Abstract
Cancers are characterized by non-random, chromosome copy number alterations that presumably contain oncogenes and tumor–suppressor genes (TSGs). The affected loci are often large, making it difficult to pinpoint which genes are driving the cancer. Here, we report a cross-species in vivo screen of 84 candidate oncogenes and 39 candidate TSGs, located within 28 recurrent chromosomal alterations in ependymoma. Through a series of mouse models we validate eight new ependymoma oncogenes and 10 ependymoma TSGs that converge on a small number of cell functions including vesicle trafficking, DNA modification and cholesterol biosynthesis, pinpointing these as potential new therapeutic targets.
DNA copy number alterations (CNAs) are a hallmark of cancer that can drive the expression of oncogenes1 or delete tumor suppressor genes (TSGs)2. Recurrent CNAs that disrupt the same gene can be highly informative, pinpointing oncogenic alterations that may serve as therapeutic targets. More often CNAs are large; making it difficult to identify which gene(s) is driving transformation. RNA silencing technologies have uncovered novel TSGs within large deletions3, but approaches to screen the oncogenic capacity of genes located within large regions of gain are less well developed.
Recently we showed that EPHB2 amplification4 or the C11orf95-RELA translocation5 – two genetic alterations in the brain tumor ependymoma – can transform embryonic, Ink4a/Arf−/− cerebral neural stem cells (cNSCsnull) to generate these tumors in mice. These mouse models are proving useful to develop new treatments for the clinic6. The identification of additional ependymoma oncogenes and TSGs might further advance understanding of the disease, but whole genome and transcriptome sequencing has failed to detect additional recurrent mutations or focal CNAs in ependymoma5, 7. Rather, these tumors are characterized by large CNAs that encompass many genes.
To identify genes that drive ependymoma, we performed a comprehensive in vivo screen of 84 candidate oncogenes and 39 candidate TSGs located within 28 recurrent CNAs. Through a series of mouse models, we identify eight new ependymoma oncogenes and eleven new ependymoma TSGs that disrupt a limited number of cell functions, including vesicle trafficking and cholesterol synthesis. We propose these cell functions are important components of ependymoma pathogenesis and potential therapeutic targets.
Results
The ependymoma CNA landscape
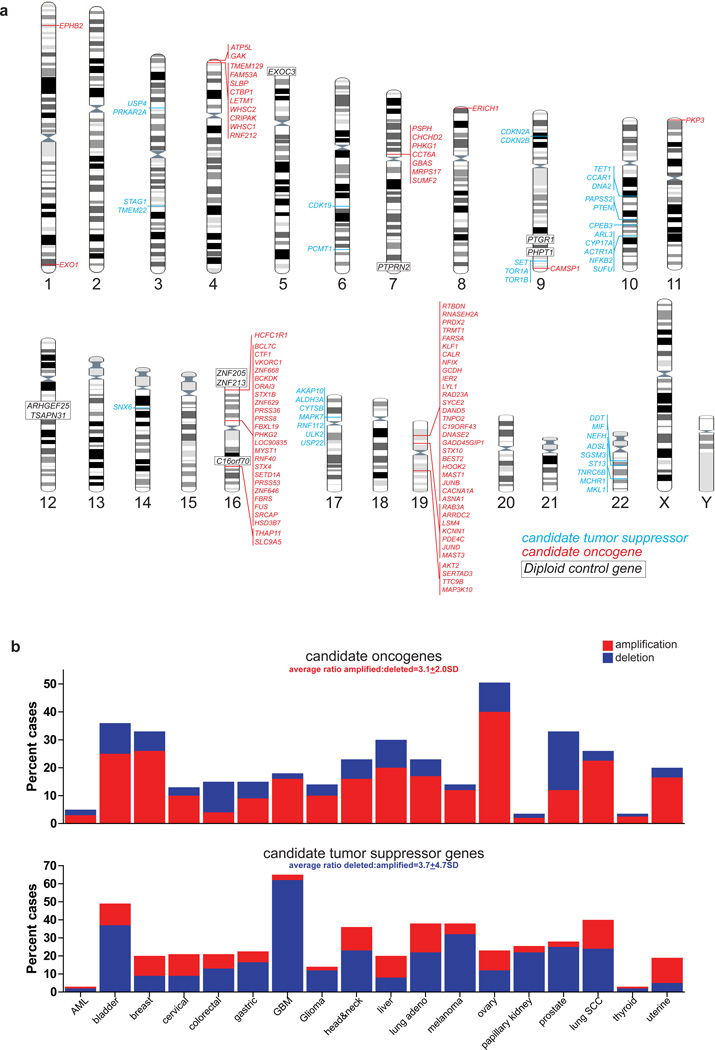
We previously generated genome-wide DNA copy number and gene expression profiles of 83 ependymomas4. These data identified 14 regions of recurrent gain (median size 509kb, range 16.6kb–2.0Mb), containing 85 candidate oncogenes, that were selectively over-expressed in tumors containing the CNA (Fig. 1a; Supplementary Table 1) and 15 recurrent deletions (average 1.1Mb, range 780kb–6.7Mb) containing 39 candidate TSGs (Fig. 1a; Supplementary Table 2; see Methods for details of candidate selection)4. A minority of these candidates are putative or known oncogenes (e.g., JUNB, AKT2, EPHB2) or TSGs (e.g., PTEN, CDKN2A/CDKN2B), but most have not been implicated in cancer.
Figure 1.
Candidate ependymoma oncogenes and tumor suppressor genes. (a) Location of copy number alterations containing candidate oncogenes (red) or tumor suppressor genes (blue). Diploid control genes for in vivo oncogensis screen are shown in black. (b) Output from the cBioPortal web resource reporting the percentage of amplifications and deletions of candidate oncogenes (top) and tumor suppressor genes (bottom) across ~4,900 samples of 18 different human cancers.
