Abstract
Understanding the dynamics of the complex interaction network of cytokines, defined as ‘‘cytokinome’’, can be useful to follow progression and evolution of hepatocellular carcinoma (HCC) from its early stages as well as to define therapeutic strategies. Recently we have evaluated the cytokinome profile in patients with type 2 diabetes (T2D) and/or chronic hepatitis C (CHC) infection and/or cirrhosis suggesting specific markers for the different stages of the diseases. Since T2D has been identified as one of the contributory cause of HCC, in this paper we examined the serum levels of cytokines, growth factors, chemokines, as well as of other cancer and diabetes biomarkers in a discovery cohort of patients with T2D, chronic hepatitis C (CHC) and/or CHC-related HCC comparing them with a healthy control group to define a profile of proteins able to characterize these patients, and to recognize the association between diabetes and HCC. The results have evidenced that the serum levels of some proteins are significantly and differently up-regulated in all the patients but they increased still more when HCC develops on the background of T2D. Our results were verified also using a separate validation cohort. Furthermore, significant correlations between clinical and laboratory data characterizing the various stages of this complex disease, have been found. In overall, our results highlighted that a large and simple omics approach, such as that of the cytokinome analysis, supplemented by common biochemical and clinical data, can give a complete picture able to improve the prognosis of the various stages of the disease progression. We have also demonstrated by means of interactomic analysis that our experimental results correlate positively with the general metabolic picture that is emerging in the literature for this complex multifactorial disease.
Introduction
Recently it has been reported that the liver cancer is the second death cause due to cancer. In particular, the hepatocellular carcinoma (HCC) is the more common form of liver cancer and are diagnosed more than 700,000 cases in each year [1]. Several risk factors have been identified to contribute to the international burden of HCC such as chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV), alcoholic liver disease, non-alcoholic steato-hepatitis (NASH), diabetes mellitus (DM), obesity, intake of aflatoxins-contaminated food, tobacco smoking, excessive alcohol drinking and genetically inherited disorders (hemochromatosis, α-1 anti-trypsin deficiency, porphyrias) [2].
The type 2 diabetes (T2D) is a metabolic disorder characterized by hyperglycemia which may predispose the liver to relative insulin resistance due to inadequate secretion or receptor insensitivity to the endogenous insulin. In recent years, type 2 diabetes has been associated with increase risk for several malignancies including breast, colon, kidney, liver, endometrium and pancreatic cancers [3]. Recently some reported showed that the T2D presence tends to increase the HCC development and induces a poor prognosis for these patients, in both presence or absence of cirrhosis [4]. Because the liver plays a crucial role in glucose metabolism, it is not surprising that T2D is an epiphenomenon of many chronic liver diseases such as chronic hepatitis, fatty liver, liver failure and cirrhosis [5]. In addition, T2D as part of the insulin resistance syndrome, has been implicated as a risk factor for non-alcoholic fatty liver disease (NAFLD), including its most severe form non-alcoholic steato-hepatitis (NASH), and has been identified as a cause of both cirrhosis and HCC [6].
An important feature of the progression of chronic liver disease as well in the early stages of cancer is the minimal presence of clinical manifestations, making subtle the disease. In this context the cytokines are known to play an important role not only in the mechanisms of insulin resistance and glucose disposal defects but also in the pathological processes occurring in the liver during viral infection. In fact, understanding in patients affected from cancers or other diseases the dynamics of the complex interaction network of cytokines [7–9], defined ‘‘cytokinome” [10], should be very useful to follow the disease progression and evolution from its early stages as well as to define therapeutic strategies by using systems biology approaches [7–9].
Recently we evaluated the serum levels of many cytokines, chemokines, adipokines and growth factors in patients with type 2 diabetes, chronic hepatitis C (CHC) infection, CHC-related cirrhosis, CHC and type 2 diabetes and CHC-related cirrhosis and type 2 diabetes by BioPlex assay [9]. Our data evidenced that the serum levels of some proteins were significantly up-regulated in all the patients, but unfortunately they were often high also in individuals affected by only one syndrome, thus this fact makes not clearly attributable the analytes when both diseases are associated. Therefore, we suggested specific markers for the different stages of the diseases, useful for the clinical monitoring of patients in regard to the progression from CHC to LC and from CHD to LCD [9].
However, since the T2D is one of the most common co-morbid illnesses found in HCC patients, which is significantly associated with the worsening of the HCC development, we have focused more efforts on the understanding of the mechanisms underlying the HCC onset as well its progression, particularly in diabetic patients to try to improve their already poor prognosis. Therefore, aim of this study is to examine the serum levels of cytokines, growth factors, chemokines, as well as of other cancer and diabetes biomarkers in the patients with T2D, CHC, CHC-related HCC alone or in presence of T2D, comparing them with a healthy control group to define a profile of proteins able to characterize these patients, also identifying in the same time some diagnostic/prognostic markers useful for recognizing the association between diabetes and HCC.
Methods
Patients
In this study we enrolled in the discovery step 17 patients with T2D (11 women, 6 men), 20 patients with CHC (10 women and 10 men), 34 patients with HCC (11 women, 23 men), 10 patients with T2D-HCC (4 women, 6 men), and 20 healthy controls (11 women, 9 men). In Table 1 we report clinical characteristics and biochemical laboratory data of all the patients. The ADA criteria were used to classify patients with the T2D [11]: i) fasting plasma glucose 126 mg/dL (7.0 mmol/L) where fasting is defined as no caloric intake for at least 8 h or ii) symptoms of hyperglycemia and a casual plasma glucose 200 mg/dL (11.1 mmol/L) where casual is defined as any time of day without regard to time since last meal whereas the classic symptoms of hyperglycemia include polyuria, polydipsia, and unexplained weight loss, or iii) 2-h plasma glucose 200 mg/dL (11.1 mmol/L) during an OGTT where the test has been performed as described by the World Health Organization, using a glucose load containing the equivalent of 75 g anhydrous glucose dissolved in water. The patients with T2D were overweight with BMI values in the range between 25–29 kg/m2. The stage of fibrosis was assessed for the CHC patients according to the Ishak index [12]. In particular, F2 corresponds to fibrosis of the majority of portal tracts, F3 to fibrosis of the majority of portal tracts with occasional port-portal septa, and F4 to fibrosis of the majority of portal tracts with port-portal and port-central septa. Moreover, all HCC patients had HCV-related cirrhosis, and were non treated with drugs. In particular, the severity of cirrhosis was defined by Child-Pugh score and liver biopsies were performed only on patients with Child-Pugh score A. The patients with HCC had higher serum transaminase alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels compared to the control patients, as evaluated in the healthy donors. Finally, the patients with HCC and T2D had hyperglycemia.
Table 1. Clinical and laboratory data of patients with type 2 diabetes (T2D), chronic HCV (CHC), HCC with HCV-related cirrhosis (HCC), and HCC with HCV-related cirrhosis and type 2 diabetes (T2D+HCC) belonging to discovery set.
The corresponding patients belonging to validation set are indicated as T2DV, CHCV, HCCV and T2D+HCCV. We report the number of patients to whom the parameters refer. The related control ranges of the clinical data evaluated for the healthy donors, are also shown.
| T2D | T2DV | CHC | CHCV | HCC | HCCV | T2D+HCC | T2D+HCCV | Control range | |
|---|---|---|---|---|---|---|---|---|---|
| Age | 61.8±5.2 | 57.8±6.1 | 62.5±9.5 | 60.0±9.0 | 71.0±6.1 | 65.1±9 | 68.3±8.3 | 67.8±7.8 | 60.92 |
| Gender | 11M-6F | 10M-10F | 10M-10F | 11M-9F | 23M-11F | 12M-8F | 6M-4F | 7M-3F | |
| Glycemia (mg/dL) | 145.9±12.3 | 154.1±8.2 | 86.0±10.1 | 98.0±4.1 | 84.2±7.1 | 90.0±11.2 | 172.3±21.4 | 164.5±16.4 | 70–105 |
| AST (IU/L) | 31±4 | 28±3 | 71.4±2.3 | 61.8±6.2 | 104±3 | 90.5±5.5 | 74±26.2 | 63.7±5.2 | 5–40 |
| ALT (IU/L) | 33±6 | 30±5 | 121.1±8.6 | 100.8±5.2 | 100±4 | 102±6.0 | 105±10.4 | 100±12 | 7–56 |
| TotBilirubin (mg/dL) | 1.01±0.06 | 1.1±0.01 | 0.94±0.08 | 1.0±0.04 | 1.51±0.05 | 1.38±0.04 | 1.3±0.6 | 1.4±0.8 | 0.20–1.30 |
| Albumin (g/dL) | 4.2±0.9 | 4.0±0.7 | 4.01±0.07 | 3.9±0.4 | 3±0.3 | 3.1±0.04 | 2.8±0.2 | 2.5±0.1 | 3.5–5 |
| PLT (mL) | 198464±10221 | 220000±112 | 187413±7315 | 191635±5256 | 124534±2341 | 126254±5211 | 148333±34239 | 150000±28684 | 150000–400000 |
| BMI (kg/m 2 ) | 24.4±0.8 | 24.3±0.2 | 24.2±0.9 | 23.8±1.2 | 23.2±3.2 | 24.0±0.4 | 23.3±1.5 | 23.9±1.3 | 18.9–24.9 |
| HCV–PCR RNA | negative | negative | positive | positive | positive | positive | positive | positive | negative |
| HCV genot | negative | negative | 1:11; 2:9 | 1:13; 2:7 | 1: 20; 2: 14 | 1:14; 2:6 | 1:4; 2: 6 | 1:8; 2:2 | |
| AFP (ng/mL) | <10 | <10 | <10 | <10 | 150±20 | 120±40 | 173±31 | 170±25 | <10 |
| Child Pügh | A:15; B:13;C:6 | A:5; B:4;C:1 | A:2; B:2;C:6 | A:3; B: 3;C:4 | |||||
| Tumor invasion | T1:10; T2:12; T3:12 | T1: 4; T2:4; T3: 2 | T1:3; T2:3; T3:4 | T1:4; T2:4; T3: 2 |
Moreover, we verified the results using a separate validation cohort of 90 age/gender matched subjects, including 20 patients with T2D, 20 patients with CHC, 20 with HCC and 10 with T2D-HCC, and 20 healthy control subjects. These subjects had clinical characteristics similar to those used in the discovery step, and no significant differences can be evidenced between two sets (Table 1).
For this study we obtained ethics approval from the ethics committee of our institution (Second University of Naples) and obtained written informed consent from all involved participants.
Bio-Plex Assay
Blood samples were collected from a peripheral vein and kept on ice. Serum was collected by centrifugation (3,000 rpm for 10 min at 4°C), aliquoted, and stored at −80°C until analyzed. A multiplex biometric ELISA-based immunoassay, containing dyed microspheres conjugated with a monoclonal antibody specific for a target protein was used according to the manufacturer’s instructions (Bio-plex, Bio-Rad Lab., Inc., Hercules, CA, USA). Soluble molecules were measured using four commercially available kits: i) 21-plex immunoassay panel: IL-1α, IL-2R, IL-3, IL-12p40, IL-16, IL-18, CCL27, CXCL1, CXCL9, CXCL12, HGF, IFN-α2, LIF, MCP-3, M-CSF, MIF, β-NGF, SCF, SCGF-β, TNF-β, TRAIL; ii) 16-Plex panel: sEGFR, FGF-basic, Follistatin, G-CSF, HGF, sHER-2/neu, sIL-6Rα, Leptin, Osteopontin, PECAM-1, PDGF-AB/BB, Prolactin (PRL), SCF, sTIE-2, sVEGFR-1 (FLT1) and sVEGFR-2 (KDR); iii) 10-Plex panel: C-peptide, ghrelin, GIP, glp-1, glucagon (GCG), insulin, leptin (LEP), PAI-1, resistin, visfatin and iv) 2-Plex panel: adiponectin (ADIPOQ) and adipsin.
Each experiment was performed in duplicate using the same procedure described in our recent papers [7–9]. Serum levels of all proteins were determined using a Bio-Plex array reader (Luminex, Austin, TX) that quantifies multiplex immunoassays in a 96-well plate with very small fluid volumes. The analytes concentration was calculated using a standard curve, with software provided by the manufacturer (Bio-Plex Manager Software).
Data Analysis and Statistics
To evaluate the differences between cytokine, chemokine adipokines, cancer biomarkers and growth factor ratios in the patients and healthy controls belonging to discovery and validation steps, we used the nonparametric Mann-Whitney U test by obtaining U test and P values, the Unparied t test by P value, t value, the number of degrees of freedom (df), the difference between the means, 95% confidence interval, and R squared, and F test by F value, degrees of freedom for the numerator (DFn) and for the denominator (Dfd) and P value. In particular p<0.05 is indicated with *, p<0.01 with **, and p<0.001 with ***. Moreover, the correlations between the cytokine levels and clinical/biochemical data were determined using the Pearson correlation coefficient. Values of p<0.05 were considered to be statistically significant. The statistical programs Prism 4 (GraphPad Software, San Diego, CA, USA) was employed.
Functional and Interactomic studies
The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to classify proteins according to their biological processes, as well as the metabolic pathways in which they are involved [13]. Moreover, the network analysis between the most significant proteins was performed by Ingenuity Pathway Analysis (IPA).
Anti-TP53 assay
Anti-p53 antibodies were detected with an ELISA test kit (Pharmacell, Paris, France) by using microtiter plates coated with recombinant wild-type human p53 protein (to detect specific anti-p53 antibodies) or with a control protein (to detect nonspecific anti-p53 interactions). A peroxidase-conjugated goat antihuman IgG bound to anti-p53 antibodies. The specific p53/anti-p53-conjugated complexes were revealed by the addition of a peroxidase substrate (TMB), resulting in a colorimetric reaction. The absorbance was read at 450 nm, and the anti-p53 levels were expressed in units/mL and categorized as positive when >0.90 units/mL and negative otherwise [14].
Results
Comparison between Patients with T2D, CHC or HCC and Healthy Donors
In Fig 1, and in Tables 2 and 3 we report the proteins that show different serum levels in T2D or CHC or HCC patients respect to controls with the related statistical evaluations; data not statistically significant are not reported. Greater amounts of HGF, IL-2R, s-IL-6Ra, IL-18, leptin, sVEGFR-1 and sVEGFR-2 were secreted by T2D, CHC and HCC patients in comparison with the healthy controls, whereas those of glucagon only by T2D and HCC patients, those of β-NGF, CXCL1, CXCL9, CXCL12, IL-16, and PECAM-1 by CHC and HCC patients, and those of IFN-α and Prolactin only in HCC patients. The most part of these data agrees with our recently published results. In fact, we have confirmed the increased amount of IL-2R, IL-18, HGF, glucagon, and leptin that were found in T2D patients [9] as well as of β-NGF, CXCL1, CXCL9, CXCL12, HGF, IL-2R, s-IL-6Ra, IL-18, IFN-α, IL-16, PECAM-1 and Prolactin found in patients with CHC as well as with HCC and CHC-related cirrhosis [8–9].
