1 contributes to cortactin deacetylation and augments the migration of melanoma cells. J Invest Dermatol. 2011;131:1710–9. doi: 10.1038/jid.2011.110. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Nakayama K, Yeasmin S, et al. Expression of a BTB/POZ protein, NAC1, is essential for the proliferation of normal cyclic endometrial glandular cells and is up-regulated by estrogen. Clin Cancer Res. 2009;15:804–11. doi: 10.1158/1078-0432.CCR-08-2134. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nakayama N, Wang TL, Shih Ie M. NAC-1 controls cell growth and survival by repressing transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer Res. 2007;67:8058–64. doi: 10.1158/0008-5472.CAN-07-1357. [DOI] [PubMed] [Google Scholar]
- Wu PH, Hung SH, Ren T, Shih Ie M, Tseng Y. Cell cycle-dependent alteration in NAC1 nuclear body dynamics and morphology. Phys Biol. 2011;8:015005. doi: 10.1088/1478-3975/8/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinawath N, Vasoontara C, Yap KL, et al. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–8. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cheng Y, Ren X, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2011;31:1055–64. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–7. doi: 10.1034/j.1600-0854.2002.30601.x. [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24:331–9. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Flotho A, Klug H, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31:371–82. doi: 10.1016/j.molcel.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–71. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Koken MH, Quignon F, et al. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:3978–83. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Puccetti E, Jensen K, et al. PIC-1/SUMO-1-modified PML-retinoic acid receptor alpha mediates arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Mol Cell Biol. 1999;19:5170–8. doi: 10.1128/mcb.19.7.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Yang Z, Privalsky ML. Arsenic trioxide is a potent inhibitor of the interaction of SMRT corepressor with Its transcription factor partners, including the PML-retinoic acid receptor alpha oncoprotein found in human acute promyelocytic leukemia. Mol Cell Biol. 2001;21:7172–82. doi: 10.1128/MCB.21.21.7172-7182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–61. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–34. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–52. [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Oberhofer E, et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12:264–9. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- Macauley MS, Errington WJ, Okon M, et al. Structural and dynamic independence of isopeptide-linked RanGAP1 and SUMO-1. J Biol Chem. 2004;279:49131–7. doi: 10.1074/jbc.M408705200. [DOI] [PubMed] [Google Scholar]
- Gao M, Wu RC, Herlinger AL, et al. Identification of the NAC1-regulated genes in ovarian cancer. Am J Pathol. 2014;184:133–40. doi: 10.1016/j.ajpath.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8:948–59. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–93. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Protocols for experiments.
Fig. S1. Confocal immunofluorescence observations of GFP-nucleus accumbens associated 1 (NACC1) and Cherry-SUMO2/3 fusion proteins.
Fig. S2. In vivo nucleus accumbens associated 1 (NACC1) SUMOylation in MCF-7. The reproducibility of NACC1 SUMOylation in vivo was confirmed in MCF-7 cells.