Abstract
The Phox homology domain (PX domain) is a phosphoinositide-binding structural domain that is critical in mediating protein and cell membrane association and has been found in more than 100 eukaryotic proteins. The abundance of PX domains in nature offers an opportunity to redesign the protein using EvoDesign, a computational approach to design new sequences based on structure profiles of multiple evolutionarily related proteins. In this study, we report the X-ray crystallographic structure of a designed PX domain from the cytokine-independent survival kinase (CISK), which has been implicated as functioning in parallel with PKB/Akt in cell survival and insulin responses. Detailed data analysis of the designed CISK-PX protein demonstrates positive impacts of knowledge-based secondary structure and solvation predictions and structure-based sequence profiles on the efficiency of the evolutionary-based protein design method. The structure of the designed CISK-PX domain is close to the wild-type (1.54 Å in Cα RMSD), which was accurately predicted by I-TASSER based fragment assembly simulations (1.32 Å in Cα RMSD). This study represents the first successfully designed conditional peripheral membrane protein fold and has important implications in the examination and experimental validation of the evolution-based protein design approaches.
Keywords: Protein design, X-ray crystallography, protein structure prediction, cytokine-independent survival kinase, Phox homology, membrane
INTRODUCTION
Conditional peripheral membrane protein domains such as C1 and C2, PH, FYVE, and PX specifically recognize cell membrane components and recruit other proteins domains to the cell membrane, driving crucial biological activity (Moravcevic et al., 2012). The ability to computationally redesign these conditional peripheral membrane protein domains with altered function to control their spatial and temporal localization has fortuitous implications for research and therapeutics; this may open up unexplored venues in the drug delivery of protein-polymer conjugates (Li et al., 2013; Liechty et al., 2010). Our recent efforts in this area focused on the computationally design and preliminary biophysical characterization of a PX domain-based on the CISK-PX scaffold (Mitra et al., 2013b). In this report, we evaluate the accuracy of the computationally designed sequence at attaining a conditional peripheral membrane protein domain fold at the atomic level by X-ray crystallography. This is the first X-ray structure of a protein designed using evolutionary principles.
The PX domains are noted in > 100 eukaryotic genes (some hypothetical) and are fused to various membrane associated proteins such as sorting nexins, phospholipase, NADPH oxidase, bud emergence, t-SNARE, and CISK proteins (Ellson et al., 2002). For example, the PX domain can regulate cytokine-independent survival kinase (CISK) localization and function by binding endosomal phosphoinositides (PI), while the CISK domain mediates cell growth and survival (Xu et al., 2001), The CISK-PX fusion is the only known member of the serum and glucocorticoid-regulated kinase family that contains an intact PX domain (Liu et al., 2000). The PX domain is attractive from a computational protein design standpoint as well because of its potential use in localizing probes and therapeutics to the endosomal cell sorting compartment and its well-characterized compact globular structure (Xing et al., 2004).
However, computationally designing complex macromolecular biologics useful in understanding and combating human disease will require a deeper understanding of the chemical determinates of macromolecular functionality. Anfinsen’s thermodynamic folding principle is central to many protein design and protein structure prediction algorithms (Anfinsen, 1973); scoring methods based on this principle are complicated by the sheer number of elements involved -- to include bulk solvent. The problem can be further broken down to force field inaccuracies and limitations in conformational sampling of large atomic ensembles (Bradley et al., 2005; Floudas et al., 2006; Onuchic and Wolynes, 2004; Zhang, 2009). While many successful protein design methodologies have been developed using physics-based scoring methods (Lauck et al., 2010; Leaver-Fay et al., 2011), various sophisticated knowledge and statistical-based potentials have found their usefulness to constrain the fold and stability of designed proteins (Mitra et al., 2013b; Socolich et al., 2005).
Notably, this follows a similar trend found in protein folding, a reverse process of protein design. The knowledge-based methods, which construct structure models utilizing homologous/analogous templates from the PDB followed by structure refinement simulations, represent by far the most reliable approach in protein structure prediction. One advantage of the knowledge-based modeling approach is the efficiency in learning global folding principles and features from large-scale experimental structure data; this helps sidestep detailed atomistic interactions that are currently unknown to us (Zhang, 2008a). A variety of effective knowledge-based structure prediction programs, including I-TASSER, Modeller, Rosetta, QUARK and others (Das and Baker, 2008; Fiser and Sali, 2003; Xu and Zhang, 2012; Yang et al., 2015), have been developed. These programs incorporate information from query-template alignments, secondary structure prediction, solvent accessibility prediction, and Phi/Psi (Φ/Ψ) torsional angle prediction to guide the assembly simulations of global folds. Some of these knowledge-based template features have been successfully employed by several protein design algorithms (Bellows-Peterson et al., 2012; Bender et al., 2007; Kuhlman et al., 2003; Poole and Ranganathan, 2006). However, EvoDesign was probably the first method to systematically explore the possibility of integrating threading-based structure profiling to constrain the sequence design search (Mitra et al., 2013a; Mitra et al., 2013b).
Most recently, EvoDesign was applied to the design of a large-scale set of >300 soluble protein folds in silico; five domains with variable fold type and sequence length (including heterogeneous nuclear ribonucleoprotein K domain, thioredoxin domain, light oxygen voltage domain, translation initiation factor 1 domain, and the CISK-PX domain) were experimentally examined through circular dichroism and NMR spectroscopy (Mitra et al., 2013b). It was found that all 5 proteins were soluble and possessed secondary structure as determined by circular dichroism. But only 3 design domains had stable folds as seen by 1D NMR data. In particular, the design based on the wild-type cytokine-independent survival kinase phox homology scaffold, or WT-CISK-PX (Liu et al., 2000; Xing et al., 2004), shows the highest stability and secondary structure consistency in our design experiments through both 1D-NMR and chemical denaturation (Mitra et al., 2013b).