Fig 1. Significant cytokines in some patient groups belonging to discovery set.
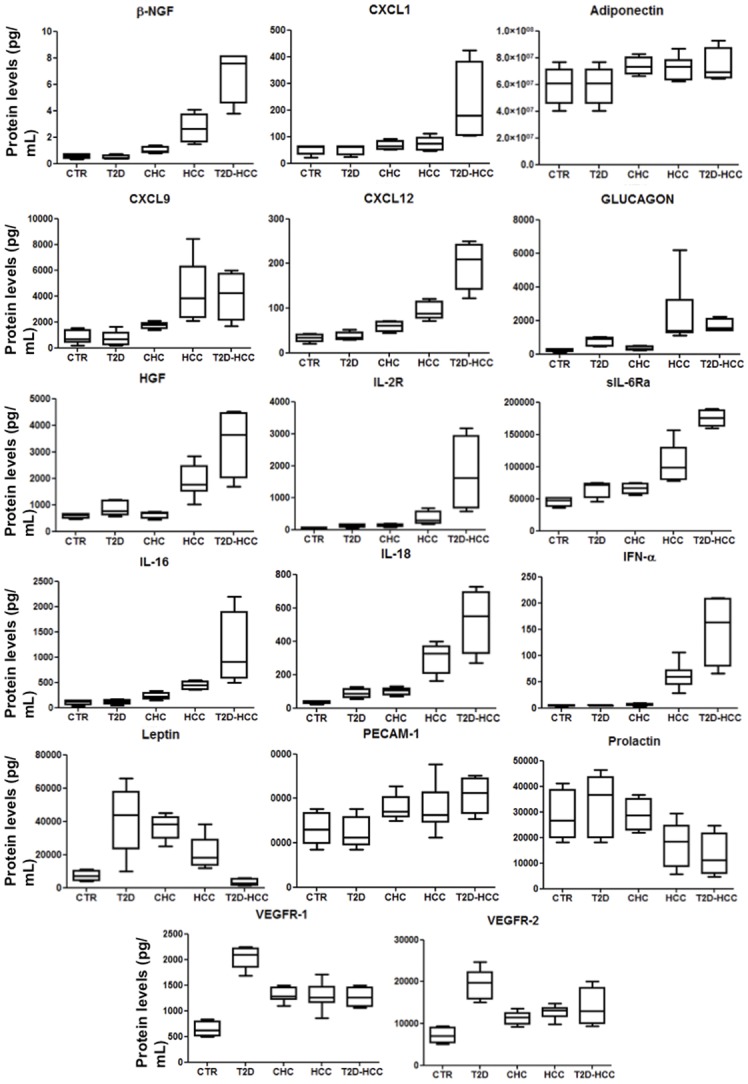
We report the significant molecule levels from controls, patients with type 2 diabetes (T2D), chronic hepatitis C (CHC), hepatocellular carcinoma (HCC) and hepatocellular carcinoma and type 2 diabetes (T2D-HCC) shown by means of box-and-whisker graphs. The boxes extend from the 25th to the 75th percentile, and the line in the middle is the median. The error bars extend down to the lowest value and up to the highest.
Table 2. Statistical evaluation on the serum levels (expressed in pg/mL) of significant cytokines in the healthy controls and in four patient groups belonging to discovery set.
We report for each cytokine the minimum and maximum values, the 25% and 75% Percentiles, the median, the mean, standard deviation, standard error, and the lower and upper 95% confidence intervals (CI).
| CTR | T2D | CHC | HCC | T2D-HCC | |
|---|---|---|---|---|---|
| ADIPOQ | |||||
| Minimum (pg/mL) | 40200000 | 40240000 | 69550000 | 62550000 | 66550000 |
| 25% Percentile (pg/mL) | 45590000 | 46240000 | 70240000 | 63180000 | 67190000 |
| Median (pg/mL) | 60980000 | 60180000 | 73180000 | 73250000 | 70683331 |
| 75% Percentile (pg/mL) | 66350000 | 67700000 | 80520000 | 78570000 | 87570000 |
| Maximum (pg/mL) | 66730000 | 68730000 | 82670000 | 87070000 | 92670000 |
| Mean (pg/mL) | 56970000 | 57610000 | 74650000 | 71880000 | 75158454 |
| Std. Deviation (pg/mL) | 11290000 | 11660000 | 5661000 | 9057000 | 8696033 |
| Std. Error (pg/mL) | 5049000 | 5216000 | 2831000 | 3423000 | 5956000 |
| Lower 95% CI (pg/mL) | 42960000 | 43130000 | 65640000 | 63500000 | 56190000 |
| Upper 95% CI (pg/mL) | 70990000 | 72100000 | 83650000 | 80260000 | 94100000 |
| GLUCAGON | |||||
| Minimum (pg/mL) | 124.0 | 453.3 | 220.0 | 1088 | 1400 |
| 25% Percentile (pg/mL) | 156.3 | 458.2 | 235.0 | 1256 | 1407 |
| Median (pg/mL) | 265.0 | 923.0 | 300.0 | 1390 | 1629 |
| 75% Percentile (pg/mL) | 303.3 | 992.5 | 450.0 | 3239 | 2093 |
| Maximum (pg/mL) | 312.0 | 1026 | 510.0 | 6177 | 2224 |
| Mean (pg/mL) | 241.5 | 764.9 | 334.0 | 2437 | 1650 |
| Std. Deviation (pg/mL) | 81.99 | 282.4 | 117.6 | 1837 | 248.9 |
| Std. Error (pg/mL) | 41.00 | 126.3 | 52.59 | 694.3 | 191.1 |
| Lower 95% CI (pg/mL) | 111.0 | 414.2 | 188.0 | 738.0 | 1079 |
| Upper 95% CI (pg/mL) | 372.0 | 1116 | 480.0 | 4136 | 2296 |
| β-NGF | |||||
| Minimum (pg/mL) | 0.3100 | 0.3700 | 0.8000 | 1.470 | 3.770 |
| 25% Percentile (pg/mL) | 0.3700 | 0.3700 | 0.8250 | 1.655 | 4.578 |
| Median (pg/mL) | 0.6000 | 0.3700 | 0.9500 | 2.610 | 7.550 |
| 75% Percentile (pg/mL) | 0.7100 | 0.6400 | 1.300 | 3.735 | 8.140 |
| Maximum (pg/mL) | 0.7300 | 0.7300 | 1.400 | 4.090 | 8.140 |
| Mean (pg/mL) | 0.5600 | 0.4600 | 1.025 | 2.678 | 6.975 |
| Std. Deviation (pg/mL) | 0.1822 | 0.1800 | 0.2630 | 1.079 | 1.493 |
| Std. Error (pg/mL) | 0.09110 | 0.0900 | 0.1315 | 0.4823 | 1.033 |
| Lower 95% CI (pg/mL) | 0.2701 | 0.1736 | 0.6065 | 1.339 | 3.475 |
| Upper 95% CI (pg/mL) | 0.8499 | 0.7464 | 1.443 | 4.017 | 10.05 |
| CXCL1 | |||||
| Minimum (pg/mL) | 22.52 | 24.47 | 50.52 | 46.10 | 104.6 |
| 25% Percentile (pg/mL) | 33.50 | 31.00 | 52.91 | 49.91 | 105.3 |
| Median (pg/mL) | 60.85 | 62.25 | 64.65 | 73.19 | 200.4 |
| 75% Percentile (pg/mL) | 63.63 | 64.93 | 83.30 | 95.78 | 380.8 |
| Maximum (pg/mL) | 65.01 | 65.01 | 90.74 | 110.7 | 424.4 |
| Mean (pg/mL) | 51.02 | 50.82 | 67.51 | 72.91 | 212.5 |
| Std. Deviation (pg/mL) | 17.84 | 18.71 | 16.48 | 25.40 | 95.21 |
| Std. Error (pg/mL) | 7.980 | 8.367 | 6.729 | 11.36 | 75.61 |
| Lower 95% CI (pg/mL) | 28.86 | 27.59 | 50.22 | 41.37 | -18.97 |
| Upper 95% CI (pg/mL) | 73.18 | 74.05 | 84.81 | 104.5 | 462.3 |
| CXCL12 | |||||
| Minimum (pg/mL) | 20.35 | 29.35 | 45.00 | 71.33 | 122.0 |
| 25% Percentile (pg/mL) | 25.18 | 29.68 | 47.50 | 76.93 | 141.4 |
| Median (pg/mL) | 34.00 | 34.00 | 61.00 | 88.00 | 205.6 |
| 75% Percentile (pg/mL) | 40.79 | 45.79 | 70.00 | 114.6 | 242.5 |
| Maximum (pg/mL) | 42.58 | 52.58 | 71.00 | 121.5 | 250.0 |
| Mean (pg/mL) | 33.19 | 36.99 | 59.50 | 94.21 | 202.6 |
| Std. Deviation (pg/mL) | 8.626 | 9.530 | 11.82 | 20.15 | 32.8 |
| Std. Error (pg/mL) | 3.858 | 4.262 | 5.909 | 9.010 | 27.33 |
| Lower 95% CI (pg/mL) | 22.48 | 25.15 | 40.69 | 69.19 | 110.9 |
| Upper 95% CI (pg/mL) | 43.90 | 48.82 | 78.31 | 119.2 | 284.9 |
| CXCL9 | |||||
| Minimum (pg/mL) | 188.4 | 188.4 | 1400 | 2062 | 1673 |
| 25% Percentile (pg/mL) | 427.4 | 230.8 | 1500 | 2346 | 2117 |
| Median (pg/mL) | 705.9 | 666.5 | 1800 | 3842 | 4073 |
| 75% Percentile (pg/mL) | 1396 | 1163 | 1950 | 6297 | 5750 |
| Maximum (pg/mL) | 1519 | 1619 | 2100 | 8443 | 6000 |
| Mean (pg/mL) | 870.6 | 690.6 | 1740 | 4421 | 4106 |
| Std. Deviation (pg/mL) | 528.4 | 567.7 | 260.8 | 2325 | 1141.5 |
| Std. Error (pg/mL) | 236.3 | 253.9 | 116.6 | 822.0 | 944.9 |
| Lower 95% CI (pg/mL) | 214.5 | -14.31 | 1416 | 2478 | 1024 |
| Upper 95% CI (pg/mL) | 1527 | 1396 | 2064 | 6365 | 7038 |
| HGF | |||||
| Minimum (pg/mL) | 459.1 | 569.1 | 450.0 | 1014 | 65.92 |
| 25% Percentile (pg/mL) | 489.6 | 614.1 | 480.0 | 1510 | 79.78 |
| Median (pg/mL) | 619.2 | 757.3 | 680.0 | 1774 | 162.7 |
| 75% Percentile (pg/mL) | 658.7 | 1165 | 720.0 | 2456 | 208.5 |
| Maximum (pg/mL) | 659.1 | 1181 | 740.0 | 2831 | 210.0 |
| Mean (pg/mL) | 589.1 | 863.1 | 616.0 | 1900 | 150.3 |
| Std. Deviation (pg/mL) | 94.03 | 283.8 | 127.8 | 591.2 | 69.32 |
| Std. Error (pg/mL) | 47.02 | 126.9 | 57.15 | 187.0 | 34.66 |
| Lower 95% CI (pg/mL) | 439.5 | 510.8 | 457.3 | 1477 | 40.04 |
| Upper 95% CI (pg/mL) | 738.8 | 1215 | 774.7 | 2323 | 260.6 |
| IFN-α | |||||
| Minimum (pg/mL) | 3.190 | 4.190 | 3.500 | 28.45 | 65.92 |
| 25% Percentile (pg/mL) | 3.393 | 4.190 | 4.250 | 45.08 | 79.78 |
| Median (pg/mL) | 4.095 | 4.595 | 7.000 | 59.34 | 169.20 |
| 75% Percentile (pg/mL) | 5.210 | 5.750 | 8.500 | 71.76 | 208.5 |
| Maximum (pg/mL) | 5.550 | 6.000 | 9.000 | 106.1 | 210.0 |
| Mean (pg/mL) | 4.233 | 4.845 | 6.500 | 60.70 | 161.81 |
| Std. Deviation (pg/mL) | 0.9795 | 0.8595 | 2.236 | 22.78 | 42.38 |
| Std. Error (pg/mL) | 0.4898 | 0.4297 | 1.000 | 7.593 | 34.66 |
| Lower 95% CI (pg/mL) | 2.674 | 3.477 | 3.724 | 43.19 | 40.04 |
| Upper 95% CI (pg/mL) | 5.791 | 6.213 | 9.276 | 78.21 | 260.6 |
| IL-16 | |||||
| Minimum (pg/mL) | 34.36 | 44.36 | 150.0 | 353.8 | 497.7 |
| 25% Percentile (pg/mL) | 61.94 | 66.94 | 175.0 | 355.2 | 579.3 |
| Median (pg/mL) | 117.6 | 117.6 | 220.0 | 444.3 | 942.2 |
| 75% Percentile (pg/mL) | 142.3 | 154.8 | 290.0 | 522.8 | 1894 |
| Maximum (pg/mL) | 151.7 | 171.7 | 330.0 | 545.7 | 2192 |
| Mean (pg/mL) | 105.2 | 112.2 | 230.0 | 440.1 | 1015.5 |
| Std. Deviation (pg/mL) | 45.67 | 48.33 | 66.71 | 85.39 | 440.6 |
| Std. Error (pg/mL) | 20.43 | 21.62 | 29.83 | 38.19 | 369.5 |
| Lower 95% CI (pg/mL) | 48.49 | 52.19 | 147.2 | 334.1 | -47.48 |
| Upper 95% CI (pg/mL) | 161.9 | 172.2 | 312.8 | 546.1 | 2304 |
| IL-18 | |||||
| Minimum (pg/mL) | 22.00 | 54.86 | 70.00 | 161.4 | 268.9 |
| 25% Percentile (pg/mL) | 24.75 | 63.64 | 79.00 | 206.7 | 326.6 |
| Median (pg/mL) | 36.50 | 88.10 | 105.0 | 326.4 | 551.2 |
| 75% Percentile (pg/mL) | 43.00 | 113.5 | 120.0 | 372.6 | 694.5 |
| Maximum (pg/mL) | 44.00 | 125.9 | 130.0 | 396.7 | 725.9 |
| Mean (pg/mL) | 34.75 | 88.46 | 100.6 | 291.6 | 532.1 |
| Std. Deviation (pg/mL) | 9.639 | 27.13 | 22.73 | 88.09 | 114.0 |
| Std. Error (pg/mL) | 4.820 | 12.13 | 10.17 | 33.29 | 96.71 |
| Lower 95% CI (pg/mL) | 19.41 | 54.78 | 72.37 | 210.2 | 215.8 |
| Upper 95% CI (pg/mL) | 50.09 | 122.1 | 128.8 | 373.1 | 831.4 |
| IL-2R | |||||
| Minimum (pg/mL) | 31.00 | 61.73 | 75.61 | 171.3 | 576.9 |
| 25% Percentile (pg/mL) | 37.00 | 67.89 | 89.83 | 195.7 | 682.7 |
| Median (pg/mL) | 50.00 | 137.0 | 137.0 | 303.1 | 1713.2 |
| 75% Percentile (pg/mL) | 65.00 | 167.0 | 175.1 | 564.5 | 2939 |
| Maximum (pg/mL) | 75.00 | 175.6 | 191.7 | 681.5 | 3174 |
| Mean (pg/mL) | 50.80 | 121.4 | 133.4 | 365.3 | 1765.1 |
| Std. Deviation (pg/mL) | 16.25 | 50.88 | 45.39 | 195.6 | 708.5 |
| Std. Error (pg/mL) | 7.269 | 22.75 | 20.30 | 69.14 | 591.5 |
| Lower 95% CI (pg/mL) | 30.62 | 58.18 | 76.99 | 201.8 | -136.6 |
| Upper 95% CI (pg/mL) | 70.98 | 184.5 | 189.7 | 528.8 | 3628 |
| Leptin | |||||
| Minimum (pg/mL) | 3820 | 10000 | 25000 | 11770 | 1274 |
| 25% Percentile (pg/mL) | 4323 | 23420 | 30000 | 13390 | 1602 |
| Median (pg/mL) | 6917 | 44020 | 38250 | 18380 | 2778.7 |
| 75% Percentile (pg/mL) | 10150 | 57910 | 42500 | 29120 | 5270 |
| Maximum (pg/mL) | 10870 | 65820 | 45000 | 38250 | 6000 |
| Mean (pg/mL) | 7131 | 41330 | 36650 | 20680 | 2967.9 |
| Std. Deviation (pg/mL) | 3022 | 20520 | 7449 | 10320 | 1182.5 |
| Std. Error (pg/mL) | 1511 | 9176 | 3331 | 4615 | 997.3 |
| Lower 95% CI (pg/mL) | 2323 | 15860 | 27400 | 7866 | 61.65 |
| Upper 95% CI (pg/mL) | 11940 | 66810 | 45900 | 33490 | 6409 |
| PECAM-1 | |||||
| Minimum (pg/mL) | 16900 | 168850 | 30030 | 22530 | 30920 |
| 25% Percentile (pg/mL) | 19600 | 19170 | 31760 | 29290 | 33190 |
| Median (pg/mL) | 26020 | 22310 | 33950 | 32480 | 43197.6 |
| 75% Percentile (pg/mL) | 33380 | 31660 | 40660 | 42670 | 49150 |
| Maximum (pg/mL) | 35300 | 35890 | 45510 | 55510 | 50440 |
| Mean (pg/mL) | 26400 | 24800 | 35810 | 35800 | 42653.7 |
| Std. Deviation (pg/mL) | 7280 | 7081 | 5680 | 9966 | 5018.9 |
| Std. Error (pg/mL) | 3256 | 3167 | 2319 | 3322 | 4167 |
| Lower 95% CI (pg/mL) | 17360 | 16000 | 29850 | 28140 | 28400 |
| Upper 95% CI (pg/mL) | 35430 | 33590 | 41770 | 43460 | 54920 |
| Prolactin | |||||
| Minimum (pg/mL) | 18140 | 18260 | 21930 | 5655 | 4550 |
| 25% Percentile (pg/mL) | 20030 | 20030 | 23030 | 8760 | 5912 |
| Median (pg/mL) | 26560 | 36560 | 28760 | 18530 | 10914.2 |
| 75% Percentile (pg/mL) | 38760 | 43760 | 35220 | 24700 | 21610 |
| Maximum (pg/mL) | 41200 | 46320 | 36560 | 29300 | 24760 |
| Mean (pg/mL) | 28830 | 32830 | 29000 | 17510 | 11859.3 |
| Std. Deviation (pg/mL) | 9701 | 12260 | 6303 | 8279 | 5109.9 |