As an initial test of their candidacy as oncogenes and TSGs, we looked at the copy number of these genes (excluding the previously validated ependymoma oncogene EPHB2, Ref. 4) in ~4,900 samples of 18 different human cancers in the cBioPortal database8. Candidate ependymoma oncogenes were 3.1 times more likely to be amplified than deleted in other human cancers, while candidate TSGs were 3.7 times more likely to be deleted than gained (Fig. 1b, net average difference in CNA between candidate oncogenes and TSGs, P<0.0001, Mann-Whitney). Similar results were obtained from analyses of human cancers reported in the COSMIC database (Supplementary Fig.1). Thus, the majority of our candidates appear to be involved in non-random CNAs in human cancer, supporting the notion that they participate in ependymoma pathogenesis.
In vivo screen of candidate ependymoma oncogenes
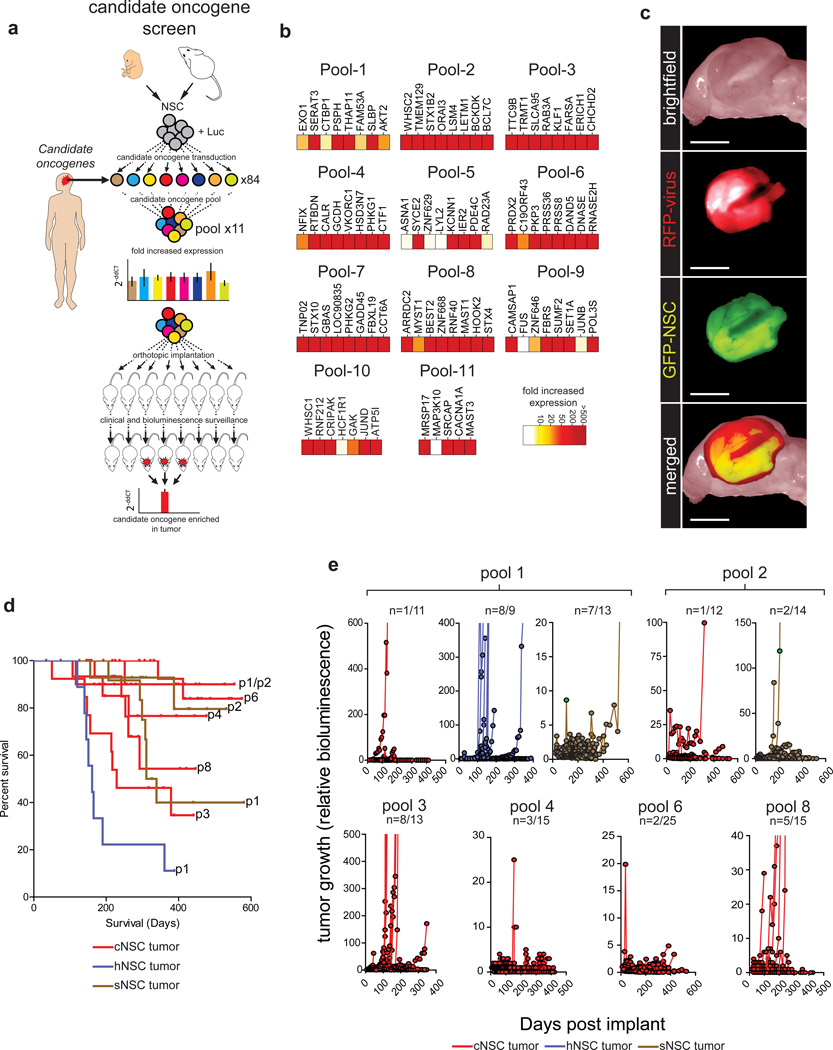
We showed previously that enhanced Green Fluorescence Protein+ (eGFP+) cNSCsnull isolated from Blbp-eGFP mouse embryos generate cerebral ependymomas when transduced with viruses encoding human cerebral ependymoma oncogenes4, 5. Since all 84 candidate oncogenes were gained in at least one human cerebral ependymoma, we adapted this approach to screen the transforming capacity of each candidate (Fig. 2a; Supplementary Table 1). Freshly isolated cNSCsnull were transduced with a Luciferase-IRES-Yellow Fluorescence Protein (YFP) retrovirus to allow bioluminescence imaging of cells in vivo. eGFP+/YFP+ cNSCsnull were then fluorescence activated cell sorted and separated into 84 fractions. Each fraction was transduced with one of 84 retroviruses encoding a different candidate oncogene upstream of an IRES-red fluorescence protein (RFP) reporter. eGFP+/YFP+/RFP+ cNSCsnull from each fraction were then randomly mixed together in equal proportions, creating 11 cell pools comprising mixtures of cells that carried up to eight different candidate oncogenes (Fig. 2a,b). Since candidate oncogenes were also gained in human posterior fossa (hindbrain) and spinal ependymomas, we reasoned that cell context might impact transformation. Therefore, we also generated a limited set of candidate-transduced hindbrain and spinal NSC pools (hNSCnull and sNSCnull). Finally, to control for transformation that might result from random retroviral insertional mutagenesis, we transduced nine additional eGFP+/YFP+ cNSCnull fractions with nine different diploid control genes that we never observed within human ependymoma CNAs (Fig. 1a; Supplementary Table 3).
Figure 2.
In vivo screen of candidate ependymoma oncogenes. (a) Cartoon depicting the overall oncogene screening approach. (b) Heatmaps reporting the fold expression of each candidate oncogene in each pool. (c) Direct fluorescence images of a pool-derived tumor in the cerebrum (scale bar=1 cm). (d) Kaplan-Meier survival curves of mice implanted with the indicated NSC pools for the nine pools that generated tumors. (e) Tumor growth reported as bioluminescence relative to day 0 for the nine pools that generated tumors.