In this work, we will focus on the X-ray crystallography structure determination of the CISK-PX designed domain protein (DS-CISK-PX) for several reasons. First, many de novo protein design methods are based on physics-based force fields where the native recapitulation or sequence identity of the designed protein to the target is relatively low. Accordingly, it is often challenging to retain the desired target fold (or even have a stable fold) in the de novo designs (Bazzoli et al., 2011; Larson et al., 2003; Saunders and Baker, 2005). Since DS-CISK-PX represents the first conditional peripheral membrane protein domain that was designed based on an alternative evolution-based protocol, the structure determination and comparative studies with the WT target will help examine the efficiency of the new protocol on specifying the global fold of protein structures. Second, the recent large-scale design studies (Bazzoli et al., 2011; Mitra et al., 2013b) have revealed a strong correlation between the foldability of protein designs and the confidence score of protein structure prediction using I-TASSER. This strong correlation raises the possibility of using the protein folding confidence scores as a potential indicator/selection feature in distinguishing between foldable and unfoldable protein designs prior to gene synthesis. I-TASSER modeling of the DS-CISK-PX sequence yields a significant confidence score (C-score=1.31). The structure determination of this design and the modeling comparisons with the WT target will provide a useful blind test of this assumption, i.e. to exploit the state of the art structure prediction methods to assist in silico validations of protein designs. Additionally, comparative studies of the solved structure with the designed sequence will enable a systematic examination of the strength and weakness of the EvoDesign protocol and the underlying principles.
MATERIALS AND METHODS
Plasmid construction
The 116 residue designed PX gene was ligation independently cloned into a variant of the popular Midwestern Center Structural Genomics over expression plasmid. The vector contains an N-terminal 6 × His tag, a Mocr solubility domain, and a recombinant Tobacco Etch Virus (rTEV) protease site followed by the designed domain (DelProposto et al., 2009).
Expression
Rosetta 2 cells were transformed with the over-expression plasmid bearing the designed PX gene. The cells were grown in Luria-Bertani media and ampicillin (100 μg ml−1) at 310 K until a 0.7 OD600nm. The temperature was lowered to 303 K and protein over-expression was induced with 0.2 mM IPTG for 4 hours. The cells were harvested by centrifugation 6000 g × 15 min using a JLA 8.1000 Beckman rotor and frozen.
Purification
Cells were resuspended in 50 mM Tris pH 7.5, 150 mM NaCl, 5 mM Imidazole and lysed by sonication using a Fisher 705 sonicator at 50% amplitude for 5 min at 277 K. The cell lysis was clarified by centrifugation using a J25.50 Beckman rotor at 30,000 × G for 30 min and the supernatant retained. Protein was bound to Ni-NTA™ resin (Qiagen) via batch binding and then washed with 100 column volumes of lysis buffer in a gravity feed column. The sample was subsequently eluted using lysis buffer plus 200 mM imidazole. The N-terminal fusion tag was removed by rTEV digestion overnight while undergoing dialysis into 50 mM Tris pH 7.5, 150 mM NaCl, and 1mM dithiothreitol using 3000 M.W.C.O. SnakeSkin™ Dialysis Tubing. The N-terminal Mocr affinity fusion tag and rTEV were removed from the sample by subtractive NI-NTA affinity purification and cation ion exchange using a Pall AcroSep™ Q anion exchanger. The protein was concentrated to 5 mg/ml in a final buffer of 50 mM Tris pH 7.5, 100 mM NaCl prior to crystallization.
Crystallization
Over 500 different conditions were initially screened using the Qiagen JCSG suites I-IV™ plus several in-house ammonium sulfate and PEG screens by sitting drop vapor diffusion using Greiner bio-one Crystalquick™ plates in 1 μl: 1 μl protein to precipitant ratios. Initial crystals were formed in a combination of low pH, ammonium sulfate, and PEGs. The condition used to grow crystal used for diffraction was 100 mM sodium acetate pH 4.6, 100 mM ammonium sulfate, and 30% w/v PEG 2000 MME. The crystals (greater than 100 μm per edge) were harvested, cryoprotected in 20% glycerol, and flash cooled in liquid nitrogen.
Protein Structure Accession Code
X-ray crystallographic data was collected at Advance Photo Source, Chicago, Ill. The designed PX homology domain structure factors and coordinates were deposited into the PDB databank under accession code 4OXW.
Data Collection
A high-resolution data set was collected at the Argonne National Laboratory Advance Photon Source (ANL-APS), Life Science Institute Collaborative Access Team (LSI-CAT) beam line 21-ID-G. A diffraction data set was collected at 1.73 Å resolution (λ= 0.978 Å). The data set consisted of 310 frames collected with 1º oscillations. Data reduction was performed with HKL2000 (Otwinowski and Minor, 1997). Crystals of the DS-CISK-PX domain belong to space group P212121 (unit-cell parameters a=36.72, b=49.26, c=68.01 Å). A single domain in the asymmetric unit yields a calculated solvent content of 44.4% (Matthews coefficient Vm=2.23 Å3Da−1) (Matthews, 1968).
We note that only one designed sequence of the CISK-PX domain from the EvoDesign application, which has the largest size in the SPICKER-based sequence clustering (Mitra et al., 2013b), was cloned and expressed; this sequence was then submitted to the subsequent purification and crystallization experiments. A summary of the crystallographic statistics is listed in Table 1.
Table 1.
Crystallographic and structure refinement statistics for the designed PX X-ray crystal structure at 1.73 Å resolution.
| Indicator | Statisticsa |
|---|---|
| Data collection | |
| Source | APS-LS-CAT 21-ID-G |
| Wavelength (Å) | 0.978 |
| Space Group | P212121 |
| Molecules per asymmetric unit | 1 |
|
| |
| Unit Cell Parameters | |
| a (Å) | 36.72 |
| b (Å) | 49.26 |
| c (Å) | 68.01 |
| Resolution (Å) | 38.9-1.73 |
| Number of Reflection | 12689 |
| Working Reflections | 12035 |
| Free set Reflections | 654 |
| Completeness (%) | 99.4 (98.2) |
| Multiplicity/Redundancy | 11.9 (11.7) |
| I/sigma(I) | 47.1 (3.8) |
| Rmerge | 0.088 |
|
| |
| Refinement | |
| Rcryst work (%) | 19.29 |
| Rcryst free (%) | 21.86 |
| RMSD bond lengths (Å) | 0.12 |
| RMSD bond angles (°) | 1.4 |
| B-factor mean (Å2) | 35.2 |
| Protein atoms | 895 |
| Compound atoms | 10 |
| Water molecules | 67 |
| Solvent Content (%) | 44.41 |
| Matthew Coefficent Å3 Da−1 | 2.23 |
|
| |
| Ramachandran plot | |
| Favored regions (%) | 97.1 |
| Allowed regions ( %) | 2.9 |
Parenthesis indicates data in the highest resolution bin
Phasing solution and structural determination
An initial phasing solution was generated by molecular replacement (Rossmann, 1990). A search model of an all alanine CISK-PX domain (residues 4-119) was used as a template by the computational molecular replacement method Phaser (McCoy et al., 2007). The model was built using a combination of ArpWarp and Coot (Cohen et al., 2008; Emsley and Cowtan, 2004) and refined using REFMAC5 to an R/Rfree of 19.3%/21.9% respectively (Murshudov et al., 2011). Sulfate molecules and waters were added either by hand or automatically. The structure was validated by Rampage (Lovell et al., 2003) (Table 1).