| Std. Error (pg/mL) | 4338 | 5481 | 3151 | 2760 | 4275 |
| Lower 95% CI (pg/mL) | 16780 | 17610 | 18970 | 11140 | -740.4 |
| Upper 95% CI (pg/mL) | 40870 | 48050 | 39030 | 23870 | 26470 |
| sIL-6Ra | |||||
| Minimum (pg/mL) | 35780 | 45780 | 55780 | 77270 | 160000 |
| 25% Percentile (pg/mL) | 38060 | 52100 | 57300 | 79320 | 162500 |
| Median (pg/mL) | 47990 | 71460 | 66460 | 98930 | 177407 |
| 75% Percentile (pg/mL) | 51650 | 74130 | 73940 | 129800 | 187700 |
| Maximum (pg/mL) | 51840 | 74900 | 74900 | 156700 | 190000 |
| Mean (pg/mL) | 45900 | 65900 | 65900 | 103400 | 176227 |
| Std. Deviation (pg/mL) | 7429 | 13520 | 8693 | 31720 | 8960.9 |
| Std. Error (pg/mL) | 3715 | 6758 | 4347 | 14180 | 6507 |
| Lower 95% CI (pg/mL) | 34080 | 44390 | 52070 | 64050 | 154500 |
| Upper 95% CI (pg/mL) | 57720 | 87410 | 79730 | 142800 | 195900 |
| VEGFR-1 | |||||
| Minimum (pg/mL) | 500.0 | 1683 | 1097 | 855.8 | 1056 |
| 25% Percentile (pg/mL) | 514.1 | 1846 | 1220 | 1159 | 1087 |
| Median (pg/mL) | 617.8 | 2096 | 1282 | 1262 | 1292 |
| 75% Percentile (pg/mL) | 799.3 | 2217 | 1460 | 1468 | 1460 |
| Maximum (pg/mL) | 839.4 | 2251 | 1492 | 1705 | 1500 |
| Mean (pg/mL) | 643.8 | 2044 | 1305 | 1288 | 1288 |
| Std. Deviation (pg/mL) | 150.4 | 221.5 | 138.8 | 233.4 | 124.8 |
| Std. Error (pg/mL) | 75.18 | 99.06 | 52.47 | 73.80 | 96.42 |
| Lower 95% CI (pg/mL) | 404.5 | 1769 | 1177 | 1121 | 961.9 |
| Upper 95% CI (pg/mL) | 883.0 | 2320 | 1434 | 1455 | 1576 |
| VEGFR-2 | |||||
| Minimum (pg/mL) | 5055 | 15150 | 9258 | 9866 | 9375 |
| 25% Percentile (pg/mL) | 5291 | 15870 | 9866 | 11660 | 10030 |
| Median (pg/mL) | 6990 | 19720 | 11450 | 13080 | 12791 |
| 75% Percentile (pg/mL) | 9027 | 22260 | 12580 | 13730 | 18540 |
| Maximum (pg/mL) | 9375 | 24750 | 13530 | 14750 | 20060 |
| Mean (pg/mL) | 7103 | 19190 | 11190 | 12760 | 14445 |
| Std. Deviation (pg/mL) | 1945 | 3695 | 1576 | 1413 | 3793.6 |
| Std. Error (pg/mL) | 972.3 | 1652 | 595.7 | 446.7 | 2272 |
| Lower 95% CI (pg/mL) | 4008 | 14610 | 9730 | 11750 | 6621 |
| Upper 95% CI (pg/mL) | 10200 | 23780 | 12650 | 13770 | 21080 |
Table 3. Comparison of cytokine serum levels between patients and healthy controls in the discovery set.
We report the results of all the performed statistical analysis obtained by the nonparametric Mann-Whitney U test in terms of U test and P values, by the Unparied t test in terms of P value, t, the number of degrees of freedom (df), the difference between the means, 95% confidence interval, and R squared, and by F test in terms of F, degrees of freedom for the numerator (DFn) and for the denominator (Dfd) and P value. In particular, we reported in bold the values of p<0.05 indicated with *, of p<0.01 with **, and of p<0.0001 with ***.
| ADIPOQ | CTR vs T2D | CTR vs CHC | CTR vs HCC | CHC vs HCC | T2D vs HCC | T2D vs T2D-HCC | HCC vs T2D-HCC |
|---|---|---|---|---|---|---|---|
| Mann-Whitney Utest | |||||||
| U test | 11.5 | 0 | 6 | 11 | 6 | 2 | 12 |
| P value | 0.44 | 0.045* | 0.038* | 0.71 | 0.035* | 0.048* | 0.15 |
| Unpaired t test | |||||||
| P value | 0.9319 | 0.0253* | 0.0292* | 0.5982 | 0.0377* | 0.0418* | 0.6181 |
| t, df | t = 0.08816 df = 8 | t = 2.832 df = 7 | t = 2.543 df = 10 | t = 0.5462 df = 9 | t = 2.393 df = 10 | t = 2.221 df = 7 | t = 0.5163 df = 9 |
| Difference between means | -640000 ± 7259000 | -17670000 ± 6241000 | -14910000 ± 5861000 | 2768000 ± 5067000 | -14270000 ± 5961000 | -17530000 ± 7896000 | -3268000 ± 6330000 |
| 95% confidence interval | -17380000 to 16100000 | -32430000 to -2914000 | -27960000 to -1847000 | -8695000 to 14230000 | -27550000 to -985100 | -36210000 to 1141000 | -17590000 to 11050000 |
| R squared | 0.0009707 | 0.5339 | 0.3928 | 0.03209 | 0.3642 | 0.4133 | 0.02876 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.068, 4, 4 | 3.976, 4, 3 | 1.554, 4, 6 | 2.559, 6, 3 | 1.659, 4, 6 | 1.043, 3, 4 | 1.730, 3, 6 |
| P value | 0.9510 | 0.2860 | 0.5989 | 0.4713 | 0.5517 | 0.9289 | 0.5196 |
| GLUCAGON | |||||||
| Mann-Whitney Utest | |||||||
| U test | 0 | 7 | 0 | 0 | 0 | 0 | 12 |
| P value | 0.041* | 0.55 | 0.011* | 0.041* | 0.0295* | 0.0159* | 0.57 |
| Unpaired t test | |||||||
| P value | 0.0094** | 0.2259 | 0.0444* | 0.0304* | 0.0245* | 0.0041** | 0.4505 |
| t, df | t = 3.544 df = 7 | t = 1.328 df = 7 | t = 2.334 df = 9 | t = 2.521 df = 10 | t = 2.991 df = 10 | t = 4.181 df = 7 | t = 0.7887 df = 9 |
| Difference between means | -523.4 ± 147.7 | -92.50 ± 69.66 | -2195 ± 940.6 | -2103 ± 834.4 | -1672 ± 839.7 | -922.6 ± 220.7 | 749.5 ± 950.3 |
| 95% confidence interval | -872.6 to -174.2 | -257.3 to 72.25 | -4323 to -67.82 | -3962 to -244.1 | -3543 to 198.8 | -1444 to -400.8 | -1400 to 2899 |
| R squared | 0.6422 | 0.2012 | 0.3771 | 0.3885 | 0.2839 | 0.7141 | 0.06465 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 11.86, 4, 3 | 2.057, 4, 3 | 502.0, 6, 3 | 244.0, 6, 4 | 42.32, 6, 4 | 1.833, 3, 4 | 23.09, 6, 3 |
| P value | 0.0699 | 0.5798 | 0.0003*** | P<0.0001 | 0.0028** | 0.5628 | 0.0263* |
| β-NGF | |||||||
| Mann-Whitney Utest | |||||||
| U test | 6.5 | 0 | 0 | 0 | 0 | 0 | 1 |
| P value | 0.32 | 0.021* | 0.0021** | 0.041* | 0.0018** | 0.0001*** | 0.010** |
| Unpaired t test | |||||||
| P value | 0.4646 | 0.0271* | 0.0064** | 0.0212* | 0.0051** | 0.0009*** | 0.0062** |
| t, df | t = 0.7809 df = 6 | t = 2.907 df = 6 | t = 3.832 df = 7 | t = 2.957 df = 7 | t = 4.014 df = 7 | t = 6.078 df = 6 | t = 3.855 df = 7 |
| Difference between means | 0.1000 ± 0.1281 | -0.4650 ± 0.1600 | -2.118 ± 0.5527 | -1.653 ± 0.5590 | -2.218 ± 0.5526 | -6.303 ± 1.037 | -4.085 ± 1.059 |
| 95% confidence interval | -0.2134 to 0.4134 | -0.8565 to -0.07354 | -3.425 to -0.8108 | -2.975 to -0.3311 | -3.525 to -0.9111 | -8.840 to -3.765 | -6.590 to -1.579 |
| R squared | 0.09225 | 0.5847 | 0.6772 | 0.5554 | 0.6971 | 0.8603 | 0.6798 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.025, 3, 3 | 2.083, 3, 3 | 35.04, 4, 3 | 16.82, 4, 3 | 35.90, 4, 3 | 131.8, 3, 3 | 3.670, 3, 4 |
| P value | 0.9845 | 0.5620 | 0.0150* | 0.0430* | 0.0145* | 0.0022** | 0.2413 |
| CXCL1 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 11 | 1.1 | 0.9 | 14.5 | 0.6 | 0 | 1.1 |
| P value | 0.88 | 0.035* | 0.039* | 0.75 | 0.041* | 0.01** | 0.0381* |
| Unpaired t test | |||||||
| P value | 0.9866 | 0.0415* | 0.0453* | 0.6800 | 0.456* | 0.0383* | 0.0438* |
| t, df | t = 0.01730 df = 8 | t = 1.953 df = 9 | t = 1.977 df = 8 | t = 0.4261 df = 9 | t = 1.966 df = 8 | t = 2.547 df = 7 | t = 2.199 df = 7 |
| Difference between means | 0.2000 ± 11.56 | -16.49 ± 10.36 | -21.89 ± 13.88 | -5.399 ± 12.67 | -22.09 ± 14.11 | -170.8 ± 67.08 | -148.7 ± 67.64 |
| 95% confidence interval | -26.46 to 26.86 | -39.92 to 6.930 | -53.91 to 10.12 | -34.06 to 23.26 | -54.63 to 10.44 | -329.5 to -12.19 | -308.7 to 11.24 |
| R squared | 0.00003740 | 0.2199 | 0.2371 | 0.01978 | 0.2346 | 0.4809 | 0.4085 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.099, 4, 4 | 1.172, 4, 5 | 2.027, 4, 4 | 2.375, 4, 5 | 1.844, 4, 4 | 65.33, 3, 4 | 35.44, 3, 4 |
| P value | 0.9291 | 0.8457 | 0.5107 | 0.3683 | 0.5680 | 0.0015** | 0.0049** |
| CXCL12 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| P value | 0.76 | 0.0364* | 0.0031** | 0.038* | 0.0011** | 0.0070** | 0.0196* |
| Unpaired t test | |||||||
| P value | 0.5272 | 0.0061** | 0.0003*** | 0.0191* | 0.0004*** | 0.0003*** | 0.0054** |
| t, df | t = 0.6611 df = 8 | t = 3.877 df = 7 | t = 6.226 df = 8 | t = 3.029 df = 7 | t = 5.741 df = 8 | t = 6.570 df = 7 | t = 3.974 df = 7 |
| Difference between means | -3.800 ± 5.748 | -26.31 ± 6.787 | -61.02 ± 9.801 | -34.71 ± 11.46 | -57.22 ± 9.967 | -160.9 ± 24.49 | -103.7 ± 26.09 |
| 95% confidence interval | -17.06 to 9.456 | -42.37 to -10.26 | -83.62 to -38.42 | -61.81 to -7.606 | -80.20 to -34.24 | -218.8 to -103.0 | -165.4 to -41.98 |
| R squared | 0.05179 | 0.6823 | 0.8289 | 0.5672 | 0.8047 | 0.8605 | 0.6929 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.221, 4, 4 | 1.877, 3, 4 | 5.455, 4, 4 | 2.906, 4, 3 | 4.469, 4, 4 | 32.91, 3, 4 | 7.364, 3, 4 |
| P value | 0.8515 | 0.5488 | 0.1291 | 0.4074 | 0.1761 | 0.0056** | 0.0835 |
| CXCL9 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 10.5 | 1 | 0 | 1 | 0 | 0 | 14 |
| P value | 0.59 | 0.041* | 0.0023** | 0.042* | 0.0011** | 0.0435* | 0.16 |
| Unpaired t test | |||||||
| P value | 0.6178 | 0.0109* | 0.0070** | 0.0281* | 0.0052** | 0.0067** | 0.7781 |
| t, df | t = 0.5189 df = 8 | t = 3.299 df = 8 | t = 3.310 df = 11 | t = 2.527 df = 11 | t = 3.470 df = 11 | t = 3.803 df = 7 | t = 0.2895 df = 10 |
| Difference between means | 180.0 ± 346.9 | -869.4 ± 263.5 | -3551 ± 1073 | -2681 ± 1061 | -3731 ± 1075 | -3340 ± 878.4 | 390.6 ± 1349 |
| 95% confidence interval | -619.9 to 979.9 | -1477 to -261.7 | -5912 to -1190 | -5017 to -345.9 | -6097 to -1364 | -5418 to -1263 | -2616 to 3397 |
| R squared | 0.03257 | 0.5764 | 0.4990 | 0.3673 | 0.5226 | 0.6738 | 0.008309 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.154, 4, 4 | 4.106, 4, 4 | 19.36, 7, 4 | 79.49, 7, 4 | 16.77, 7, 4 | 11.08, 3, 4 | 1.514, 7, 3 |
| P value | 0.8927 | 0.2001 | 0.0123** | 0.0008*** | 0.0162** | 0.0417* | 0.7962 |
| HGF | |||||||
| Mann-Whitney Utest | |||||||
| U test | 2.5 | 7 | 2 | 0 | 2 | 0 | 5 |
| P value | 0.045* | 0.89 | 0.01** | 0.0013** | 0.027* | 0.0016** | 0.036* |
| Unpaired t test | |||||||
| P value | 0.0489* | 0.7369 | 0.0010** | 0.0004*** | 0.0029** | 0.0038** | 0.0109* |
| t, df | t = 1.990 df = 7 | t = 0.3497 df = 7 | t = 4.309 df = 12 | t = 4.716 df = 13 | t = 3.665 df = 13 | t = 4.254 df = 7 | t = 3.006 df = 12 |
| Difference between means | -274.0 ± 149.7 | -26.87 ± 76.84 | -1311 ± 304.2 | -1284 ± 272.2 | -1037 ± 282.9 | -2510 ± 590.2 | -1474 ± 490.3 |
| 95% confidence interval | -628.0 to 80.06 | -208.6 to 154.9 | -1973 to -647.8 | -1872 to -695.8 | -1648 to -425.6 | -3906 to -1115 | -2542 to -405.4 |