Reverse transcription and Real-Time quantitative polymerase chain reaction analysis (RT-qrtPCR) of RNA isolated from each cell pool demonstrated that 89% (n=75/84) of candidate oncogenes were expressed ≥10 fold in cNSCnull pools relative to mock transduced controls (Fig. 2b), while 4% (n=3/84) of candidates displayed between 5 and 10 fold increased expression, and 7% (n=6/84) of candidates were expressed ≤2 fold that of controls. Similar increases in candidate expression were observed in hNSCnull and sNSCnull pools, as well as cNSCnull harboring diploid-control genes (data not shown).
1.5×106 eGFP+/YFP+/RFP+ cells from each cNSCnull, hNSCnull or sNSCnull pool were separately implanted into the cerebrum of individual immunocompromised mice, exactly as described previously4 (average of 15 mice per pool [range 11–25], total=213 mice; Supplementary Table 4). Separate cohorts of mice were similarly implanted with eGFP+/YFP+/RFP+ diploid control gene transduced cNSCsnull (average of 12 mice per gene [range 9–18], total=111 mice; Supplementary Table 5). All 324 mice were imaged twice-monthly using bioluminescence to detect tumor development. None of 111 mice carrying diploid control gene transduced cNSCsnull or 86 mice implanted with cNSCnull and/or sNSCnull pools 5, 7, 9, 10 or 11, developed tumors (median follow up 284 days; Supplementary Tables 4 and 5). Thus viral transduced NSCsnull are not intrinsically tumorigenic. Conversely, cNSCnull pools 1, 2, 3, 4, 6 and 8 generated eGFP+/YFP+/RFP+ tumors with variable penetrance, ranging from 8% (n=2/25) for pool 6 to 62% (n=8/13) for pool 3 (Fig. 2c-e). sNSCsnull and hNSCsnull containing the same candidates as cNSCnull pool 1, and sNSCsnull containing the same candidates as cNSCnull pool 2, also generated tumors (Fig. 2d,e). Therefore, we concluded that the 48 candidates within pools 1–4, 6 and 8 likely included oncogenes.
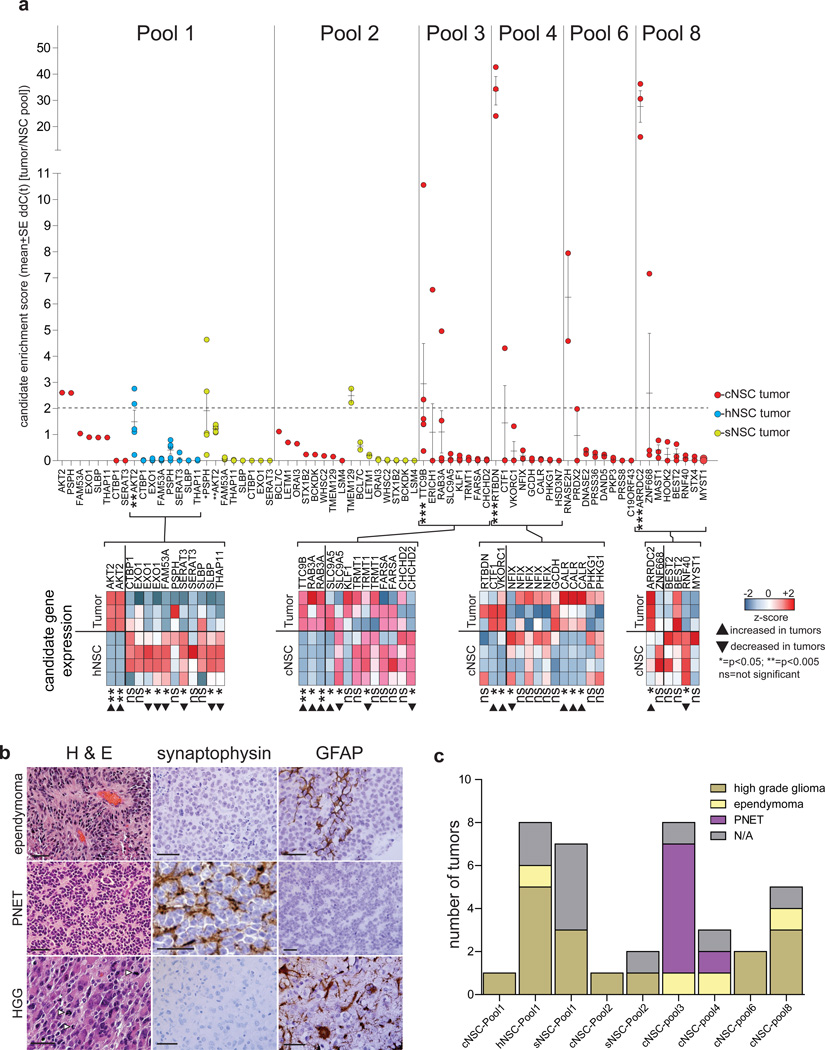
To detect which of these 48 candidate oncogenes might have driven transformation, we used RT-qPCR to quantify candidate DNAs in tumors relative to the corresponding, pre-implantation cell pool (Fig. 3a). Seventy-one percent (n=34/48) of candidate DNAs were present in significantly reduced amounts in tumors, supporting the notion that NSCsnull harboring these candidates were outgrown by those containing oncogenes (average ≤10-fold [range <1.3 to <1,000 fold]; Fig. 3a). In stark contrast, one or two candidate DNAs were selectively and recurrently enriched ≥2-fold in tumors, indicating these candidates had likely driven NSCnull proliferation in vivo and might be oncogenes (average 11.6 [range >2.2 to >42.7 fold]): AKT2, PSPH (both pool-1), TMEM129 (pool-2), TTC9B (pool-3), RTBDN (pool-4); RNASEH2 (pool-6), and ARRDC2 (pool-8). In addition, ERICH1, RAB3A (both pool-3), CTF1 (pool-4), PRDX2 (pool-6) and ZNF668 (pool-8) were enriched ≥ 2 fold in a single tumor each; while BCL7C (pool-2) and VKORC1 (pool-4) were enriched between 1 and 2 fold in a single tumor each. Analysis of tumors with sufficient material showed that 75% (n=6/8) of enriched candidates were also expressed at higher levels in tumors than the corresponding pre-implantation NSC pool, whereas 95% (n=18/19) of non-enriched candidates displayed unchanged or decreased expression (P<0.005, Fishers Exact; Fig. 3a). Together, these data identified 14 of our initial 84 candidate oncogenes as leads for validation studies.