RESULTS
Structural analysis of the designed PX domain
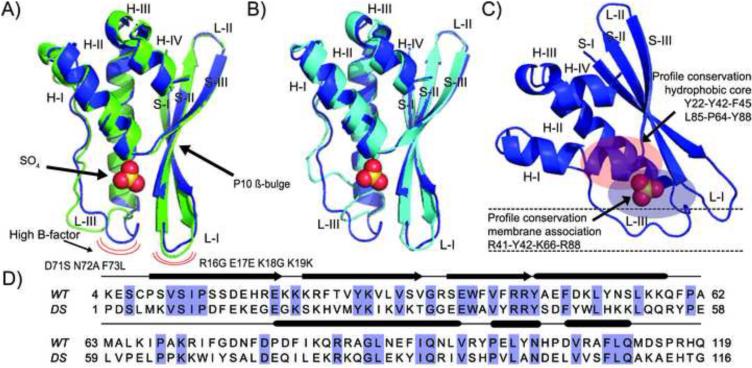
The designed PX domain X-ray structure consists of a single monomer in the asymmetric unit forming a compact globular domain that consists of an antiparallel β-sheet meander comprised of 3 β-strands and 4 helices. Residues 5-110 are in good electron density, except for loop residues (16-19 & 71-73), which display high B-factors. The first strand in the DS-CISK-PX X-ray structure has a β-bulge at residue P10, similar to the WT-CISK-PX structure, and a sulfate anion is reported in a cleft where phosphoinositides are known to bind to the WT-CISK-PX protein (Xing et al., 2004). A second sulfate molecule close to the first facilitates stabilization of the crystal lattice. The design has a high degree of structure complexity (7 different secondary structure elements) for a designed sequence, with 32% sequence identity to the wild-type. See Figure 1 for fold similarity and sequence alignment between the two proteins.
Figure 1.
Structural and sequence comparison between DS-CISK-PX X-ray structure, WT-CISK-PX X-ray structure and DS-CISK-PX I-TASSER model. (A) Superposition of WT-CISK-PX X-ray structure (1XTE) (green) and DS-CISK-PX X-ray structure (blue). Sulfate from DS-CISK-PX shown in spheres to highlight binding cleft. Major structural differences in loop regions having a high B-factor are marked with the red curves. (B) Superposition of DS-CISK-PX X-ray structure (blue) and the I-TASSER DS-CISK-PX model (cyan). C) Predicted orientation of the PX domain relative to the cell membrane (black dash lines) to bind phosphoinositide. D) Sequence alignment between WT-CISK-PX and DS-CISK-PX. Identical residues are highlighted in blue. Secondary structure elements features shared between the two sequences are annotated (strands-arrows, helices-rods, and coils-thin lines).
In Table 2, we compare the fold similarity between the WT-CISK-PX and DS-CISK-PX X-ray structures as well as that between the X-ray structure and the I-TASSER model of DS-CISK-PX. All the similarity matrices are calculated for Cα-traces by the TM-score superposition matrix (Zhang and Skolnick, 2004). Because TM-score scales smaller distance stronger than larger distances, the value of TM-score is more sensitive to the global fold rather than the local structure derivation compared to conventionally used root mean square deviation (RMSD). TM-score value ranges from [0, 1]. Based on a large-scale all-to-all comparative study on unrelated protein structures from the PDB, it was demonstrated that a TM-score < 0.17 corresponds to the similarity of two random structures, TM-scores > 0.5 to the same SCOP/COTH fold, and 1 to a perfect match (Xu and Zhang, 2010). The data in Table 2 show that the designed sequence recapitulated the overall PX domain fold with a Cα RMSD=1.54 Å and TM-score=0.90. Moreover, I-TASSER modeling recognized the fold of the design sequence with a Cα RMSD=1.32 Å and TM-score=0.91, despite the fact that the sequence similarity between the designed and wild-type sequences is at the border of the twilight-zone for protein structure prediction (Rost, 1999).
Table 2.
X-ray structure and I-TASSER model comparisons between WT-CISK-PX (WT) and DS-CISK-PX (DS).
| Comparison (Cα RMSD Å/TM-score) | X-ray (WT) | I-TASSER model (DS) |
|---|---|---|
| I-TASSER model (WT) | 1.36 Å/0.91 | |
| X-ray (DS) | 1.54 Å/0.90 | 1.32 Å/0.91 |
The most notable structure differences between DS-CISK-PX and WT-CISK-PX proteins occur in the loop regions (16-19 &71-73) that display high B-factors and are known to facilitate phosphoinositide binding at the membrane (Figure 1). This is expected as these loops are highly mobile as seen in multiple PX domain NMR structures (Lu et al., 2002; Zhong et al., 2005). In the WT-CISK-PX X-ray structure, the loops are in crystal contacts and well ordered. The WT-CISK-PX loops are thought to undergo conformational changes upon binding phosphoinositide, similar to the structurally homologous p40-PX co-crystal structure with bound phosphoinositide (PDB ID: 1H6H) (Bravo et al., 2001); herein referred to as WT-p40PX(PI). Further, the polyproline helix capping motif seen in many PX domains is missing in the design structure (residue Gln76 vs Pro79), which likely influences loop positioning, and can possibly explain the structural variation between the two sequences.