| R squared | 0.3237 | 0.01717 | 0.6074 | 0.6311 | 0.5081 | 0.7210 | 0.4295 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 9.107, 4, 3 | 1.847, 4, 3 | 39.53, 9, 3 | 21.40, 9, 4 | 4.341, 9, 4 | 21.10, 3, 4 | 4.860, 3, 9 |
| P value | 0.1001 | 0.6415 | 0.0117* | 0.0098** | 0.1711 | 0.0130* | 0.0562 |
| IFN-α | |||||||
| Mann-Whitney Utest | |||||||
| U test | 4 | 4 | 0 | 0 | 0 | 0 | 2 |
| P value | 0.72 | 0.66 | 0.035* | 0.041* | 0.0069** | 0.0040** | 0.0152* |
| Unpaired t test | |||||||
| P value | 0.3835 | 0.1037 | 0.0005*** | 0.0002*** | 0.0006*** | 0.0057** | 0.0039** |
| t, df | t = 0.9400 df = 6 | t = 1.870 df = 7 | t = 4.836 df = 11 | t = 5.212 df = 12 | t = 4.784 df = 11 | t = 4.198 df = 6 | t = 3.631 df = 11 |
| Difference between means | -0.6125 ± 0.6516 | -2.268 ± 1.213 | -56.47 ± 11.68 | -54.20 ± 10.40 | -55.85 ± 11.68 | -145.5 ± 34.66 | -89.64 ± 24.69 |
| 95% confidence interval | -2.207 to 0.9819 | -5.136 to 0.6007 | -82.17 to -30.76 | -76.86 to -31.54 | -81.55 to -30.15 | -230.3 to -60.68 | -144.0 to -35.31 |
| R squared | 0.1284 | 0.3331 | 0.6801 | 0.6936 | 0.6753 | 0.7460 | 0.5452 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.299, 3, 3 | 5.211, 4, 3 | 540.7, 8, 3 | 103.8, 8, 4 | 702.4, 8, 3 | 6505, 3, 3 | 9.261, 3, 8 |
| P value | 0.8349 | 0.2063 | 0.0002*** | 0.0005*** | 0.0002*** | P<0.0001*** | 0.0111* |
| IL-16 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 11 | 1 | 0 | 0 | 0 | 0 | 2 |
| P value | 0.89 | 0.031* | 0.004** | 0.048* | 0.0013** | 0.0059** | 0.0132* |
| Unpaired t test | |||||||
| P value | 0.8198 | 0.0087** | P<0.0001 | 0.0467* | 0.0346* | 0.0168* | 0.0436* |
| t, df | t = 0.2354 df = 8 | t = 3.452 df = 8 | t = 7.733 df = 8 | t = 1.937 df = 12 | t = 2.382 df = 12 | t = 3.123 df = 7 | t = 2.102 df = 7 |
| Difference between means | -7.000 ± 29.74 | -124.8 ± 36.16 | -334.9 ± 43.31 | -516.0 ± 266.5 | -633.8 ± 266.0 | -1016 ± 325.5 | -688.4 ± 327.4 |
| 95% confidence interval | -75.58 to 61.58 | -208.2 to -41.42 | -434.8 to -235.0 | -1097 to 64.56 | -1214 to -54.13 | -1786 to -246.5 | -1463 to 85.96 |
| R squared | 0.006878 | 0.5983 | 0.8820 | 0.2381 | 0.3211 | 0.5821 | 0.3871 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.120, 4, 4 | 2.133, 4, 4 | 3.495, 4, 4 | 76.43, 8, 4 | 145.6, 8, 4 | 233.8, 3, 4 | 74.91, 3, 4 |
| P value | 0.9153 | 0.4811 | 0.2529 | 0.0008*** | 0.0002*** | 0.0001*** | 0.0011** |
| IL-18 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| P value | 0.041* | 0.023* | 0.0021** | 0.039* | 0.030* | 0.0001*** | 0.034* |
| Unpaired t test | |||||||
| P value | 0.0073** | 0.0011** | 0.0003*** | 0.0009*** | 0.0006*** | 0.0015** | 0.0212* |
| t, df | t = 3.732 df = 7 | t = 5.362 df = 7 | t = 5.682 df = 9 | t = 4.679 df = 10 | t = 4.932 df = 10 | t = 5.057 df = 7 | t = 2.786 df = 9 |
| Difference between means | -53.71 ± 14.39 | -65.85 ± 12.28 | -256.9 ± 45.21 | -191.0 ± 40.83 | -203.2 ± 41.20 | -435.1 ± 86.04 | -232.0 ± 83.25 |
| 95% confidence interval | -87.74 to -19.67 | -94.89 to -36.81 | -359.2 to -154.6 | -282.0 to -100.1 | -295.0 to -111.4 | -638.6 to -231.6 | -420.3 to -43.64 |
| R squared | 0.6655 | 0.8042 | 0.7820 | 0.6865 | 0.7087 | 0.7851 | 0.4631 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 7.919, 4, 3 | 5.562, 4, 3 | 83.51, 6, 3 | 15.01, 6, 4 | 10.55, 6, 4 | 50.84, 3, 4 | 4.821, 3, 6 |
| P value | 0.1206 | 0.1902 | 0.0040** | 0.0205* | 0.0391* | 0.0024** | 0.0973 |
| IL-2R | |||||||
| Mann-Whitney Utest | |||||||
| U test | 0 | 0 | 0 | 2 | 1 | 0 | 2 |
| P value | 0.049* | 0.031* | 0.003** | 0.036* | 0.023* | 0.016* | 0.0132* |
| Unpaired t test | |||||||
| P value | 0.0183* | 0.0050** | 0.0047** | 0.0261* | 0.0210* | 0.0168* | 0.0071** |
| t, df | t = 2.954 df = 8 | t = 3.829 df = 8 | t = 3.529 df = 11 | t = 2.568 df = 11 | t = 2.691 df = 11 | t = 3.123 df = 7 | t = 3.374 df = 10 |
| Difference between means | -70.55 ± 23.89 | -82.55 ± 21.56 | -314.5 ± 89.11 | -231.9 ± 90.29 | -243.9 ± 90.64 | -1625 ± 520.2 | -1381 ± 409.3 |
| 95% confidence interval | -125.6 to -15.47 | -132.3 to -32.83 | -510.6 to -118.3 | -430.7 to -33.18 | -443.4 to -44.42 | -2855 to -394.3 | -2293 to -468.8 |
| R squared | 0.5217 | 0.6469 | 0.5310 | 0.3749 | 0.3970 | 0.5822 | 0.5323 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 9.797, 4, 4 | 7.799, 4, 4 | 144.8, 7, 4 | 18.56, 7, 4 | 14.77, 7, 4 | 540.7, 3, 4 | 36.60, 3, 7 |
| P value | 0.0483* | 0.0716 | 0.0002*** | 0.0134* | 0.0205* | P<0.0001*** | 0.0002*** |
| Leptin | |||||||
| Mann-Whitney Utest | |||||||
| U test | 1 | 0 | 0 | 2.5 | 6 | 0 | 0 |
| P value | 0.011* | 0.035* | 0.002** | 0.041* | 0.012* | 0.0016** | 0.0112* |
| Unpaired t test | |||||||
| P value | 0.0138* | 0.022* | 0.0404* | 0.0017** | 0.0059** | 0.0082** | 0.0133* |
| t, df | t = 3.261 df = 7 | t = 2.373 df = 7 | t = 2.510 df = 7 | t = 4.031 df = 12 | t = 3.335 df = 12 | t = 3.649 df = 7 | t = 3.288 df = 7 |
| Difference between means | -34200 ± 10490 | -29520 ± 4004 | -13550 ± 5399 | 23720 ± 5886 | 28410 ± 8517 | 38100 ± 10440 | 17440 ± 5306 |
| 95% confidence interval | -59010 to -9397 | -38990 to -20050 | -26320 to -780.5 | 10900 to 36550 | 9848 to 46970 | 13400 to 62790 | 4895 to 29990 |
| R squared | 0.6030 | 0.8859 | 0.4736 | 0.5752 | 0.4811 | 0.6554 | 0.6069 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 46.11, 4, 3 | 6.078, 4, 3 | 11.67, 4, 3 | 2.510, 8, 4 | 3.023, 4, 8 | 105.8, 4, 3 | 26.77, 4, 3 |
| P value | 0.0100* | 0.1700 | 0.0715 | 0.3903 | 0.1713 | 0.0029** | 0.0221* |
| PECAM-1 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 10.5 | 4 | 9 | 36.5 | 7 | 1 | 11 |
| P value | 0.84 | 0.041* | 0.032* | 0.77 | 0.029* | 0.0377* | 0.33 |
| Unpaired t test | |||||||
| P value | 0.7337 | 0.0390* | 0.0405* | 0.6774 | 0.0157* | 0.0133* | 0.3290 |
| t, df | t = 0.3523 df = 8 | t = 2.413 df = 9 | t = 1.991 df = 12 | t = 0.4233 df = 17 | t = 2.702 df = 16 | t = 3.290 df = 7 | t = 1.021 df = 11 |
| Difference between means | 1600 ± 4542 | -9410 ± 3900 | -9405 ± 5109 | -1798 ± 4248 | -12810 ± 4741 | -16870 ± 5127 | -5861 ± 5738 |
| 95% confidence interval | -8873 to 12070 | -18230 to -589.6 | -20540 to 1726 | -10760 to 7165 | -22860 to -2758 | -28990 to -4741 | -18490 to 6768 |
| R squared | 0.01528 | 0.3929 | 0.2203 | 0.01043 | 0.3133 | 0.6072 | 0.08664 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.057, 4, 4 | 1.643, 4, 5 | 1.874, 8, 4 | 2.837, 12, 5 | 1.825, 12, 4 | 1.385, 3, 4 | 1.430, 8, 3 |
| P value | 0.9585 | 0.5940 | 0.5690 | 0.2581 | 0.5917 | 0.7374 | 0.8433 |
| Prolactin | |||||||
| Mann-Whitney Utest | |||||||
| U test | 9.5 | 9.5 | 9 | 4 | 8 | 2 | 12 |
| P value | 0.77 | 0.62 | 0.041* | 0.047* | 0.045* | 0.043* | 0.57 |
| Unpaired t test | |||||||
| P value | 0.5829 | 0.9764 | 0.0393* | 0.0319* | 0.0158* | 0.0285* | 0.3751 |
| t, df | t = 0.5722 df = 8 | t = 0.03066 df = 7 | t = 2.313 df = 12 | t = 2.456 df = 11 | t = 2.807 df = 12 | t = 2.749 df = 7 | t = 0.9244 df = 11 |
| Difference between means | -4000 ± 6990 | -173.1 ± 5644 | 11320 ± 4897 | 11500 ± 4681 | 15320 ± 5458 | 19960 ± 7261 | 4641 ± 5020 |
| 95% confidence interval | -20120 to 12120 | -13520 to 13180 | 654.1 to 21990 | 1193 to 21800 | 3430 to 27220 | 2792 to 37140 | -6409 to 15690 |
| R squared | 0.03932 | 0.0001343 | 0.3083 | 0.3541 | 0.3964 | 0.5192 | 0.07208 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 1.596, 4, 4 | 2.369, 4, 3 | 1.373, 4, 8 | 1.726, 8, 3 | 2.191, 4, 8 | 2.055, 4, 3 | 1.067, 3, 8 |
| P value | 0.6616 | 0.5046 | 0.6499 | 0.7106 | 0.3206 | 0.5805 | 0.8317 |
| sIL-6Ra | |||||||
| Mann-Whitney Utest | |||||||
| U test | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| P value | 0.032* | 0.039* | 0.0037** | 0.032* | 0.021* | 0.0001*** | 0.024* |
| Unpaired t test | |||||||
| P value | 0.0410* | 0.0129* | 0.0099** | 0.0119* | 0.0125* | P<0.0001*** | 0.0040** |
| t, df | t = 2.594 df = 6 | t = 3.498 df = 6 | t = 3.505 df = 7 | t = 3.010 df = 11 | t = 2.981 df = 11 | t = 11.65 df = 6 | t = 4.205 df = 7 |
| Difference between means | -20000 ± 7711 | -20000 ± 5718 | -57530 ± 16410 | -69420 ± 23060 | -69420 ± 23290 | -109300 ± 9381 | -71760 ± 17070 |
| 95% confidence interval | -38870 to -1131 | -33990 to -6009 | -96340 to -18720 | -120200 to -18660 | -120700 to -18160 | -132200 to -86340 | -112100 to -31400 |
| R squared | 0.5286 | 0.6710 | 0.6371 | 0.4517 | 0.4468 | 0.9577 | 0.7163 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 3.309, 3, 3 | 1.369, 3, 3 | 18.22, 4, 3 | 26.43, 8, 3 | 10.93, 8, 3 | 1.078, 3, 3 | 5.939, 4, 3 |
| P value | 0.3519 | 0.8024 | 0.0384 | 0.0212* | 0.0746 | 0.9519 | 0.1751 |
| VEGFR-1 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 0 | 0 | 0 | 32.5 | 1 | 0 | 18.5 |
| P value | 0.0004*** | 0.0043** | 0.0031** | 0.71 | 0.0013** | 0.0016** | 0.89 |
| Unpaired t test | |||||||
| P value | P<0.0001*** | P<0.0001*** | 0.0003*** | 0.8609 | 0.0011** | 0.0009*** | 0.8882 |
| t, df | t = 10.75 df = 7 | t = 7.395 df = 9 | t = 5.048 df = 12 | t = 0.1782 df = 15 | t = 6.012 df = 13 | t = 5.515 df = 7 | t = 0.1436 df = 12 |
| Difference between means | -1401 ± 130.3 | -661.7 ± 89.48 | -644.0 ± 127.6 | 17.65 ± 99.04 | 756.7 ± 125.9 | 775.7 ± 140.7 | 19.03 ± 132.5 |
| 95% confidence interval | -1709 to -1093 | -864.1 to -459.3 | -922.0 to -366.0 | -193.4 to 228.7 | 484.8 to 1029 | 443.0 to 1108 | -269.7 to 307.7 |
| R squared | 0.9429 | 0.8587 | 0.6799 | 0.002114 | 0.7355 | 0.8129 | 0.001716 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 2.171, 4, 3 | 1.173, 3, 6 | 2.410, 9, 3 | 2.827, 9, 6 | 1.110, 9, 4 | 1.319, 4, 3 | 1.465, 9, 3 |
| P value | 0.5504 | 0.7904 | 0.5071 | 0.2188 | 0.9950 | 0.8537 | 0.8307 |
| VEGFR-2 | |||||||
| Mann-Whitney Utest | |||||||
| U test | 0 | 1 | 0 | 15.5 | 0 | 4 | 18 |
| P value | 0.0005*** | 0.045* | 0.039* | 0.68 | 0.0007*** | 0.036* | 0.89 |
| Unpaired t test | |||||||
| P value | 0.0006*** | 0.0041** | 0.0010** | 0.482 | 0.0003*** | 0.042* | 0.4866 |
| t, df | t = 5.872 df = 7 | t = 3.816 df = 9 | t = 2.116 df = 12 | t = 2.151 df = 15 | t = 4.975 df = 13 | t = 1.951 df = 7 | t = 0.7179 df = 12 |