Figure 3.
Enrichment of candidate oncogenes in NSC pool–derived tumors. (a) Top, Enrichment of each candidate measured by quantitative Real Time-PCR in pool-derived tumors. Bottom, heatmaps reporting the expression of candidates in tumors derived from the indicated NSC pools and the corresponding pre-implantation NSCs. (b) Hematoxylin and eosin (H&E) stains and synaptophysin and glial fibrillary acidic protein (GFAP) immunostains of the various morphologic types of pool-derived tumors (scale bars=50 µm). (c) Numbers of the different morphological forms of tumor arising from each pool.
Pool-derived tumors displayed a variety of morphologies that approximated human high-grade glioma (59%, n=16/27), primitive neuroectodermal tumor (PNET; 26%, n=7/27) and ependymoma (15%, n=4/27; Figure 3b,c; Supplementary Table 4). These data are compatible with the recurrent gain of candidates in a broad range of human cancers (Fig. 1b). Indeed, AKT2 that was selectively enriched high-grade glioma-like pool 1 tumors, promotes the growth, invasion and treatment resistance of human high-grade glioma9; while TTC9B that was selectively enriched in pool 3-derived PNET-like tumors is located on human chromosome 19q that is recurrently gained in human PNETs.
In vivo validation of ependymoma oncogenes
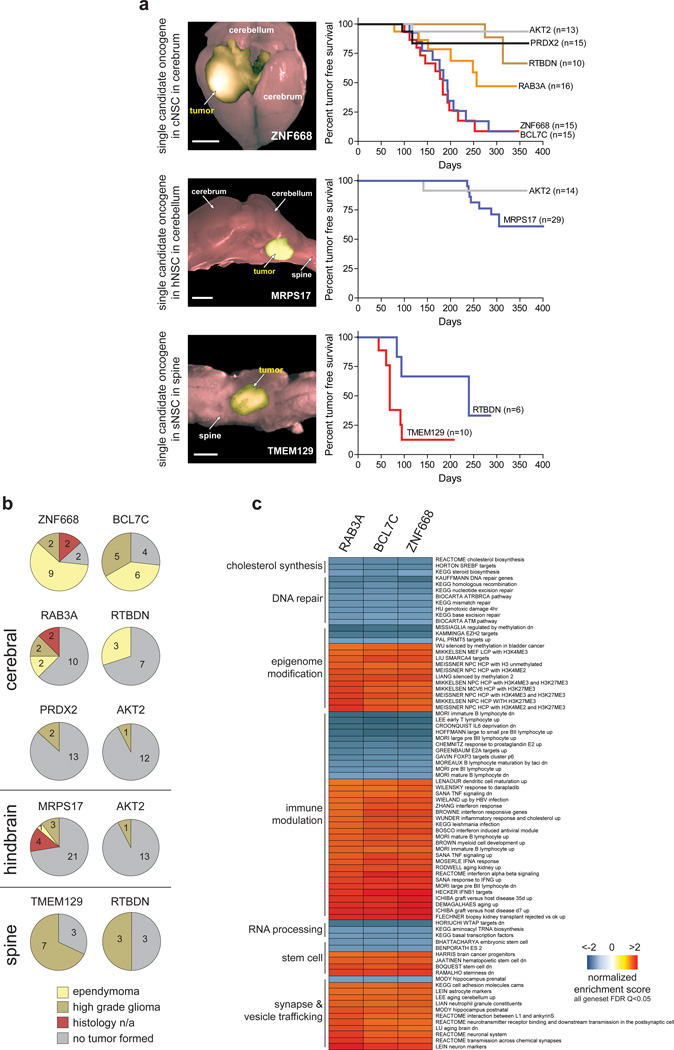
To validate our lead candidate oncogenes, we transduced 14 new fractions of eGFP+/YFP+ cNSCsnull with a single lead candidate each and implanted 1.5×106 of these cells into the cerebrum of separate immunocompromised mice (average of 16 mice per candidate [range 10–30], total=209 mice; Supplementary Table 6; Fig. 4a). Additionally, to further test the role of central nervous system topography in ependymoma tumorigenesis, we implanted the cerebella of 14 immunocompromised mice with fresh fractions of hNSCsnull harbouring AKT2 that was enriched in hNSCnull pool-1 tumors, and the spines of 10 mice with TMEM129-transduced sNSCsnull that was enriched in sNSCnull pool-2 tumors (Figs 3a, 4a; Supplementary Table 6). Two other regional tumorigenesis assays were performed: 29 cerebella implants of MRPS17-transduced hNSCsnull that is highly expressed in posterior fossa ependymoma4 but was not tested in a hNSCsnull pool; and six spinal implants of RTBDN–transduced sNSCnull cells – RTBDN was the most enriched candidate in all cNSCsnull pool tumors.