Impact of the threading-based structure profiles on EvoDesign
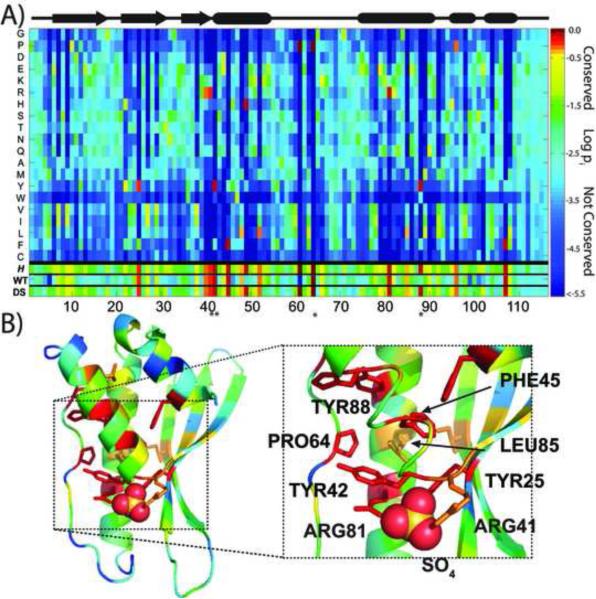
The tendency of the designed sequence to follow the EvoDesign structure profile can be seen in the form of a position specific scoring matrix that was mapped on the DS-CISK-PX structure (Figure 2). The construction of the profile was guided by a Henikoff-Henikoff BLOSSUM-62 matrix weighting scheme to provide coverage for all amino acids at all positions (Henikoff and Henikoff, 1992). The evolutionary-based structure profile reports on the favorability of a residue at a given position for a protein fold. Nineteen templates were used in the construction of the structure profile with TM-scores to the target template ranging from 0.7-0.8. All the templates were collected by the structural alignment program, TM-align (Zhang and Skolnick, 2005). Under the matrix is listed the Shannon entropy that is defined by , where p(i, j) is the probability of the jth type of amino acid appearing at the ith position in the structure profile. The highly conserved residue positions in the matrix display low Shannon entropy, while non-conserved residues positions display high Shannon entropy (Shannon, 1997).
Figure 2.
Mapping the evolutionary profile onto the DS-CISK-PX X-ray structure. (A) X-axis indicates residue position in the structure profile and Y-axis represents amino acid type. Matrix elements are rainbow color-coded based on the conservation of a specific amino acid for a given position. The secondary structure of the designed sequence is shown at top. At the bottom of the map, the Shannon entropy (H) and the WT-CISK-PX (WT)/DS-CISK-PX (DS) sequences are highlighted using the same color-code. The 4 “*” represent residues that are proximal to the sulfate molecule in the crystal structure and help define the cleft. (B) The DS-CISK-PX structure is color-coded based on the structure profiles, with the well-conserved residues in the profile matrix shown as sticks in the X-ray structure. Figure on the right highlights the well-conserved TYR/PHE aromatic residue cage (red) that encircles Leu85 (orange); the first helix ribbon is partially removed for clarity.
As seen in Figure 2A, the key low-entropic residue positions in the profile (bright orange-red matrix elements) are generally conserved with a high fidelity in the DS-CISK-PX sequence. Although the structure profile was constructed based on WT-CISK-PX, the DS-CISK-PX sequence tends to resemble the profile more closely than the WT-CISK-PX sequence (see bottom of Figure 2A). This tendency should be favorable from a foldability perspective.
In Figure 2B, we mapped the structure profile onto the DS-CISK-PX X-ray structure to track the positional relationship between the low-entropy, well-conserved residue positions. These positions are tightly clustered and can be broken down into two features. First, the profile plot easily identified a conserved aromatic/proline cage of residues around L85, consisting of Y25, Y42, F45, P64, and Y88 (0.8 Å mean Cα RMSD from WT-CISK-PX). From an evolutionary perspective, these residues represent the core of the PX domain fold and are appropriately captured in the flexibility of the structure profile. Second, the positively charged surface residues known to be crucial for wild-type membrane association and phosphate binding are identified in the DS-CISK-PX sequence, consisting of R41, Y42, K66, and R88 (0.9 Å mean Cα RMSD from WT-CISK-PX). This is a likely result of the structure profile, while other nearby residues are allowed to vary (Xing et al., 2004).
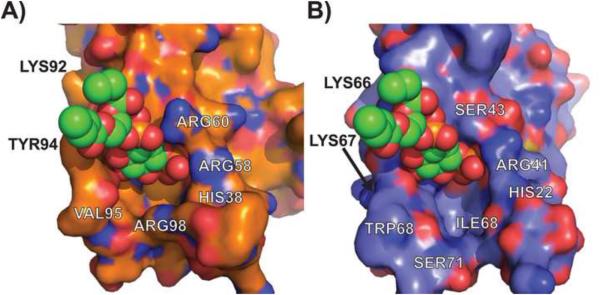
In Figure 3, we examine the surface features of the DS-CISK-PX proteins in comparison with the WT-p40PX(PI). It is shown that the phosphoinositide molecule from the WT-p40PX(PI) complex can be structurally aligned to the DS-CISK-PX cleft reasonably well, despite the overall structural variation witnessed in the loops. The phosphate from phosphoinositide can be superposed to cleft bound sulfate in the DS-CISK-PX structure with a deviation of 0.7 Å. This confirms that in general the algorithm is capable of potentially creating a binding cleft suitable for binding various phosphoinositides, which might enhance the medical potential of the design proteins since the WT-p40PX domain is known to bind phosphoinositide weakly (dissociation constant 5 μM) (Bravo et al., 2001). However, there are structural differences within the binding pockets. The WT-p40PX(PI) binding pocket cleft more tightly surrounds the phosphoinositide molecule versus DS-CISK-PX; and the WT-p40PX(PI) residues, ARG60 and TYR94, that contact phosphoinositide and stabilize the interaction at a more solvent exposed position do not have corresponding DS-CISK-PX residues that could serve the same function. This is understandable because the binding interaction was not specified in the original EvoDesign design.
Figure 3.
Surface cleft-site comparisons between WT-p40PX(PI) and DS-CISK-PX X-ray structures. Expanded view of the phosphoinositide binding sites in WT-p40PX(PI) (A) and DS-CISK-PX (B) domains, respectively, with the phosphoinositide molecule from p40-PX superposed onto the DS-CISK-PX cleft to highlight surface feature similarity.