| Difference between means | -12090 ± 2059 | -4085 ± 1070 | -5654 ± 924.5 | -1569 ± 729.5 | 6438 ± 1294 | 5342 ± 2737 | -1096 ± 1527 |
| 95% confidence interval | -16960 to -7222 | -6506 to -1663 | -7668 to -3639 | -3124 to -14.66 | 3643 to 9233 | -1132 to 11820 | -4423 to 2231 |
| R squared | 0.8313 | 0.6180 | 0.7571 | 0.2358 | 0.6556 | 0.3523 | 0.04118 |
| F test to compare variances | |||||||
| F,DFn, Dfd | 3.610, 4, 3 | 1.522, 3, 6 | 1.895, 3, 9 | 1.245, 6, 9 | 6.841, 4, 9 | 1.513, 3, 4 | 10.35, 3, 9 |
| P value | 0.3199 | 0.6044 | 0.4019 | 0.7365 | 0.0164 | 0.6803 | 0.0056 |
Comparing the serum levels in CHC and HCC patients we can underline that the concentrations of β-NGF, CXCL9, CXCL12, IL-16, IL-18, IL-2R, Leptin, sIL-6Ra were higher in HCC patients and indicated as possible index of the chronic inflammation leading in CHC patients to the HCC development. Moreover, since the stage of fibrosis in CHC patients has been determined by Ishak index (Table 1), we divided these patients into three subgroups corresponding to stages F2, F3 and F4. No significant difference was observed in CHC patients matching F3 and F4 grades probably because they corresponded to two stages of fibrosis, already well advanced. The comparison of F2 and F4 patients showed that the concentrations of IL-2R, CXCL9, CXCL12 and sIL-6Ra were statistically higher (with p<0.05) in CHC patients with F4 grade. In overall, we find that these results are in agreement with those recently published by our group [15].
Finally, we have also compared the serum levels of these proteins in patients with T2D and HCC evidencing that glucagon, HGF, β-NGF, CXCL1, CXCL9, CXCL12, IFN-α, IL-2R, IL-16, IL-18, PECAM-1 and Prolactin are higher in HCC patients, whereas leptin, sVEGFR-1 and sVEGFR-2 are lower than in patients with T2D. No difference of sIL-6R levels is evident between T2D and HCC patients.
Comparison between Patients with T2D-HCC and those with T2D or HCC
Since our aim is to identify new markers specific for the association between diabetes and HCC, we compared the levels of all the 49 proteins evaluated in T2D-HCC patients and in those with T2D or HCC alone. From the Fig 1 and the Tables 2 and 3, we can underline that: i) the levels of ADIPOQ, β-NGF, CXCL1, CXCL12, HGF, IL-2R, sIL-6Ra, IL-16, IL-18, IFN-α were higher in T2D-HCC patients in comparison with those with T2D or HCC, ii) the levels of LEP were lower in T2D-HCC patients in comparison with those with T2D or HCC, iii) the levels of CXCL9, PECAM-1, Prolactin, glucagon, sVEGFR-1 and sVEGFR-2 presented similar levels in patients with only HCC and with both T2D and HCC, iv) the levels of CXCL9, PECAM-1, Prolactin, and glucagon were higher in T2D-HCC patients than in those with only T2D, and v) the levels of sVEGFR-1 and sVEGFR-2 were lower in T2D-HCC patients than in those with only T2D.
Then, we have correlated the serum levels of all the significant proteins in T2D-HCC patients with clinical/biochemical data by means of the Pearson correlation coefficient. In these patients, IL-18 showed a significant correlation with alpha-fetoprotein (AFP) and glycemic levels while HGF only with AFP. This suggests that IL-18 can be used as an index of the co-presence of type 2 diabetes and liver cancer whereas HGF is specific only for the cancer. Moreover, CXCL9 and Prolactin resulted to be correlated with the transaminases (AST and ALT), thus confirming that these proteins can be considered as predictors of inflammatory activation during the progression of T2D and HCV-related cirrhosis, which leads to the cancer.
Functional and network analysis
In general, epidemiological studies show that the liver carcinogenesis has very complex etiologies and, in addition to being associated with viral infection, is also connected with other risk factors such as obesity and T2D. In this context the availability of large amounts of molecular data, as the ones we have collected, can give rise to computational analyses aimed at creating new concepts and statistical and computational models. In this context, it is not easy to correlate those clinical and molecular data, which may be the most representative and sensitive to distinguish the stages of progression of the different syndromes, considered individually. So, to understand in which metabolic pathways all the major proteins that we identified were involved, we conducted a functional analysis using the DAVID tool [13]. This analysis was also supplemented by a network study with the Ingenuity Pathway Analysis (IPA). Table 4 shows how these seventeen proteins, based on what is known in literature, may be involved in six metabolic pathways.
Table 4. Metabolic pathways showing the constitutive proteins considered as significantly involved.
| Pathways | Proteins |
|---|---|
| Cytokine-cytokine receptor interaction | CXCL1, CXCL9, CXCL12, HGF, IL-2RA, IL-6R, IL-18, LEP, PRL, VEGFR-1, VEGFR-2 |
| JAK-STAT signaling | IL-2RA, IL-6R, LEP, PRL |
| Hepatic fibrosis/Hepatic Stellate Cell Activation | CXCL9, HGF, IL-6R, LEP, VEGFR-1, VEGFR-2 |
| Granulocyte Adhesion and Diapedesis | CXCL1, CXCL9, CXCL12, IL-18, PECAM-1 |
| NFkB signaling | IL-18, NGF, VEGFR-1, VEGFR-2 |
| Role of macrophages, fibroblasts, and endothelial cells | CXCL12, IL-6R, IL-16, IL-18 |
The interactomic analysis shows that all the significant cytokines are involved in a network named “Cellular movement, Hematological System Development and Function, Immune Cell Trafficking” on the basis of the function associated with them and of data mining from the experimental studies reported in the literature (Fig 2). This network reveals that these proteins are connected by six HUB nodes, such as EP300 (E1A binding protein p300), NR4A1 (nuclear receptor subfamily 4, group A, member 1), NR2F1 (nuclear receptor subfamily 2, group F, member 1), RELA (nuclear factor NF-kappa-B p65 subunit), STAT3 (signal-transducer-and-activator-of-transcription 3) and TP53 (tumor protein p53), which are closely related between them. The hub nodes, representing the centers of metabolic correlation that exercise a direct control over the coordinated proteins and often through the formation of a complex, have a strategic value, both because they centralize the control and because they are the best targets for each project aimed at creating specific drugs. In details we can underline that: i) EP300 is connected with ADIPOQ, Glucagon (GCG), sVEGFR-2 (KDR), Leptin (LEP), and Prolactin (PRL), ii) NR2F1 with HGF, iii) NR4A1 with ADIPOQ, CXCL12, IL-16, Leptin (LEP), and Prolactin (PRL), iv) RELA with CXCL9, CXCL12, IL-2RA, PECAM-1, and VEGF that interacts with its two receptors, VEGFR-1 (FLT1) and VEGFR-2 (KDR), v) STAT3 with ADIPOQ, CXL9, HGF, IL-2RA, sVEGFR-2 (KDR), Leptin (LEP), and PECAM-1, and vi) TP53 with CXCL1, CXCL12, IL-2RA, PECAM-1, Prolactin, and VEGF as in the case of RELA.
Fig 2. Interactomic analysis of the significant molecules performed by means of the Ingenuity Pathway Analysis (IPA).
The interactome shows the close functional association between significant cytokines (evidenced by yellow symbols) as well as the paths in which other functionally relevant molecules are also involved (evidenced by white symbols). Moreover, the six HUB nodes are evidenced by cyan symbols. On the left side the cellular localization of the molecules in the graph is shown.
To experimentally verify the putative interactions found between our significant cytokines and the HUB nodes by means of the network analysis, we have determined the serum concentrations of the TP53 protein in the patients with HCC and T2D-HCC. As shown in S1 Table, 19 patients with HCC and 6 with T2D-HCC resulted negative to TP53 antibody while 15 patients with HCC and 4 with T2D-HCC were positive. In addition, we have also correlated the concentrations of TP53 with those of CXCL1, CXCL12, IL-2RA, PECAM-1, and Prolactin because they have been found associated with TP53 from the network analysis. TP53 showed no correlation with CXCL1, IL-2RA, PECAM-1, and Prolactin whereas a significant correlation (with p-values <0.05) has been found with CXCL12 in HCC as well as in T2D-HCC patients.
Bio-Plex Assay on validation set
To validate all the results we have evaluated the serum levels of cytokines, growth factors, chemokines, as well as of other cancer and diabetes biomarkers in a validation set, including 20 patients with T2D, 20 patients with CHC, 20 with HCC, 10 with T2D-HCC, and 20 healthy control subjects (Fig 3 and S2 Table). Then, we compared the obtained serum levels between the patients and healthy controls by the Mann Whitney U-test, the Unparied t test and F test and obtained that the same proteins, already resulted in the discovery set, were significant also in the patients groups belonging to the validation set (S3 Table).
Fig 3. Significant cytokines in some patient groups belonging to validation set.

We report the significant molecule levels from controls, patients with type 2 diabetes (T2D), chronic hepatitis C (CHC), hepatocellular carcinoma (HCC) and hepatocellular carcinoma and type 2 diabetes (T2D-HCC) shown by means of box-and-whisker graphs. The boxes extend from the 25th to the 75th percentile, and the line in the middle is the median. The error bars extend down to the lowest value and up to the highest.
Furthermore, we compared also the serum levels for the significant proteins obtained for this validation cohort with those evaluated in the discovery set. As shown in Table 5, we have verified that all the P values were higher than 0.05 and, hence, no statistically significant difference exists between the two subject groups (“discovery set” and “validation set”). This evidences the reliability of our results, and the possibility to use them for discriminating the different patient groups.
Table 5. Comparison of cytokine serum levels between discovery and validation sets in patients and healthy controls.