Figure 4.
Validation of ependymoma oncogenes. (a) Left, direct fluorescence images of single candidate-driven cerebral, hindbrain and spinal tumors (scale bars=5 mm). Right, Kaplan-Meier survival curves of mice implanted with the indicated NSCsnull transduced with single oncogene candidates and implanted in the corresponding CNS site. (b) Numbers of the different morphological forms of tumor driven by the indicate candidate oncogene in the indicated site. (c) Gene set enrichment analyses of BCL7C, RAB3A and ZNF668 driven tumors.
All 268 mice were imaged twice a month using bioluminescence. 53% (n=8/15) of candidates drove tumors and are therefore validated as neural oncogenes (Fig. 4a,b; Supplementary Table 6). Oncogenes validated within the cerebrum included: ZNF668 that links chromatin relaxation state to DNA repair10 and BCL7C a member of the SWI/SNF chromatin regulatory complex11 (both among 23 candidates in the 16p11.2 amplicon; Supplementary Table 1); RAB3A that controls late stage vesicle trafficking and exocytosis in neuronal cells12, 13 (among 7 candidates in the 19p13.11 amplicon); the putative oncogene PRDX2 that regulates oxidation-induced apoptosis14, and RTBDN of unknown function (both among 25 candidates in the 19p13.13 amplicon); and the AKT2 oncogene (among four candidates in the 19q13.2 amplicon; Fig. 4a). AKT2 and RTBDN also drove tumorigenesis in the hindbrain and spine, respectively, indicating that these genes are broadly oncogenic in the CNS. TMEM129 of unknown function (among 9 candidates in a 4p16.3 amplicon) and the mitochondrial ribosomal protein MRPS17 (among seven candidates in the 7p11.2 amplicon) were validated as spinal and hindbrain oncogenes, respectively (Fig. 4a). Conversely, ARRDC2, CTF1, ERICH1, PSPH, RNASEH2, TTC9B and VKORC1 failed to drive tumors and were therefore not validated as neural oncogenes. Interestingly, validated oncogenes were ten-fold more likely to be gained across human cancers than non-validated candidates, although this difference did not reach significance (Supplementary Fig. 2).
In contrast to pool-derived tumors, single candidate-driven tumors never displayed a PNET-like morphology and were three times more likely to be ependymomas (62.5% [n=20/32] single-candidate vs. 18.8% [n=3/16] pool-derived tumors; p<0.01, Fisher exact test; Figs. 3c and 4b). Thus our validated candidates are ependymoma oncogenes and the genetic heterogeneity of pooled tumors may have contributed to their morphologic diversity. Conversely, 93% (n=14/15) of single candidate-driven cerebellar and spinal tumors were high-grade glioma-like (Fig. 4b). Thus cell context and location may dictate tumor morphology in the CNS.
BCL7C, RAB3A and ZNF668 oncogenes and ependymoma subtype
Human supratentorial ependymomas comprise two molecular subtypes defined by the presence or absence of the C11orf95-RELA translocation5. To test which of these subtypes is modelled by our validated oncogene-driven mouse ependymomas, we compared the transcriptomes of human and mouse C11orf95-RELA positive and negative cerebral ependymomas using our AGDEX algorithm that tests transcriptomic similarities between tissues from different species4. BCL7C, RAB3A and ZNF668 driven mouse ependymomas displayed a significant match with human C11orf95-RELA-negative ependymoma, confirming these as faithful models of this subtype of the human disease (Supplementary Fig. 3).
To begin to understand the cell processes that are disrupted by ependymoma oncogenes, we used Gene Set Enrichment Analysis (GSEA) to compare the transcriptomes of parent cNSCsnull with those of daughter RAB3A (n=6), BCL7C (n=9) and ZNF668 (n=9) tumors (Fig. 4c; Supplementary Table 7). As expected, almost one half of the top 200 most positively or negatively enriched gene sets in mouse ependymomas included regulators of cancer processes (n=52/200), the cell cycle (n=22/200), or cell signaling (n=16/200). The remaining enriched gene sets were dominated by a handful of cellular functions that included the immune response (23%, n=23/100), synaptogenesis and vesicle trafficking (11%, n=11/100), modifications to the epigenome (11%, n=11/100) and stem cell regulation (4%, n=4/100). These data are in keeping with RAB3A’s function as a regulator of exocytosis in neural cells12, 13 and the roles that BCL7C11 and ZNF66810 play in chromatin modification. Additionally, we and others have previously reported significant upregulation of vesicle trafficking and immune response regulators in human ependymoma4, 15. Regulators of DNA repair (8%, n=8/100) and cholesterol biosynthesis (3%, n=3/100) were among the most downregulated gene sets in our mouse tumors (Fig. 4c).
Together, these data identify eight new ependymoma oncogenes located within six recurrent ependymoma CNAs; provide three new models of C11orf95-RELA-negative human ependymoma; and implicate aberrant vesicle trafficking, DNA modification and repair, immune response and cholesterol biosynthesis in ependymoma tumorigenesis.