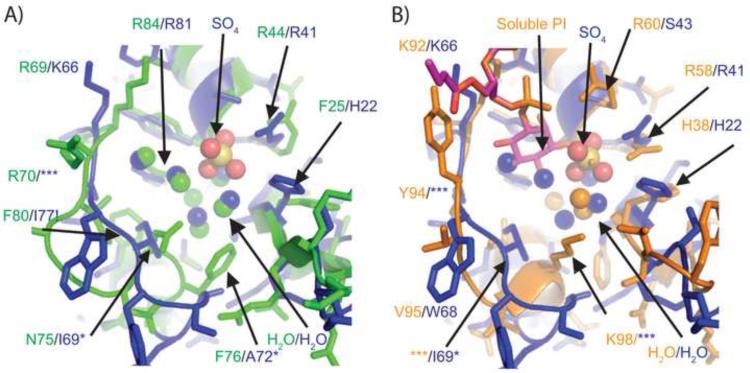
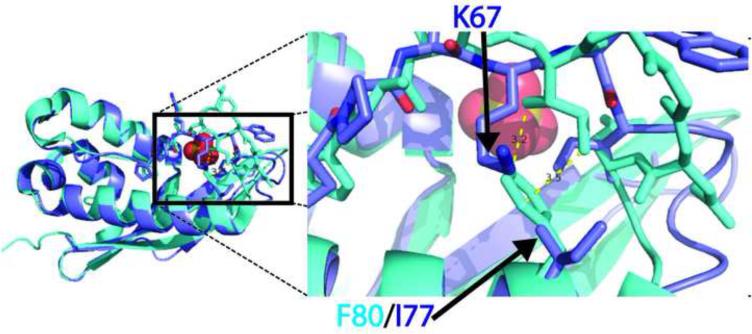
In Figure 4, we highlight the atomic differences of the binding pockets between WT-CISK-PX versus DS-CISK-PX and WT-p40PX(PI) versus DS-CISK-PX structures. The sequence identity between WT-CISK-PX and DS-CISK-PX is 40% in the binding pocket regions. The residues that line the binding pocket for phosphoinositide (spatially defined by the WT-p40PX(PI) structure) are different in the WT-CISK-PX versus the DS-CISK-PX structures with the exception of 4 residues, R44/R41, Y45/Y42, R69/K66 (very similar), and R84/R81, respectively. The rest of the residues in the binding pocket are noticeably different between the WT-CISK-PX and DS-CISK-PX structures (F25/H22, T26/V23, A46/S43, N75/I69, F76/A72, and F80/I77, where the last 3 are actually in distinct spatial positions due to main chain deviations). The F80/I77 WT-CISK-PX/DS-CISK-PX residue difference appears to affect loop positioning.
Figure 4.
Atomic structural comparison of phosphoinositide and sulfate binding cleft sites from WT-CISK-PX, DS-CISK-XP and WT-p40PX(PI) X-ray structures. (A) Binding pocket comparison between the WT-CISK-PX (Green carbon atoms) and the DS-CISK-PX X-ray structures in stick mode (light blue carbon atoms-sulfate anion yellow with two water molecules in red dots); (B) comparison of the WT-p40PX(PI) structure (orange carbon atoms with phosphoinositide in green) and the DS-CISK-PX X-ray structures (light blue). Additionally nitrogen atoms are in dark blue, oxygen atoms in red, phosphorous atoms in orange, and sulfur atoms in yellow. Green residue numbers correspond to WT-CISK-PX X-ray residues and blue numbers to DS-CISK-PX X-ray residues that are adjacent to the WT-CISK-PX residues. Residues that are spatially proximal but cannot be aligned in a gapless sequence alignment are denoted by ‘*’. DS-CISK-PX residue I77 is denoted with “!” to highlight that the residue is putative driving the loop confirmation variation versus WT-CISK-PX. The ‘***’ indicates that main chain movements are too large for positional residue side chain comparison.
The chemical environments around the F80/I77 residues are examined in Figure 5. It seems likely that the design scoring function did not recognize the effect of the WT-CISK-PX bulky aromatic F80 side chain on the stability of the opposing loop. The aligned residue in DS-CISX-PX, I77, is unable to stabilize the neighboring loop in the same manner as F80 does in WT-CISK-PX. Thus, the neighboring loop compensates and moves down towards residue DS-CISK-PX I77 to keep the domain more compact. One explanation for the structural variance is that the WT-CISK-PX F80 residue sterically excludes the binding pocket loop conformation seen in the DS-CISK-PX X-ray structure. Another possibility is that crystal contacts that exist in both structures, involving this loop, are different; this is also causing the structural variation. Importantly, the helix and opposing loop are involved in forming the binding pocket for the WT-CISK-PX phosphoinositide ligand. This highlights where improvements can be made regarding loop positioning around a potential ligand-binding site to help control ligand affinity. The residue is not highly conserved in the PX domain folds, as seen in figure 2, and thus might modulate loop dynamics and help drive ligand specificity for WT-CISK-PX for phosphoinositide. Despite the loop differences, several water molecules and a sulfate anion are bound well in the DS-CISK-PX cleft. Further, WT-p40PX(PI) and DS-CISK-PX also share similarities at the atomic level. The residues in similar spatial positions and of identical residue type include H38/H22, R58/R41, K92/K66, and R105/R81. These results suggest that the DS-CISK-PX domain has attributes derived from multiple known PX domain templates through the structural profiles, rather than a single template.
Figure 5.
Unintended structural effects of amino acid selection on the design simulation. The WT-CISK-PX and DS-CISK-PX structures are aligned by Cα-superposition with F80/I77 from WT/DS highlighted. The loss of the aromatic F80 side-chain is filed by the K67 side-chain group for DI-XIAP-PX on the opposing loop.
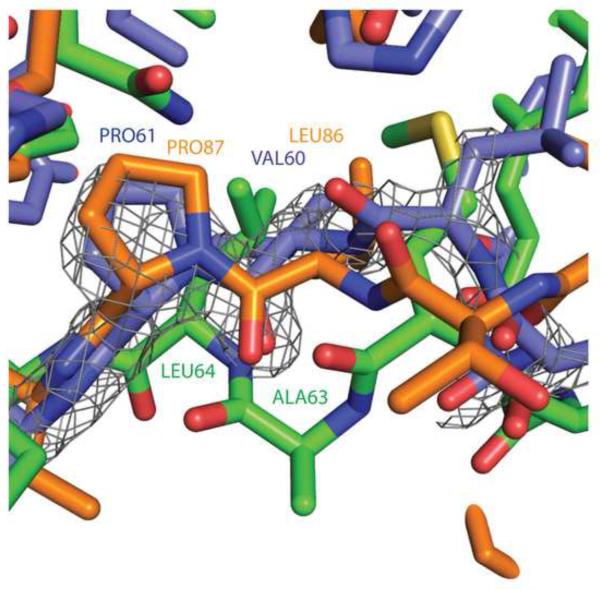
The threading-based structure profile provided an assessment of the complex cleft sequence energy landscape during the design simulation to avoid unfavorable sequence trajectories that would be difficult to fold. Here, the EvoDesign structure profile identified key residues involved in both the foldability and the general membrane associative function of the domain near the cleft. Figure 6 demonstrates that subtle sequence variation in the structure profile (DS-CISK-PX residue position P61) can lead to a designed sequence with a conformation that is more similar to another PX domain other than the target template. At residue position 61, the structure profile actually favors Pro over Leu (see also Figure 2). Interestingly, DS-CISK-PX P61 aligns itself more favorably with the P87 in WT-p40PX(PI) X-ray structure (with a Cα RMSD=0.8 Å) than the target template WT-CISK-PX L64 residue (Cα RMSD=1.9 Å). The structural inflexibility of P61 apparently drives the DS-CISK-PX structure towards greater similarity with WT-px40(PI) over the WT-CISK-PX structure at residue positions V60 and P61. Despite the design being based on a fixed backbone, EvoDesign has evolved a protein that is different from the target scaffold structurally, but still considered a PX domain; this data partly highlights the efficiency of conformation evaluation through the empirical threading-based structure profile.