We report the results of all the performed statistical analysis obtained by the nonparametric Mann-Whitney U test in terms of U test and P values, by the Unparied t test in terms of P value, t, the number of degrees of freedom (df), the difference between the means, 95% confidence interval, and R squared, and by F test in terms of F, degrees of freedom for the numerator (DFn) and for the denominator (Dfd) and P value.
| CTR vs CTRV | T2D vs T2DV | CHC vs CHCV | HCC vs HCCV | T2D-HCC vs T2D-HCCV | |
|---|---|---|---|---|---|
| ADIPOQ | |||||
| Mann-Whitney Utest | |||||
| U-test | 12.5 | 11 | 8.5 | 23.5 | 6.5 |
| P-value | 0.9166 | 0.8413 | 0.8057 | 0.949 | 0.7715 |
| Unpaired t test | |||||
| P value | 0.7829 | 0.7086 | 0.8682 | 1.0000 | 0.5762 |
| t, df | t = 0.2850 df = 8 | t = 0.3874 df = 8 | t = 0.1722 df = 7 | t = 0.0000 df = 12 | t = 0.5909 df = 6 |
| Difference between means | -1888000 ± 6623000 | -2480000 ± 6402000 | 933200 ± 5420000 | 0.0000 ± 4178000 | -4861000 ± 8227000 |
| 95% confidence interval | -17160000 to 13390000 | -17240000 to 12280000 | -11880000 to 13750000 | -9103000 to 9103000 | -24990000 to 15270000 |
| R squared | 0.01005 | 0.01841 | 0.004218 | 0.0000 | 0.05499 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.386, 4, 4 | 1.975, 4, 4 | 2.814, 4, 3 | 2.043, 6, 6 | 1.101, 3, 3 |
| P value | 0.7592 | 0.5260 | 0.4217 | 0.4059 | 0.9386 |
| GLUCAGON | |||||
| Mann-Whitney Utest | |||||
| U-test | 5.5 | 10 | 11 | 22.5 | 5.5 |
| P-value | 0.5614 | 0.6905 | 0.8335 | 0.8478 | 0.5614 |
| Unpaired t test | |||||
| P value | 0.4629 | 0.8907 | 0.7513 | 0.6242 | 0.6009 |
| t, df | t = 0.7840 df = 6 | t = 0.1419 df = 8 | t = 0.3280 df = 8 | t = 0.5027 df = 12 | t = 0.5519 df = 6 |
| Difference between means | -41.25 ± 52.62 | -24.52 ± 172.8 | -25.40 ± 77.44 | 376.4 ± 748.8 | -199.8 ± 361.9 |
| 95% confidence interval | -170.0 to 87.50 | -423.0 to 373.9 | -204.0 to 153.2 | -1255 to 2008 | -1085 to 685.8 |
| R squared | 0.09292 | 0.002511 | 0.01327 | 0.02063 | 0.04832 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.545, 3, 3 | 1.147, 4, 4 | 1.168, 4, 4 | 6.139, 6, 6 | 2.585, 3, 3 |
| P value | 0.7294 | 0.8974 | 0.8839 | 0.0441 | 0.4560 |
| β-NGF | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 7.5 | 6.5 | 10.5 | 10 |
| P-value | 0.8857 | 1 | 0.7715 | 0.753 | 0.9009 |
| Unpaired t test | |||||
| P value | 0.8544 | 0.8710 | 0.4273 | 0.8926 | 0.8335 |
| t, df | t = 0.1915 df = 6 | t = 0.1695 df = 6 | t = 0.8513 df = 6 | t = 0.1394 df = 8 | t = 0.2182 df = 7 |
| Difference between means | -0.02500 ± 0.1305 | -0.02250 ± 0.1328 | -0.2375 ± 0.2790 | -0.08800 ± 0.6312 | 0.3105 ± 1.423 |
| 95% confidence interval | -0.3444 to 0.2944 | -0.3473 to 0.3023 | -0.9202 to 0.4452 | -1.544 to 1.368 | -3.054 to 3.675 |
| R squared | 0.006075 | 0.004765 | 0.1078 | 0.002424 | 0.006758 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.053, 3, 3 | 1.176, 3, 3 | 3.501, 3, 3 | 1.403, 4, 4 | 1.094, 4, 3 |
| P value | 0.9670 | 0.8973 | 0.3308 | 0.7508 | 0.9804 |
| CXCL1 | |||||
| Mann-Whitney Utest | |||||
| U-test | 11 | 10.5 | 17 | 15 | 10 |
| P-value | 0.8335 | 0.7533 | 0.6161 | 0.7436 | 0.9017 |
| Unpaired t test | |||||
| P value | 0.5960 | 0.7636 | 0.8059 | 0.7940 | 0.9462 |
| t, df | t = 0.5520 df = 8 | t = 0.3112 df = 8 | t = 0.2518 df = 11 | t = 0.2683 df = 10 | t = 0.06993 df = 7 |
| Difference between means | -5.574 ± 10.10 | -3.814 ± 12.26 | -2.150 ± 8.538 | 3.846 ± 14.34 | -6.450 ± 92.23 |
| 95% confidence interval | -28.86 to 17.71 | -32.08 to 24.45 | -20.94 to 16.64 | -28.10 to 35.79 | -224.6 to 211.7 |
| R squared | 0.03669 | 0.01196 | 0.005729 | 0.007145 | 0.0006981 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.663, 4, 4 | 1.146, 4, 4 | 1.323, 5, 6 | 1.134, 4, 6 | 1.435, 3, 4 |
| P value | 0.6342 | 0.8983 | 0.7344 | 0.8465 | 0.7142 |
| CXCL12 | |||||
| Mann-Whitney Utest | |||||
| U-test | 12.5 | 11.5 | 7 | 11 | 7 |
| P-value | 0.9161 | 0.916 | 0.883 | 0.8335 | 0.8839 |
| Unpaired t test | |||||
| P value | 0.9604 | 0.8203 | 0.7749 | 0.8126 | 0.8651 |
| t, df | t = 0.05122 df = 8 | t = 0.2348 df = 8 | t = 0.2991 df = 6 | t = 0.2450 df = 8 | t = 0.1773 df = 6 |
| Difference between means | -0.3040 ± 5.936 | 1.374 ± 5.852 | -2.738 ± 9.151 | -3.382 ± 13.80 | 6.730 ± 37.96 |
| 95% confidence interval | -13.99 to 13.38 | -12.12 to 14.87 | -25.13 to 19.66 | -35.21 to 28.44 | -86.16 to 99.62 |
| R squared | 0.0003278 | 0.006844 | 0.01469 | 0.007450 | 0.005211 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.367, 4, 4 | 1.130, 4, 4 | 1.399, 3, 3 | 1.347, 4, 4 | 1.077, 3, 3 |
| P value | 0.7690 | 0.9088 | 0.7894 | 0.7801 | 0.9529 |
| CXCL9 | |||||
| Mann-Whitney Utest | |||||
| U-test | 11.5 | 10 | 11 | 30 | 7 |
| P-value | 0.9166 | 0.674 | 0.834 | 0.8745 | 0.8857 |
| Unpaired t test | |||||
| P value | 0.9226 | 0.8130 | 0.9399 | 0.8860 | 0.8300 |
| t, df | t = 0.1003 df = 8 | t = 0.2446 df = 8 | t = 0.07785 df = 8 | t = 0.1459 df = 14 | t = 0.2242 df = 6 |
| Difference between means | -34.34 ± 342.5 | -85.41 ± 349.2 | 14.07 ± 180.8 | 149.2 ± 1022 | -302.3 ± 1348 |
| 95% confidence interval | -824.2 to 755.5 | -890.7 to 719.9 | -402.8 to 431.0 | -2044 to 2342 | -3601 to 2997 |
| R squared | 0.001255 | 0.007420 | 0.0007570 | 0.001519 | 0.008309 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.101, 4, 4 | 1.121, 4, 4 | 1.403, 4, 4 | 1.829, 7, 7 | 1.036, 3, 3 |
| P value | 0.9281 | 0.9144 | 0.7508 | 0.4440 | 0.9776 |
| HGF | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 11 | 13.5 | 49 | 9.5 |
| P-value | 0.8857 | 0.8335 | 0.569 | 0.9698 | 1 |
| Unpaired t test | |||||
| P value | 0.9206 | 0.9427 | 0.5776 | 0.8967 | 0.8874 |
| t, df | t = 0.1039 df = 6 | t = 0.07416 df = 8 | t = 0.5756 df = 10 | t = 0.1317 df = 18 | t = 0.1469 df = 7 |
| Difference between means | 7.380 ± 71.01 | -13.33 ± 179.7 | 44.51 ± 77.33 | 33.27 ± 252.6 | 113.0 ± 769.1 |
| 95% confidence interval | -166.4 to 181.1 | -427.8 to 401.2 | -127.8 to 216.8 | -497.4 to 564.0 | -1706 to 1932 |
| R squared | 0.001797 | 0.0006870 | 0.03207 | 0.0009631 | 0.003073 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.281, 3, 3 | 1.006, 4, 4 | 1.113, 6, 4 | 1.211, 9, 9 | 1.655, 3, 4 |
| P value | 0.8435 | 0.9954 | 0.9608 | 0.7797 | 0.6239 |
| IFN-α | |||||
| Mann-Whitney Utest | |||||
| U-test | 8 | 7 | 11.5 | 40 | 7.5 |
| P value | 1 | 0.8846 | 0.9166 | 0.708 | 1 |
| Unpaired t test | |||||
| P value | 0.7778 | 0.7616 | 0.8668 | 0.7240 | 0.6738 |
| t, df | t = 0.2952 df = 6 | t = 0.3175 df = 6 | t = 0.1732 df = 8 | t = 0.3590 df = 17 | t = 0.4423 df = 6 |
| Difference between means | -0.2625 ± 0.8892 | -0.2225 ± 0.7007 | 0.2240 ± 1.294 | 3.109 ± 8.660 | -18.77 ± 42.42 |
| 95% confidence interval | -2.438 to 1.913 | -1.937 to 1.492 | -2.759 to 3.207 | -15.16 to 21.38 | -122.6 to 85.05 |
| R squared | 0.01432 | 0.01653 | 0.003734 | 0.007524 | 0.03158 |
| F test to compare variances | |||||
| F,DFn, Dfd | 2.297, 3, 3 | 1.659, 3, 3 | 1.485, 4, 4 | 2.473, 8, 9 | 2.007, 3, 3 |
| P value | 0.5125 | 0.6878 | 0.7111 | 0.1992 | 0.5817 |
| IL-16 | |||||
| Mann-Whitney Utest | |||||
| U-test | 10.5 | 12.5 | 11.5 | 7 | 10.5 |
| P-value | 0.753 | 0.916 | 0.916 | 0.9095 | 0.8307 |
| Unpaired t test | |||||
| P value | 0.7320 | 0.9652 | 0.9012 | 0.3622 | 0.6969 |
| t, df | t = 0.3547 df = 8 | t = 0.04500 df = 8 | t = 0.1281 df = 8 | t = 0.9662 df = 8 | t = 0.4038 df = 8 |
| Difference between means | -10.68 ± 30.12 | 1.378 ± 30.63 | 5.136 ± 40.08 | -53.80 ± 55.68 | 142.5 ± 352.9 |
| 95% confidence interval | -80.15 to 58.78 | -69.24 to 72.00 | -87.30 to 97.57 | -182.2 to 74.60 | -671.3 to 956.4 |
| R squared | 0.01548 | 0.0002530 | 0.002048 | 0.1045 | 0.01998 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.175, 4, 4 | 1.007, 4, 4 | 1.242, 4, 4 | 1.126, 4, 4 | 3.626, 3, 5 |
| P value | 0.8797 | 0.9944 | 0.8388 | 0.9113 | 0.1994 |
| IL-18 | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 12 | 9 | 17.5 | 9 |
| P-value | 0.8857 | 1 | 0.576 | 0.6678 | 0.6095 |
| Unpaired t test | |||||
| P value | 0.8273 | 0.7391 | 0.4978 | 0.6730 | 0.7028 |
| t, df | t = 0.2279 df = 6 | t = 0.3448 df = 8 | t = 0.7101 df = 8 | t = 0.4335 df = 11 | t = 0.3955 df = 8 |
| Difference between means | 2.123 ± 9.312 | -7.288 ± 21.14 | 12.01 ± 16.91 | 19.34 ± 44.62 | 40.02 ± 101.2 |
| 95% confidence interval | -20.66 to 24.91 | -56.03 to 41.45 | -26.99 to 51.00 | -78.86 to 117.5 | -193.3 to 273.4 |
| R squared | 0.008585 | 0.01464 | 0.05930 | 0.01680 | 0.01918 |
| F test to compare variances | |||||
| F,DFn, Dfd | 2.733, 3, 3 | 2.036, 4, 4 | 1.766, 4, 4 | 1.604, 6, 5 | 2.216, 3, 5 |
| P value | 0.4309 | 0.5081 | 0.5951 | 0.6207 | 0.4087 |
| IL-2R | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 9 | 9 | 25.5 | 11.5 |
| P-value | 0.3095 | 0.5476 | 0.5476 | 0.5283 | 1 |
| Unpaired t test | |||||
| P value | 0.1398 | 0.6507 | 0.4295 | 0.7792 | 0.9263 |
| t, df | t = 1.639 df = 8 | t = 0.4703 df = 8 | t = 0.8321 df = 8 | t = 0.2858 df = 14 | t = 0.09544 df = 8 |
| Difference between means | -16.95 ± 10.34 | -17.21 ± 36.60 | -29.05 ± 34.91 | -23.12 ± 80.90 | 55.71 ± 583.7 |
| 95% confidence interval | -40.79 to 6.893 | -101.6 to 67.20 | -109.5 to 51.45 | -196.7 to 150.4 | -1290 to 1402 |
| R squared | 0.2515 | 0.02690 | 0.07965 | 0.005800 | 0.001137 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.023, 4, 4 | 1.588, 4, 4 | 1.957, 4, 4 | 2.709, 7, 7 | 2.987, 3, 5 |
| P value | 0.9828 | 0.6650 | 0.5314 | 0.2119 | 0.2695 |
| Leptin | |||||
| Mann-Whitney Utest | |||||
| U-test | 12 | 9 | 14 | 9 | 5 |
| P-value | 1 | 0.5476 | 0.9307 | 0.5476 | 0.8571 |
| Unpaired t test | |||||
| P value | 0.9622 | 0.3256 | 0.6686 | 0.7404 | 0.5868 |
| t, df | t = 0.04894 df = 8 | t = 1.047 df = 8 | t = 0.4424 df = 9 | t = 0.3431 df = 8 | t = 0.5804 df = 5 |
| Difference between means | 80.00 ± 1635 | -10500 ± 10020 | -3057 ± 6910 | -1818 ± 5298 | -831.8 ± 1433 |
| 95% confidence interval | -3690 to 3850 | -33610 to 12620 | -18690 to 12570 | -14040 to 10400 | -4516 to 2853 |
| R squared | 0.0002993 | 0.1206 | 0.02128 | 0.01450 | 0.06312 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.908, 3, 5 | 5.182, 4, 4 | 3.425, 5, 4 | 3.146, 4, 4 | 1.403, 3, 2 |
| P value | 0.4927 | 0.1401 | 0.2565 | 0.2929 | 0.8836 |
| PECAM-1 | |||||
| Mann-Whitney Utest | |||||
| U-test | 9 | 10 | 8 | 10 | 7 |
| P-value | 0.9048 | 1 | 0.4762 | 0.2601 | 0.8857 |
| Unpaired t test | |||||
| P value | 0.8756 | 0.9450 | 0.4509 | 0.3406 | 0.6969 |
| t, df | t = 0.1624 df = 7 | t = 0.07152 df = 7 | t = 0.7926 df = 8 | t = 0.9962 df = 11 | t = 0.4088 df = 6 |
| Difference between means | 823.6 ± 5072 | -391.1 ± 5468 | -3212 ± 4053 | -5560 ± 5581 | -2317 ± 5669 |
| 95% confidence interval | -11170 to 12820 | -13320 to 12540 | -12560 to 6134 | -17850 to 6725 | -16190 to 11550 |
| R squared | 0.003752 | 0.0007301 | 0.07280 | 0.08276 | 0.02709 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.184, 3, 4 | 1.759, 3, 4 | 1.593, 3, 5 | 1.931, 8, 3 | 1.175, 3, 3 |
| P value | 0.8424 | 0.5871 | 0.6048 | 0.6369 | 0.8975 |
| Prolactin | |||||
| Mann-Whitney Utest | |||||
| U-test | 10 | 8 | 6 | 15 | 9 |
| P-value | 1 | 0.7302 | 0.6857 | 0.7105 | 0.9048 |
| Unpaired t test | |||||
| P value | 0.9318 | 0.7506 | 0.7559 | 0.7391 | 0.6458 |
| t, df | t = 0.08866 df = 7 | t = 0.3307 df = 7 | t = 0.3255 df = 6 | t = 0.3416 df = 11 | t = 0.4801 df = 7 |
| Difference between means | 552.7 ± 6235 | 2335 ± 7063 | -1538 ± 4724 | 1524 ± 4461 | -2299 ± 4789 |
| 95% confidence interval | -14190 to 15300 | -14370 to 19040 | -13100 to 10020 | -8294 to 11340 | -13630 to 9028 |
| R squared | 0.001122 | 0.01538 | 0.01735 | 0.01050 | 0.03187 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.237, 4, 3 | 2.574, 4, 3 | 1.247, 3, 3 | 3.565, 8, 3 | 2.127, 3, 4 |
| P value | 0.8970 | 0.4633 | 0.8603 | 0.3237 | 0.4790 |
| sIL-6Ra | |||||
| Mann-Whitney Utest | |||||
| U-test | 6 | 7 | 6 | 12 | 5 |
| P-value | 0.648 | 0.556 | 0.6857 | 1 | 0.1091 |
| Unpaired t test | |||||
| P value | 0.6318 | 0.5560 | 0.7160 | 0.8730 | 0.1084 |
| t, df | t = 0.5046 df = 6 | t = 0.6181 df = 7 | t = 0.3815 df = 6 | t = 0.1651 df = 8 | t = 1.782 df = 9 |
| Difference between means | 2888 ± 5723 | 4804 ± 7772 | -2557 ± 6704 | 2837 ± 17180 | 39530 ± 22180 |
| 95% confidence interval | -11120 to 16890 | -13580 to 23190 | -18960 to 13850 | -36790 to 42460 | -10640 to 89690 |
| R squared | 0.04071 | 0.05176 | 0.02368 | 0.003395 | 0.2609 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.373, 3, 3 | 1.865, 3, 4 | 1.379, 3, 3 | 2.138, 4, 4 | 10.59, 6, 3 |
| P value | 0.8005 | 0.5525 | 0.7979 | 0.4798 | 0.0793 |
| VEGFR-1 | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 12 | 7 | 15 | 7 |
| P-value | 0.8857 | 1 | 0.2303 | 0.5395 | 0.8857 |
| Unpaired t test | |||||
| P value | 0.6152 | 0.9469 | 0.1429 | 0.4231 | 0.6452 |
| t, df | t = 0.5299 df = 6 | t = 0.06876 df = 8 | t = 1.605 df = 9 | t = 0.8293 df = 12 | t = 0.4845 df = 6 |
| Difference between means | -50.67 ± 95.63 | 11.83 ± 172.1 | 143.8 ± 89.57 | 110.3 ± 133.0 | -74.32 ± 153.4 |
| 95% confidence interval | -284.7 to 183.3 | -384.9 to 408.6 | -58.83 to 346.4 | -179.5 to 400.1 | -449.7 to 301.1 |