In vivo screen of candidate ependymoma TSGs
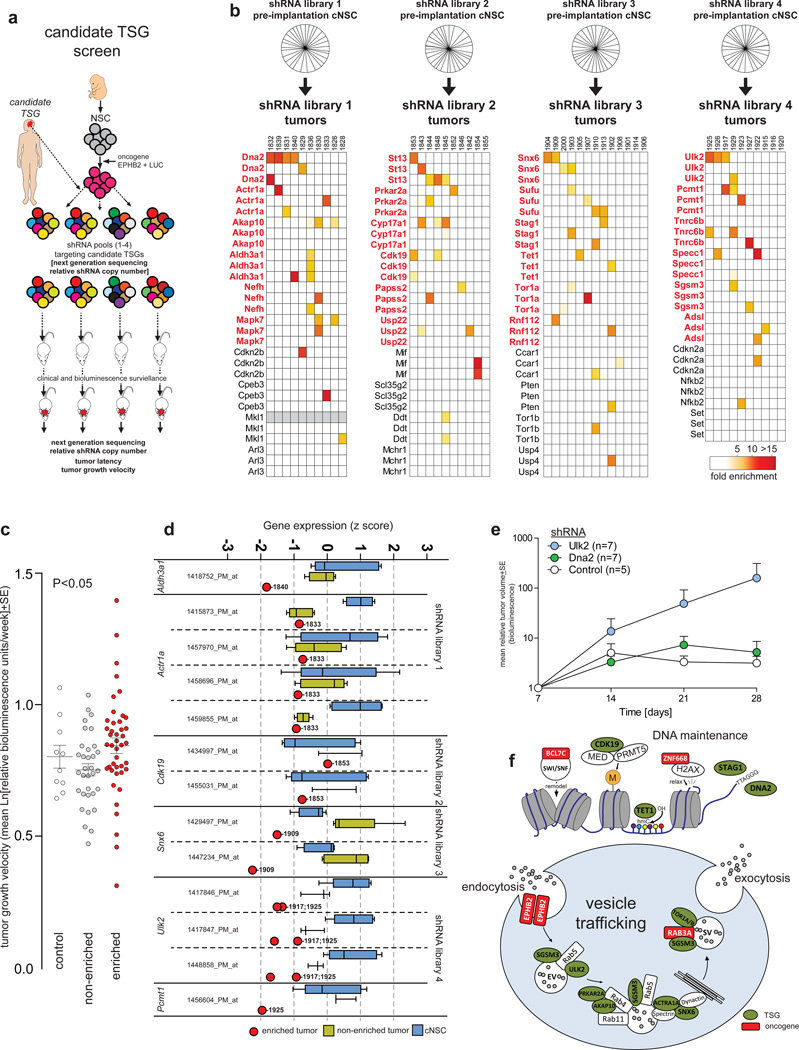
To determine if our 39 TSG candidates possess tumor–suppressor function, we assessed whether shRNAs targeting these genes could cooperate with EPHB2 to transform cNSCsnull (Fig. 5a). We previously validated EPHB2 as an ependymoma oncogene4
Figure 5.
In vivo ependymoma tumor suppressor gene screen. (a) Cartoon depicting the overall TSG screening approach. (b) Top, pie charts reporting the proportion of sequences of each individual shRNA detected by Illumina sequencing of the indicated pre-implantation cell pool. Individual shRNAs are not marked but were observed in equal proportions. Below, heatmaps reporting the fold enrichment of each shRNA in pool-derived tumors (tumor numbers are shown above each column). Genes in red are those with enriched shRNAs. (c) Tumor growth velocity of shRNA control transduced, non-shRNA enriching and shRNA-enriching tumors. Lines mark mean±SE. (d) Affymetrix gene expression profile analysis of target genes of six enriched shRNAs in tumors depicted in (b) (numbers indicate tumor identity in [b]) relative to cNSCs and non-shRNA enriching tumors (line, median; box, 25/75th percentiles; whiskers, outliers). (e) Tumor initiation by EPHB2–cNSCsnull transduced with control, Ulk2 or Dna2 shRNAs. (f) Cartoons depicting the role of validated ependymoma oncogenes (red boxes) and TSGs (green ovals) within DNA maintenance and vesicle trafficking pathways.
First, we generated 117 shRNA-IRES-cyan fluorescence protein (CFP) lentiviruses targeting our 39 candidate TSGs (three shRNA per candidate; Supplementary Table 2). Lentivirus triplets targeting a single candidate were then mixed together randomly to create four libraries: libraries 1, 2 and 3 each contained 30 lentiviruses targeting 10 candidate TSGs; library 4 included 27 lentiviruses targeting nine candidate TSGs. Each lentivirus library was then used to transduce separate fractions of EPHB2-cNSCsnull at a multiplicity of infection (MOI) of 1 or 5. Illumina sequencing confirmed equal representation of shRNA sequences within transduced cNSCsnull (Fig. 5b). 1.5×106 eGFP+/YFP+/RFP+/CFP+ EPHB2-cNSCsnull were then sorted from each of the eight fractions and implanted into the cerebrum of separate immunocompromised mice (average of 11 mice per fraction [range 9–13], total=84 mice; Supplementary Table 8). A separate cohort of 10 mice was implanted with eGFP+/YFP+/RFP+/CFP+ EPHB2-cNSCsnull transduced with scrambled control shRNA. Mice were imaged weekly using bioluminescence.
As expected all mice developed tumors4. To detect TSGs among our 39 candidates, we first looked in tumors for shRNA sequences that were enriched relative to the corresponding, pre-implantation, parent EPHB2-cNSCnull fraction (average of 3.04x105 shRNA illumina sequence reads per tumor; Fig. 5b; Supplementary Table 8). TSG candidates were judged to have been ‘targeted’ if at least one shRNA was enriched ≥4 fold in ≥2 tumors (Fig. 5b): 62% (n=24/39) of candidates achieved this targeting threshold (hereon ‘targeted TSG candidates’; Fig. 5b). Targeted TSG candidates did not differ significantly between tumors derived from MOI 1 or MOI 5 transduced EPHB2-cNSCsnull (data not shown). One quarter (n=6/24) of all targeted TSG candidates are established or putative TSGs (Dna216, Sufu, Stag117, Tet118, Aldh3a119 and St13) suggesting that the other 18 targeted candidates might also be TSGs. In keeping with this notion, tumors that enriched shRNAs of targeted candidates grew significantly faster than non-enriching tumors (Fig. 5c; P<0.05, Mann-Whitney). As expected, shRNAs targeting Cdkn2a and Cdkn2b that are known ependymoma TSGs but already deleted from cNSCsnull were not recurrently enriched.