Figure 6.
Low entropy residues in structure profile guide the DS-CISK-PX structure to alternate PX domain conformations. WT-CISK-PX, WT-p40PX(PI), and DS-CISK-PX X-ray structures are in green, orange and blue, respectively. Residue numbers follow a similar coloring scheme. Electron density map from the DS-CISK-PX X-ray structure shown in grey mesh (2Fo-Fc map contoured at 2.0 sigma).
Assessment of statistical potentials in EvoDesign
In addition to the structural profiles, EvoDesign also uses knowledge-based potentials, including secondary structure and solvent accessibility predictions, to guide the local structure packing in the design simulations. To generate the local structure predictions, traditional bioinformatics methods use neural network approaches that are trained on the multiple sequence alignment (MSA) searched by PSI-BLAST through large-scale sequence databases (Jones, 1999). Because the local structure predictions are needed at each step of EvoDesign Monte Carlo simulation and a PSI-BLAST search is computationally too expensive, EvoDesign exploited a simplified neural network that was trained on a single target sequence, which can quickly generate the local feature prediction but with a compromised accuracy (Mitra et al., 2013b).
As part of the assessment of the effectiveness of the simplified potentials on EvoDesign, in Table 3 we listed the accuracy of the secondary structure and solvent accessibility predictions by PSSpred, Solvant and EvoDesign, respectively, compared to the X-ray structures of DS-CISK-PX and WT-CISK-PX. PSSpred and Solvant are newly developed (full-version) neural networks trained on PSI-BLAST MSAs, which achieved a Q3 accuracy 85% for secondary structure and expose-bury accuracy 87% for solvent accessibility in the large-scale benchmark tests (Yang et al., 2015), while the single-sequence based neural network predictor used in EvoDesign has only accuracy of ~70% for both secondary structure and solvent accessibility (Mitra et al., 2013a). As expected, the full-version PSSpred and Solvant predictors generated predictions of much higher accuracy than the single-sequence based EvoDesign predictors in both DS-CISK-PX and WT-CISK-PX proteins. Interestingly, both secondary structure and solvent accessibility predictions from all the predictors have a higher accuracy in DS-CISK-PX than in WT-CISK-PX, including those from EvoDesign. The differences in accuracy between the DS-CISK-PX and WT-CISK-PX are statistically significant for all the predictors (except for Solvant) with a p-value <10−6. Further, the differences in prediction between DS-CISK-PX and WT-CISK-PX were even more pronounced when we focused only on residues where transitions in secondary structure occurred, i.e. from strand to coil, helix to coil, etc (second set of values in Table 3).
Table 3.
Secondary structure and solvent accessibility predictions on WT-CISK-PX and DS-CISK-PX sequences. Accuracies for the entire chain are given first and that for the translational regions of secondary second are given second.
| Proteins | Secondary structure | Solvent accessibility | ||
|---|---|---|---|---|
| PSSpred | EvoDesign | Solvant | EvoDesign | |
| DS-CISK-PX | 90.6%/87.5% | 74.2%/63.7% | 86.2%/83.0% | 75.3%/66.6% |
| WT-CISK-PX | 85.8%/79.2% | 65.7%/43.8% | 85.4%/83.0% | 69.6%/58.3% |
This striking consistency indicates that, despite the relatively low accuracy of the single-sequence neural network prediction, the knowledge-based potentials indeed helped guide the EvoDesign simulations towards the sequences with general local structure and solvation patterns, as correctly recognized by the full-version bioinformatics predictors. Physically, these residue patterns were recognized by an integrated potential in EvoDesign that combines secondary structure and solvent accessibility predictions with structural profiles. Here the structural profiles were generated from multiple sequence alignments of homologous protein structures collected by TM-align from the PDB (Zhang and Skolnick, 2005). The consensus structural profile reinforced the selection of the designed proteins with the general local structure features (Mitra et al., 2013b).
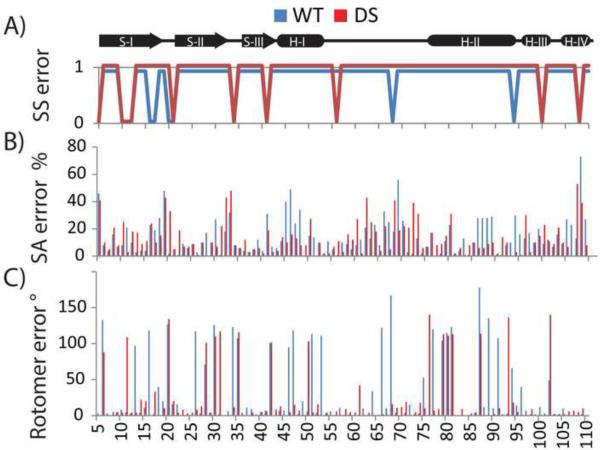
In Figures 7A and 7B, we present the local error distribution of the secondary structure and solvent accessibility predictions along the designed and WT sequences. ~2/3 of the errors in secondary structure and solvent accessibility predictions occur at strand to coil transitions and the beta-bulge of the DS-CISK-PX and WT-CISK-PX sequences. A closer look at the EvoDesign secondary structure and solvent accessibility prediction data shows that the prediction accuracy in the transitional regions is significantly lower than that along the entire sequence. In particular, the extent of the accuracy reduction by EvoDesign is much higher than that by PSSpred and Solvant (Table 3). This partly explains the relatively lower accuracy of secondary structure and solvent accessibility predictions in the transitional regions. The data is also consistent with the observations that PSSpred and Solvant have on average 11% (5%) higher errors in beta-strand (coil) than in helix predictions in the large-scale benchmark test (Yang et al., 2015). Structurally, helices are constrained by short-range intra-element hydrogen bonding that are easier to predict from local residue conservation patterns, whereas strands from β-sheets occur via long-range inter-strand hydrogen bonding, and coils have usually no regular hydrogen binding patterns. Thus, strands and coils display greater structural diversity and complexity, providing an additional challenge in designing effective statistical potentials for these regions.