| R squared | 0.04470 | 0.0005907 | 0.2226 | 0.05421 | 0.03765 |
| F test to compare variances | |||||
| F,DFn, Dfd | 1.618, 3, 3 | 2.017, 4, 4 | 1.180, 3, 6 | 1.405, 9, 3 | 1.531, 3, 3 |
| P value | 0.7023 | 0.5135 | 0.7863 | 0.8620 | 0.7348 |
| VEGFR-2 | |||||
| Mann-Whitney Utest | |||||
| U-test | 7 | 6 | 9 | 25 | 6 |
| P-value | 0.8857 | 0.4127 | 0.4121 | 1 | 0.6857 |
| Unpaired t test | |||||
| P value | 0.6770 | 0.5581 | 0.4362 | 0.9312 | 0.5172 |
| t, df | t = 0.4376 df = 6 | t = 0.6148 df = 7 | t = 0.8148 df = 9 | t = 0.08796 df = 13 | t = 0.6880 df = 6 |
| Difference between means | 483.4 ± 1105 | -1604 ± 2608 | -860.8 ± 1056 | 71.61 ± 814.1 | 1672 ± 2430 |
| 95% confidence interval | -2220 to 3187 | -7772 to 4565 | -3251 to 1529 | -1687 to 1830 | -4274 to 7618 |
| R squared | 0.03093 | 0.05124 | 0.06870 | 0.0005948 | 0.07312 |
| F test to compare variances | |||||
| F,DFn, Dfd | 3.435, 3, 3 | 1.251, 3, 4 | 1.431, 3, 6 | 1.348, 4, 9 | 6.965, 3, 3 |
| P value | 0.3379 | 0.8053 | 0.6472 | 0.6495 | 0.1452 |
Discussion
In this paper we report a simultaneous and comparative analysis of the serum levels of a large panel of cytokines, growth factors, chemokines, as well as of other cancer and diabetes biomarkers in patients with T2D, CHC, HCC and T2D-HCC by means of BioPlex assays. Our interest for these diseases depends from the fact that Southern Italy shows a high mortality trend for liver cancer in CHC patients [16] concomitantly with very high rates of T2D [17]. Recently we evaluated the cytokinome profile in patients with T2D and/or CHC infection or with CHC-related HCC suggesting some specific markers for the different stages of the diseases [8–9, 15, 18]. Since both T2D and CHC have been identified as contributory causes of HCC [2], our aim is to identify new possible diagnostic/prognostic markers useful for recognizing the features of the association between T2D and HCC.
A general view of the results in Table 3 shows that IL-2R, IL-18, Leptin, sIL-6Ra, sVEGFR-1 and sVEGFR-2 are up-expressed in all the patient groups, suggesting that these proteins are involved in those chronic inflammation processes leading to T2D through the metabolic syndrome and, often concomitantly, to cancer, and in particular, IL-18 has shown a significant correlation with AFP and glycemic levels. On the other hand, we have to underline that the presence of some of these proteins is also due to the necro-inflammatory activity of the liver. Indeed, IL-2R and sIL-6Ra show a significant correlation with the fibrotic stage of our CHC patients, as well the elevated levels of leptin indicate that immune response and host defense, active during infection and inflammation, are acting as paracrine modulator of the hepatic fibrogenesis [19].
It is also interesting to note that ADIPOQ, β-NGF, CXCL1, CXCL9, CXCL12, IL-16, and PECAM-1 are up-expressed only in those CHC and HCC patients who present a liver failure (Table 3), and, hence, linkable to the necro-inflammatory activity of the liver. In fact, it is known that in CHC patients ADIPOQ was found related to the severity of the fibrosis and suggested as HCC marker when the carcinogenesis is concomitantly supported by CHC infection [20] while β-NGF and IL-16 are involved in cancer growth and metastasis and also detected in diseased liver tissues [9, 21]. Similarly, CXCL1 and CXCL9 have chemotactic activities and roles in angiogenesis, inflammation and tumor genesis [9], CXCL12 is related to the HCC metastatic network by recruiting endothelial cell tumor progenitors [22], and PECAM-1 reflects the liver disease progression [23]. However, we have also found that CXCL9 and CXCL12 resulted statistically higher in CHC patients with F4 grade in respect to those with F2 grade, thus confirming their important role in the liver necro-inflammation.
In Table 3 we also show that the levels of HGF and glucagon were higher in T2D and HCC patients but not in those with CHC, suggesting that these two proteins can be related to the pro-inflammatory condition. In details, elevated HGF levels suggest atherosclerotic complications in T2D patients [9] whereas those of glucagon confirm its role in the dysregulated hepatic glucose production, which is characteristic of the abnormal glucose homeostasis of these patients [24]. However, also in our previous studies [8, 15, 18], HGF was found significantly up-regulated in HCC patients but not in patients with CHC and always correlated with AFP, thus supporting our proposal that this growth factor could be used as an index of cellular growth and of HCC development in patients with chronic inflammation.
Moreover, since IFN-α is a lymphokine with a wide range of biological effects and found up-expressed in pre-operative samples of HCC patients [25] while Prolactin is commonly attributed to an impaired hepatic metabolism of estrogens and associated to liver cirrhosis [8], the fact that we have found both up-expressed only in patients with HCC and that Prolactin results to correlate with the transaminase levels, leads us to think that Prolactin might be used as a severity index of liver disease.
Other points to discuss are the serum levels of β-NGF, CXCL1, CXCL12, HGF, IFN-α, IL-16, IL-18, IL-2R, Leptin and sIL-6Ra found for the T2D-HCC patients. These levels were found to be higher than those of patients with only T2D or HCC suggesting that these proteins are concomitantly involved in both diseases. On the other hand the serum levels of ADIPOQ, CXCL9, PECAM-1, Prolactin, sVEGFR-1 and sVEGFR-2 in the T2D-HCC patients were higher than those of patients with only T2D while they were similar to those of HCC patients, confirming that these proteins are specific for the cancer presence.
We have also attempted to understand how these proteins could be correlated between them on the basis of their known metabolic functions and of all the experimental data reported in the literature.
To this end, we have performed an interactomic analysis which calculated how these proteins are significantly connected in a common metabolic network where they are modulated through six HUB nodes, such as EP300, NR4A1, NR2F1, RELA, STAT3 and TP53. In detail, the transcriptional cofactor, EP300, is involved in several biological phenomena, such as cell proliferation, differentiation and apoptosis; it functions as a pleiotropic coactivator and regulates p53-dependent transcription [26]. It was demonstrated that the levels of EP300 protein expression in HCCs were strongly associated with vascular invasion, intrahepatic metastasis and poor prognosis of HCC patients [26]. In fact, the evaluation of EP300 expression was proposed in a new prognostic model based on high EP300 expression, AFP levels and vascular invasion [27]. Moreover, recently, some authors showed that high glucose levels increased the activity of the transcriptional EP300, which increases TGF-β activity via Smad2 acetylation. However its activation increases both the transactivation of glucagon gene by PAX2A protein [28] and the transcription of leptin gene by p42 C/EBP alpha protein [29]. Moreover, EP300 binds the promoter fragment containing a E2F binding site from human VEGFR-2 gene [30] and the DNA endogenous promoter from the human prolactin gene [31]. These data highlighted the importance of the role played by EP300 in both T2D and HCC and its correlation with ADIPOQ, glucagon, sVEGFR2, Leptin and Prolactin.
Moreover, NR4A1 and NR2F1 are soluble nuclear hormone receptors that regulate liver development, differentiation and function, and are implicated in the modulation of the hepatocyte priming and proliferation in regenerating liver, chronic hepatitis and HCC development. All the early changes essential for the liver regeneration, such as the activation of transcription factors (NF-kB and STAT3), as well as the increased levels of cytokines and growth factors (HGF), can be modulated by members of the NRs superfamily [32]. However, an ever-growing body of evidence suggests that members of this family of nuclear receptors (NRs) could play a pivotal role in glucose homeostasis and the development of T2D [33]. In fact, in fasting mouse, a mutant mouse Nur77 [NR4A1] gene resulted to modulate the expression of ADIPOQ, Leptin, IL-16, Prolactin and CXCL12.
RELA is the major component of NF- κB is activated constitutively in human HCC, and plays a key role in controlling apoptosis in HCC cells, suggesting that the RELA may be an important targets for novel therapeutic approaches in the treatment of the human HCC [34–35]. Recent studies have highlighted the role of NF-kB in the pathogenesis of the insulin resistance and T2D as an independent risk factor for the development of the HCC. In particular, the malignant transformation of the hepatocytes may occur through a pathway of an increased liver cell turnover induced by the chronic liver injury and regeneration in a context of inflammation, immune response and oxidative DNA damage. In fact, RELA is involved in the expression of CXCL1, CXCL9, and IL-2RA [36–37], as well increases the transactivation of a DNA endogenous promoter through the PECAM-1 gene [38]. STAT3 is implicated in the signal transduction by different cytokines, growth factors and oncogenes, and plays an important role in tumorigenesis and, in particular, in HCC through the up-regulation of genes involved in anti-apoptosis, proliferation and angiogenesis [39]. Moreover, its signal-pathway was suggested as a therapeutic target for the T2D and drug discovery [40]. In general, STAT3 decreases the expression of CXCL9 [41] and PECAM-1 [42] and is activated by HGF [43], sIL-6R [44] and Leptin [45] whereas ADIPOQ induces its decreasing [46]. Finally, TP53 is a tumor suppressor that initiates cell-cycle arrest, apoptosis, and senescence in response to the cellular stress to maintain the integrity of the genome. Single base substitutions in TP53 occur in approximately 25% of HCC, suggesting a relevant role for TP53 in this cancer [47]. Moreover, it has been found that an excessive calorie intake led to the accumulation of oxidative stress in the adipose tissue of mice with T2D–like disease and promoted the increased expression of TP53 with an associated increased production of pro-inflammatory cytokines [48]. In particular, it is reported that the mutant TP53 increases the expression of CXCL1 [49] and decreases that of PECAM-1 [50], PRL [51] and IL-2RA [52], and its inactivation decreases expression of CXCL12 that involves oncogenic mutant HRAS protein [53]. To support the conclusion of the metabolic network analysis, we have experimentally evaluated the serum levels of TP53 in HCC and T2D patients, showing how they are correlated with the CXCL12 levels and thus confirming that TP53 can suppress the production of this chemokine as already reported in cultured fibroblasts of both human and mouse origin [54]. However, this result confirm the predictive validity of the interactomic analysis because the data that are used for the calculation of the metabolic networks are based exclusively on experimental results from which we can extract complex relationships, not easily detectable but made perceptible through the representation of a mathematical graph that visually illustrates the metabolic net.
Conclusions
What appears evident from the analysis of our data is that the HCC itself is a disease with a very complex and multifactorial etiology. When associated with other syndromes, the difficulty of being able to follow the onset of the cancer and its progression in time, becomes much more difficult and often elusive. In fact, it is well known that the onset of this cancer is often clinically silent, therefore, when the cancer is discovered it is already too late, and this often determines a poor prognosis. For this reason many laboratories seek direct or indirect biomarkers that can be validly correlated with HCC progression and prognosis. What is clear from our results, is the lack of individual markers, suitable to follow validly this cancer because of the clear and extensive metabolic correlations existing between the molecular species, which generate and control it. We think that only through a large, simple and reliable omics approach, such as the cytokinome analysis, supplemented by the common biochemical/clinical data, we can take into account the correlations between the different molecular partners generating a framework that allows us to make a metabolic acceptable prognosis of the various stages of the disease progression. This is particularly important when we have the superposition of chronic liver disease and T2D. Our approach has also been validated by the interactomic analysis that has clearly shown how our results correlate well with the overall picture so far experimentally known for metabolic associations that exist for a complex and multifactorial disease as HCC. In addition, our data also suggest that the best strategy to design new drugs against HCC must have as a specific target the HUB molecules, that is, those metabolic nodes that coordinate and control the functions of many other metabolically relevant molecules.