To further assess targeted TSG candidates, we compared their expression in six tumors that enriched the corresponding shRNAs with their expression in non-enriching tumors, and the original implanted cNSCnull pool (Fig. 5b,d). Five targeted TSG candidates were expressed in tumors at levels ≥2SD below that observed in parent cNSCsnull (majority of probes reporting expression of: Aldh3a1, Actr1a, Snx6, Ulk2, Pcmt1; Fig. 5d), while four candidates were expressed at levels ≥2SD below that observed in control tumors (majority of probes reporting expression of: Aldh3a1, Snx6, Ulk2, Pcmt1; Fig. 5d). Interestingly, targeted TSG candidates were 1.2-fold more likely to be deleted across human cancers than non-validated TSGs candidates, although this difference did not reach significance (Supplementary Fig. 2).
To further validate our TSG screen, we repeated our in vivo tumorigenesis assays but this time compared tumor initiation by EPHB2-cNSCsnull cells transduced with control shRNAs, Ulk2-shRNAs or Dna2-shRNA alone. Knock-down of Ulk2 dramatically accelerated the initiation of brain tumors relative to control cells (Fig. 5e). Dna2-shRNA transduced EPHB2-cNSCsnull displayed a marginal increase in tumor growth relative to control cells. Together, these data strongly support ALDH3A1, ACTR1A, SNX6, ULK2, and PCMT1 as new ependymoma TSGs. Since DNA2, SUFU, STAG1, TET1, and ST13 have established TSG function, we also propose that these are also ependymoma TSGs.
Ependymoma oncogenes and TSGs disrupt common cell functions
Remarkably, 46% (n=11/24) of targeted candidates identified in our TSG screen regulate vesicle trafficking (ULK220, SGSM321, SNX622, ACTR1A23, AKAP1024, PRKRA2A25, TOR1A25) or DNA modification and repair (STAG126, CDK1927, DNA216, TET118) that were also disrupted in our oncogene screens (Fig. 5f; Supplementary Table 9). The convergence of our independent oncogene and TSG screens on these common cell functions suggest they are critical for ependymoma tumorigenesis.
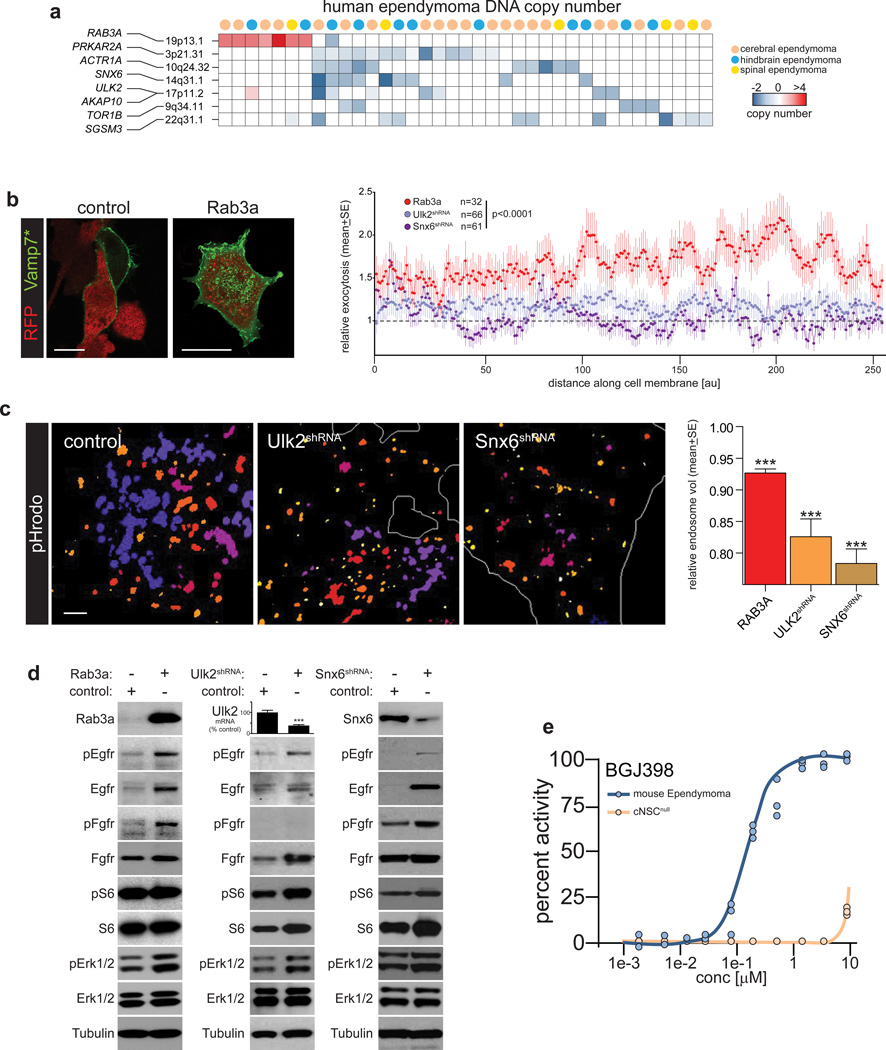
Since gain of RAB3A (19p13.1) that promotes exocytosis12, 13 and deletion of multiple targeted TSG candidates that regulate endocytosis (e.g., ULK2 and SNX6) were mutually exclusive in human ependymoma, we reasoned that these CNAs might cooperate to disrupt vesicle trafficking (Figs. 5, 6a). Indeed, both excessive exocytosis and defective endocytosis have been implicated in malignant transformation by mis-trafficking growth factor receptors to the cell membrane28, 29. Therefore, we tested if upregulation of Rab3a or downregulation of Ulk2 or Snx6 similarly disrupts vesicle trafficking and Epidermal Growth Factor Receptor (Egfr) and Fibroblast Growth Factor Receptor (Fgfr) signaling in cNSCs, since these receptors have been implicated in ependymoma6, 30, 31. Significantly more exocytosis – measured using a pH-sensitive exosome-associated fluorescent protein – was detected in Rab3a overexpressing cells than controls, but exocytosis was unaffected by either Ulk2 or Snx6 knockdown (Mann-Whitney p<0.0001; Fig. 6b). Conversely, mean endosome volume – measured using an endosome-specific fluorescent dye – was significantly decreased in Snx6–shRNA or Ulk2–shRNA transduced cells relative to controls (p<0.0005, Mann-Whitney; Fig. 6c). Interestingly, overexpression of Rab3a also reduced endosomal volume, albeit to a lesser degree. In concert with this disruption in vesicle trafficking, upregulation of Rab3a or depletion of either Ulk2 or Snx6, prolonged both Egfr and Fgfr signaling via the Mek/Erk pathway in cNSCs (Fig. 6d). Furthermore, mouse cerebral ependymoma cells proved selectively sensitive to growth inhibition by the pan-Fgfr inhibitor BGJ398 relative to cNSCsnull (Fig. 6e). Thus, we propose that ependymoma CNAs cooperate to dysregulate the trafficking of oncogenic growth factor receptors.