Figure 7.
Differences between predicted and observed local structural features in the WT-CISK-PX (blue) and DS-CISK-PX (red) sequences. (A) Differences between observed (X-ray) and PSSpred-predicted secondary structure (strand, helix, coil) on the WT-CISK-PX and DS-CISK-PX sequences. Y-axis represents if secondary structure is correctly predicted (Yes-1, No-0). (B) Percent difference in residue solvent accessibility between X-ray structure and prediction by Solvant. (C) Rotamer χ1 angle error in degrees is shown between X-ray structure and predicted side-chain position by Scwrl 4.0.
Accuracy of the side-chain fitting algorithm
The goodness of amino acid side-chain packing is a key to the folding of successful sequence designs. To examine the side-chain packing of EvoDesign, we apply Scwrl V4.0 (Krivov et al., 2009) to rebuild the rotamer conformations of the designed sequence and then use the modeling accuracy as a partial examination of the goodness of the side-chain packing. The underlying assumption is that Scwrl correctly reflects the general correlation of backbone and side-chain conformations, since the library of Scrwl is built on the large-scale statistics of rotamers observed in high-resolution PDB structures. Meanwhile, because the Scwrl rotamer potential has been integrated in the EvoDesign simulations, a closer match of the Scwrl side-chain conformation with the experiment should be a minimum request to ensure the correct implementation of the EvoDesign algorithm.
In Figure 7C, we showed the rotamer positioning error by Scwrl 4.0 for both DS-CISK-PX and WT-CISK-PX structures. The average Scwrl side-chain rotamer error on the χ1 angles for the DS-CISK-PX and WT-CISK-PX sequences against the respective X-ray structures was 23º and 32º, respectively, where the 9º difference in the modeling is statistically significant with a p-value < 4.5×10−19. As expected, the Scwrl predictions indeed showed a more favorable accuracy of side-chain modeling and therefore the goodness of side-chain packing in the DS-CISK-PX structure compared to WT-CISK-PX. These data also confirm the effectiveness of the side-chain fitting program in the EvoDesign sequence, since the DS-CISK-PX sequence has been specifically optimized based on the Scwrl rotamer conformations in the EvoDesign simulation.
CONCLUDING REMARKS
The designed PX domain X-ray crystallographic structure was solved to examine the process and efficiency of the structure-profile based evolutionary protein design approach. The method was applied to design a conditional peripheral membrane protein fold that is known to localize other proteins to the endosomal membrane. This initial X-ray structure provides a foundation from which to guide our efforts in designing a controlled localization fusion partner for a therapeutic/diagnostic. The EvoDesign procedure is fully automated, which arose from the desire to meet the growing demand for large-scale macromolecular engineering. This strategy is built on the efficiency of the template profile-based structure predictions, which are relatively unaffected by domain size, as witnessed in protein fold-recognition and template-based structure prediction experiments (Huang et al., 2014; Zhang, 2008a).
The designed PX domain protein DS-CISK-PX in this study has 116 residues with a sequence identity 32% to the wild-type scaffold WT-CISK-PX. The X-ray crystallographic structure shows that the DS-CISK-PX protein adopts a similar globule PX like fold with 7 secondary structure elements (Figure 1), which is 1.54 Å to WT-CISK-PX with a TM-score=0.91. The result shows the efficiency of evolution-based approaches in specifying global fold of designed proteins.
Importantly, the experiment also demonstrated the ability of the protein structure prediction algorithm I-TASSER to recognize the foldability of designed proteins. The I-TASSER simulations on the designed protein showed a high folding confidence with C-score=1.31 which corresponds to a TM-score of the I-TASSER modeling above 0.89 (Zhang, 2008b). In a previous study (Mitra et al., 2013b), we have found that there is a strong correlation between the confidence score of the I-TASSER modeling and the foldability of the designed sequences from several leading design programs such as Rosetta design (Koga et al., 2012) (see Figure 2 of Ref (Mitra et al., 2013b)), and therefore concluded that the I-TASSER prediction and the confidence estimation might be used as a partial in silico prediction of the success/failure of the computational designs. This work represents a blind experimental validation of the assumption based on a single design on the CISK-PX domain where the I-TASSER prediction was made before the structural solution of the sequence (Mitra et al., 2013b).
The use of the evolution-based structural profile potential helps increase the foldability of the designed domain. Information regarding conserved residues from multiple sequence and structural alignments is often critical to recognize the structure and function of the protein families. This information has been naturally integrated in the EvoDesign simulations. As demonstrated by Figure 2, the structure profile, represented by position-specific entropy matrix, helps couple long-distance restraints regarding protein domain stability and dynamics. Here the entropy matrix was calculated by sum of Shannon entropy at each residue position. Through the entropy matrix, the two specific low-entropic features were easily identified and accurately constructed in the PX design; these include a hydrophobic core of residues (Y25, Y42, F45, P64, and Y88) central to the stability of the fold, and the cleft residues known to facilitate membrane association in the wild-type protein. In theory, the structure-based profile potential can be readily tailored & tuned to the challenges of the target fold/function and can potentially overcome limitations in force field functions used to evaluate challenging protein interactions with bulk solvent and a membrane bilayer. In this case, EvoDesign created a designed domain with many latent features associated with a conditional peripheral membrane PX domain, as demonstrated in Figures 3 and 4. Additionally, the structure profile can be used to create sequence/structural variation within the design sequence (Figure 6); thus, creating more opportunities for designing functional novelty without sacrificing efficiency.
The use of the knowledge-based statistical potentials was previously shown to be important for EvoDesign through in silico means (Mitra et al., 2013b). In this report, the accuracy of the bioinformatics predictions on secondary structure (PSSpred), solvent accessibility (Solvant) and rotamer position (SCWRL) were assessed on both the wild-type and design structures. Despite the relative lower accuracy of the single-sequence feature predictions exploited in the EvoDesign, the final designed sequence was favored by the all the full-version bioinformatics feature predictions. This data demonstrates that the knowledge-based potentials introduced by the single-sequence based feature predictions did help EvoDesign to pick up the general local structure patterns, as correctly recognized by the sophisticated neural-network predictors.