Supporting Information
In details, we report the number of patients resulted negative or positive to TP53 antibody and the p-values determined between TP53 levels and those of CXCL1, CXCL12, IL-2RA, PECAM1, and PRL using the Pearson correlation coefficient. The statistically significant p-values are reported in bold and underlined.
(DOC)
We report for each cytokine the minimum and maximum values, the 25% and 75% Percentiles, the median, the mean, standard deviation, standard error, and the lower and upper 95% confidence intervals (CI).
(DOC)
We report the results of all the performed statistical analysis obtained by the nonparametric Mann-Whitney U test in terms of U test and P values, by the Unparied t test in terms of P value, t, the number of degrees of freedom (df), the difference between the means, 95% confidence interval, and R squared, and by F test in terms of F, degrees of freedom for the numerator (DFn) and for the denominator (Dfd) and P value. In particular, we reported in bold the values of p<0.05 indicated with *, of p<0.01 with **, and of p<0.0001 with ***.
(DOC)
Data Availability
Data are available from FigShare: http://dx.doi.org/10.6084/m9.figshare.1486373.
Funding Statement
The authors have no support or funding to report.
References
- 1. Sherman M. Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis. 2010;30: 3–16 10.1055/s-0030-1247128 [DOI] [PubMed] [Google Scholar]
- 2. Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42: S206–S214. 10.1016/S1590-8658(10)60507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herman WH, Zimmet P. Type 2 diabetes: an epidemic requiring global attention and urgent action. Diabetes Care. 2012;35: 943–944. 10.2337/dc12-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379: 1245–55 10.1016/S0140-6736(11)61347-0 [DOI] [PubMed] [Google Scholar]
- 5. Baffy G. Editorial: hepatocellular carcinoma in type 2 diabetes: more than meets the eye. Am J Gastroenterol. 2012;107: 53–55. 10.1038/ajg.2011.390 [DOI] [PubMed] [Google Scholar]
- 6. El-Serag H, Rudolph K. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132: 2557–2576 [DOI] [PubMed] [Google Scholar]
- 7. Guerriero E, Capone F, Rusolo F, Colonna G, Castello G, Costantini S. Dissimilar cytokine patterns in different human liver and colon cancer cell lines. Cytokine. 2013;64: 584–9. 10.1016/j.cyto.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 8. Capone F, Guerriero E, Colonna G, Maio P, Mangia A, Castello G, et al. Cytokinome profile evaluation in patients with hepatitis C virus infection. World J Gastroenterol. 2014;20: 9261–9 10.3748/wjg.v20.i28.9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costantini S, Capone F, Guerriero E, Marfella R, Sorice A, Maio P, et al. Cytokinome profile of patients with type 2 diabetes and/or chronic hepatitis C infection. PLoS One. 2012;7: e39486 10.1371/journal.pone.0039486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Costantini S, Castello G, Colonna G. Human Cytokinome: a new challenge for systems biology. Bioinformation. 2010;5: 166–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Standard of Medical Care in Diabetes. Diabetes Care. 2009;32: S62–S67.19118289 [Google Scholar]
- 12. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22: 696–699. [DOI] [PubMed] [Google Scholar]
- 13. Huang DW, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 14. Parasole R, Izzo F, Perrone F, Pignata S, Galati MG, Leonardi E, et al. Prognostic value of serum biological markers in patients with hepatocellular carcinoma. Clin Cancer Res. 2001;7: 3504–9. [PubMed] [Google Scholar]
- 15. Costantini S, Capone F, Guerriero E, Maio P, Colonna G, Castello G. Serum cytokine levels as putative prognostic markers in the progression of chronic HCV hepatitis to cirrhosis. Eur Cytokine Netw. 2010;21: 251–6. 10.1684/ecn.2010.0214 [DOI] [PubMed] [Google Scholar]
- 16. Montella M, Malvezzi M, Grimaldi M, Nocerino F, Frigeri F, Pinto A, et al. Mortality trend for tumor correlated immune system in hyperendemic area of HCV infection in southern Italy: joinpoint analysis. Hepat Mon. 2013;13: e12725 10.5812/hepatmon.12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Classen JB. Italian pediatric data support hypothesis that simultaneous epidemics of type 1 diabetes and type 2 diabetes/metabolic syndrome/obesity are polar opposite responses (i.e., symptoms) to a primary inflammatory condition. J Pediatr Endocrinol Metab. 2011;24: 455–6. [DOI] [PubMed] [Google Scholar]
- 18. Capone F, Costantini S, Guerriero E, Calemma R, Napolitano M, Scala S, et al. Serum cytokine levels in patients with hepatocellular carcinoma. Eur Cytokine Netw. 2010;21: 99–104. 10.1684/ecn.2010.0192 [DOI] [PubMed] [Google Scholar]
- 19. Otte C, Otte JM, Strodthoff D, Bornstein SR, Fölsch UR, Mönig H, et al. Expression of leptin and leptin receptor during the development of liver fibrosis and cirrhosis. Exp Clin Endocrinol Diabetes. 2004;112:10–17. [DOI] [PubMed] [Google Scholar]
- 20. Marra F, Bertalani C. Adipokines in liver diseases. Hepatology. 2009;50:957–969 10.1002/hep.23046 [DOI] [PubMed] [Google Scholar]
- 21. Li S, Deng Y, Chen ZP, Huang S, Liao XC, Lin LW, et al. Genetic polymorphism of interleukin-16 influences susceptibility to HBV-related hepatocellular carcinoma in a Chinese population. Infect Genet Evol. 2011;11: 2083–8. 10.1016/j.meegid.2011.09.025 [DOI] [PubMed] [Google Scholar]
- 22. Liu H, Pan Z, Li A, Fu S, Lei Y, Sun H, et al. Roles of chemokine receptor 4 (CXCR4) and chemokine ligand 12 (CXCL12) in metastasis of hepatocellular carcinoma cells. Cell Mol Immunol 2008;5: 373–8. 10.1038/cmi.2008.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katz SC, Pillarisetty VG, Bleier JI, Shah AB, DeMatteo RP. Liver sinusoidal endothelial cells are insufficient to activate T cells. J Immunol. 2004;173: 230–235. [DOI] [PubMed] [Google Scholar]
- 24. D’Alessio D. The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obes Metab. 2011;13: 126–32. 10.1111/j.1463-1326.2011.01449.x [DOI] [PubMed] [Google Scholar]
- 25. Zhang P, Liu XK, Chu Z, Ye JC, Li KL, Zhuang WL, et al. Detection of interleukin-33 in serum and carcinoma tissue from patients with hepatocellular carcinoma and its clinical implications. J Int Med Res. 2012;40: 1654–61. [DOI] [PubMed] [Google Scholar]
- 26. Yokomizo C, Yamaguchi K, Itoh Y, Nishimura T, Umemura A, Minami M, et al. High expression of p300 in HCC predicts shortened overall survival in association with enhanced epithelial mesenchymal transition of HCC cells. Cancer Lett. 2011;310: 140–7. 10.1016/j.canlet.2011.06.030 [DOI] [PubMed] [Google Scholar]
- 27. Li M, Luo RZ, Chen JW, Cao Y, Lu JB, He JH, et al. High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J Transl Med. 2011;9: 5 10.1186/1479-5876-9-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffmeister A, Ropolo A, Vasseur S, Mallo GV, Bodeker H, Ritz-Laser B, et al. The HMG-I/Y-related protein p8 binds to p300 and Pax2 trans-activation domain-interacting protein to regulate the trans-activation activity of the Pax2A and Pax2B transcription factors on the glucagon gene promoter. J Biol Chem. 2002;277: 22314–9. [DOI] [PubMed] [Google Scholar]
- 29. Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem. 2001;276: 16348–55. [DOI] [PubMed] [Google Scholar]
- 30. Pillai S, Kovacs M, Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70: 4931–40. 10.1158/0008-5472.CAN-10-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manfroid I, Martial JA, Muller M. Inhibition of protein phosphatase PP1 in GH3B6, but not in GH3 cells, activates the MEK/ERK/c-fos pathway and the human prolactin promoter, involving the coactivator CPB/p300. Mol Endocrinol. 2001;15: 625–37. [DOI] [PubMed] [Google Scholar]
- 32. Vacca M, Degirolamo C, Massafra V, Polimeno L, Mariani-Costantini R, Palasciano G, et al. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol Cell Endocrinol. 2013;368: 108–19. 10.1016/j.mce.2012.06.025 [DOI] [PubMed] [Google Scholar]
- 33. Duszka K, Bogner-Strauss JG, Hackl H, Rieder D, Neuhold C, Prokesch A, et al. Nr4a1 is required for fasting-induced down-regulation of Pparγ2 in white adipose tissue. Mol Endocrinol. 2013;27: 135–49. 10.1210/me.2012-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson SM, Mann DA. Role of nuclear factor kappaB in liver health and disease. Clin Sci (Lond). 2010;118: 691–705. [DOI] [PubMed] [Google Scholar]
- 35. Chiao PJ, Na R, Niu J, Sclabas GM, Dong Q, Curley SA. Role of Rel/NF-kappaB transcription factors in apoptosis of human hepatocellular carcinoma cells. Cancer. 2002;95: 1696–705. [DOI] [PubMed] [Google Scholar]
- 36. Hiroi M, Ohmori Y. The transcriptional coactivator CREB-binding protein cooperates with STAT1 and NF-kappa B for synergistic transcriptional activation of the CXC ligand 9/monokine induced by interferon-gamma gene. J Biol Chem. 2003;278: 651–60. [DOI] [PubMed] [Google Scholar]
- 37. Kavandi L, Collier MA, Nguyen H, Syed V. Progesterone and calcitriol attenuate inflammatory cytokines CXCL1 and CXCL2 in ovarian and endometrial cancer cells. J Cell Biochem. 2012;113: 3143–52. 10.1002/jcb.24191 [DOI] [PubMed] [Google Scholar]
- 38. Botella LM, Puig-Kröger A, Almendro N, Sánchez-Elsner T, Muñoz E, Corbí A, et al. Identification of a functional NF-kappa B site in the platelet endothelial cell adhesion molecule-1 promoter. J Immunol. 2000;164: 1372–8. [DOI] [PubMed] [Google Scholar]
- 39. Subramaniam A, Shanmugam MK, Perumal E, Li F, Nachiyappan A, Dai X, et al. Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochim Biophys Acta. 2013;1835: 46–60. 10.1016/j.bbcan.2012.10.002 [DOI] [PubMed] [Google Scholar]
- 40. Mashili F, Chibalin AV, Krook A, Zierath JR. Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes. 2013;62: 457–65. 10.2337/db12-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ho HH, Ivashkiv LB. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem. 2006;281: 14111–8. [DOI] [PubMed] [Google Scholar]
- 42. McCormick ME, Goel R, Fulton D, Oess S, Newman D, Tzima E. Platelet-endothelial cell adhesion molecule-1 regulates endothelial NO synthase activity and localization through signal transducers and activators of transcription 3-dependent NOSTRIN expression. Arterioscler Thromb Vasc Biol. 2011;31: 643–9. 10.1161/ATVBAHA.110.216200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wojcik EJ, Sharifpoor S, Miller NA, Wright TG, Watering R, Tremblay EA, et al. A novel activating function of c-Src and Stat3 on HGF transcription in mammary carcinoma cells. Oncogene. 2006;25: 2773–84. [DOI] [PubMed] [Google Scholar]
- 44. Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8: 1237–47. 10.7150/ijbs.4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Lv L, Xiao W, Gong C, Yin J, Wang D, et al. Leptin activates STAT3 and ERK1/2 pathways and induces endometrial cancer cell proliferation. J Huazhong Univ Sci Technolog Med Sci. 2011;31: 365–70. 10.1007/s11596-011-0382-7 [DOI] [PubMed] [Google Scholar]
- 46. Macis D, Guerrieri-Gonzaga A, Gandini S. Circulating adiponectin and breast cancer risk: a systematic review and meta-analysis. Int J Epidemiol. 2014;43: 1226–36. 10.1093/ije/dyu088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meng X, Franklin DA, Dong J, Zhang Y. MDM2-p53 Pathway in Hepatocellular Carcinoma. Cancer Res. 2014. December 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Minamino T, Orimo M, Shimizu I, Kunieda T, Yokoyama M, Ito T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15: 1082–1087. 10.1038/nm.2014 [DOI] [PubMed] [Google Scholar]
- 49. Yan W, Chen X. Identification of GRO1 as a critical determinant for mutant p53 gain of function. J Biol Chem. 2009;284: 12178–87. 10.1074/jbc.M900994200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. García-Tello A, Angulo JC, Andrés G, Ramón de Fata F, Sánchez-Chapado M, López JI. Impact of p53, MIB-1 and PECAM-1 expression on the prognosis of urothelial carcinoma of the renal pelvis. Actas Urol Esp. 2014;38: 506–14. 10.1016/j.acuro.2014.02.015 [DOI] [PubMed] [Google Scholar]
- 51. Tan D, Tan S, Zhang J, Tang P, Huang J, Zhou W, et al. Histone trimethylation of the p53 gene by expression of a constitutively active prolactin receptor in prostate cancer cells. Chin J Physiol. 2013;56: 282–90. 10.4077/CJP.2013.BAB139 [DOI] [PubMed] [Google Scholar]
- 52. Zhao T, Yasunaga J, Satou Y, Nakao M, Takahashi M, Fujii M, et al. Human T-cell leukemia virus type 1 bZIP factor selectively suppresses the classical pathway of NF-kappaB. Blood. 2009;113: 2755–64. 10.1182/blood-2008-06-161729 [DOI] [PubMed] [Google Scholar]
- 53. Kojima K, McQueen T, Chen Y, Jacamo R, Konopleva M, Shinojima N, et al. p53 activation of mesenchymal stromal cells partially abrogates microenvironment-mediated resistance to FLT3 inhibition in AML through HIF-1α-mediated down-regulation of CXCL12. Blood. 2011;118: 4431–9. 10.1182/blood-2011-02-334136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moskovits N, Kalinkovich A, Bar J, Lapidot T, Oren M. p53 Attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 2006;66: 10671–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In details, we report the number of patients resulted negative or positive to TP53 antibody and the p-values determined between TP53 levels and those of CXCL1, CXCL12, IL-2RA, PECAM1, and PRL using the Pearson correlation coefficient. The statistically significant p-values are reported in bold and underlined.
(DOC)
We report for each cytokine the minimum and maximum values, the 25% and 75% Percentiles, the median, the mean, standard deviation, standard error, and the lower and upper 95% confidence intervals (CI).
(DOC)
We report the results of all the performed statistical analysis obtained by the nonparametric Mann-Whitney U test in terms of U test and P values, by the Unparied t test in terms of P value, t, the number of degrees of freedom (df), the difference between the means, 95% confidence interval, and R squared, and by F test in terms of F, degrees of freedom for the numerator (DFn) and for the denominator (Dfd) and P value. In particular, we reported in bold the values of p<0.05 indicated with *, of p<0.01 with **, and of p<0.0001 with ***.
(DOC)
Data Availability Statement
Data are available from FigShare: http://dx.doi.org/10.6084/m9.figshare.1486373.