We note that this study has been mainly focused on structural characteristics of the designed PX domain and their implication on evolution-based design principles. Nevertheless, the highly stable fold of the protein and the structural similarity to the target scaffold, as designed, builds a solid base for the next step functional protein design. For this purpose we are currently working on the design of several PX domains with high affinity and specificity for phosphoinositides useful for controlling fusion protein localization for in vivo cellular localization studies. Here the structural variations witnessed between WT- and DS-CISK-PX proteins, especially the variation near the residue F80/I77 involved in forming the ligand binding pocket, are found particularly useful to guide the design simulation for affinity of a synthetic analog to phosphoinositide.
In conclusion, the X-ray crystallographic structure of the designed CISK-PX domain demonstrates the feasibility of systematically implementing an evolutionary profile based protein design paradigm. Data was collected and analyzed on the accuracy of the design algorithm using the designed PX X-ray crystallographic structure in conjunction with bioinformatics predictors. The results provide a wealth of information that can be used as a guide in the computational construction of future conditional peripheral membrane associated PX domains with augmented membrane affinity, membrane specificity, or altered protein interaction partners.
ACKNOWLEDGEMENTS
The authors thank Dr. Jeffrey Brender for critically reading the manuscript. This work is supported in part by the awards from the National Institutes of Health (GM083107 and GM084222).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
DS and YZ conceived the project; GD and DS generated the reagents; DS and GD collected the X-ray diffraction data and solved the structure; DS, GD, and YZ wrote the paper.
REFERENCES
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Bazzoli A, Tettamanzi AG, Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. Journal of molecular biology. 2011;407:764–776. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows-Peterson ML, Fung HK, Floudas CA, Kieslich CA, Zhang L, Morikis D, Wareham KJ, Monk PN, Hawksworth OA, Woodruff TM. De novo peptide design with C3a receptor agonist and antagonist activities: theoretical predictions and experimental validation. J Med Chem. 2012;55:4159–4168. doi: 10.1021/jm201609k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender GM, Lehmann A, Zou H, Cheng H, Fry HC, Engel D, Therien MJ, Blasie JK, Roder H, Saven JG, DeGrado WF. De novo design of a single-chain diphenylporphyrin metalloprotein. J Am Chem Soc. 2007;129:10732–10740. doi: 10.1021/ja071199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Cohen SX, Ben Jelloul M, Long F, Vagin A, Knipscheer P, Lebbink J, Sixma TK, Lamzin VS, Murshudov GN, Perrakis A. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr Purif. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson CD, Andrews S, Stephens LR, Hawkins PT. The PX domain: a new phosphoinositide-binding module. J Cell Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Floudas CA, Fung HK, McAllister SR, Monnigmann M, Rajgaria R. Advances in Protein Structure Prediction and De Novo Protein Design: A Review. Chemical Engineering Science. 2006;61:966–988. [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Mao B, Aramini JM, Montelione GT. Assessment of template-based protein structure predictions in CASP10. Proteins. 2014;82(Suppl 2):43–56. doi: 10.1002/prot.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivov GG, Shapovalov MV, Dunbrack RL., Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Larson SM, Garg A, Desjarlais JR, Pande VS. Increased detection of structural templates using alignments of designed sequences. Proteins. 2003;51:390–396. doi: 10.1002/prot.10346. [DOI] [PubMed] [Google Scholar]
- Lauck F, Smith CA, Friedland GF, Humphris EL, Kortemme T. RosettaBackrub--a web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 2010;38:W569–575. doi: 10.1093/nar/gkq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci U S A. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Polymers for drug delivery systems. Annu Rev Chem Biomol Eng. 2010;1:149–173. doi: 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Yang X, Songyang Z. Identification of CISK, a new member of the SGK kinase family that promotes IL-3-dependent survival. Curr Biol. 2000;10:1233–1236. doi: 10.1016/s0960-9822(00)00733-8. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Lu J, Garcia J, Dulubova I, Sudhof TC, Rizo J. Solution structure of the Vam7p PX domain. Biochemistry. 2002;41:5956–5962. doi: 10.1021/bi020050b. [DOI] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Shultis D, Zhang Y. EvoDesign: De novo protein design based on structural and evolutionary profiles. Nucleic acids research. 2013a;41:W273–280. doi: 10.1093/nar/gkt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Shultis D, Brender JR, Czajka J, Marsh D, Gray F, Cierpicki T, Zhang Y. An Evolution-Based Approach to De Novo Protein Design and Case Study on Mycobacterium tuberculosis. PLoS computational biology. 2013b;9:e1003298. doi: 10.1371/journal.pcbi.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Poole AM, Ranganathan R. Knowledge-based potentials in protein design. Curr Opin Struct Biol. 2006;16:508–513. doi: 10.1016/j.sbi.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Rossmann MG. The molecular replacement method. Acta Crystallogr A. 1990;46:73–82. doi: 10.1107/s0108767389009815. Pt 2. [DOI] [PubMed] [Google Scholar]
- Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Saunders CT, Baker D. Recapitulation of protein family divergence using flexible backbone protein design. Journal of molecular biology. 2005;346:631–644. doi: 10.1016/j.jmb.2004.11.062. [DOI] [PubMed] [Google Scholar]
- Shannon CE. The mathematical theory of communication. M.D. computing : computers in medical practice. 1997;14:306–317. 1963. [PubMed] [Google Scholar]
- Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- Xing Y, Liu D, Zhang R, Joachimiak A, Songyang Z, Xu W. Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX) J Biol Chem. 2004;279:30662–30669. doi: 10.1074/jbc.M404107200. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu D, Gill G, Songyang Z. Regulation of cytokine-independent survival kinase (CISK) by the Phox homology domain and phosphoinositides. J Cell Biol. 2001;154:699–705. doi: 10.1083/jcb.200105089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008a;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008b;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Protein structure prediction: when is it useful? Curr Opin Struct Biol. 2009;19:145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic. Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Watson MJ, Lazar CS, Hounslow AM, Waltho JP, Gill GN. Determinants of the endosomal localization of sorting nexin 1. Mol Biol Cell. 2005;16:2049–2057. doi: 10.1091/mbc.E04-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]