Figure 6.
Ependymoma oncogenes and tumor–suppressor genes trafficking of growth factor receptors. (a) Heatmap reporting CNAs in human ependymomas affecting the validated ependymoma oncogenes and TSGs involved in vesicle trafficking. (b) Left, direct fluorescence photomicrographs of cells transduced with control or RAB3A plasmids (both RFP) and VAMP7* (green). Note the increased occupation of the cell membrane by VAMP7* protein in Rab3a transduced cells (scale bars=10 µm). Right: graph reporting relative exocytosis recorded as VAMP7* membrane fluorescence in cells transduced with Rab3a or shRNAS targeting Ulk2 or Snx6. (c) Left, direct fluorescence photomicrographs of endosomes in cells transduced with scrambled control, Ulk2 or Snx6 shRNAs. Color denotes the z–stack plane in which endosomes are located. Note the decreased size and number of endosomes in Ulk2 and Snx6-targeted cells quantified in the graph, right (endosomes scored: Rab3a=15,950; Ulk2=5,575; Snx6=7,297). Scale bar=1µm (d) Western blots of cNSCsnull transduced with control, Rab3a, or the indicated shRNA followed by 10nm EGF and FGF stimulation for 120 mins. The graph reports expression of Ulk2 mRNA for which reliable antibodies were not available (***=p<0.0005, Mann-Whitney). (e) Dose response-growth inhibition curve measured as percent drug activity of the pan-FGFR inhibitor BGJ398 against mouse RTBDN-driven cerebral ependymoma and parental cNSCsnull in vitro.
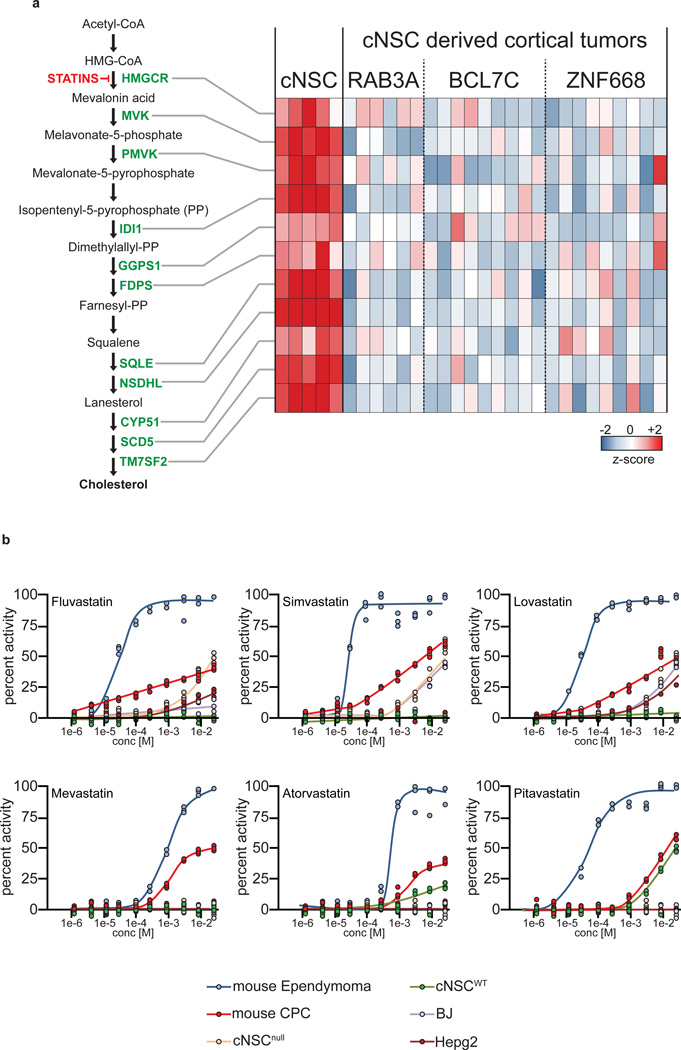
As well as disrupting vesicle trafficking, our validated, CNA-associated oncogenes generated ependymomas with diminished cholesterol biosynthesis machinery (Fig. 4c, 7a). Whether this is a cause or consequence of cNSC transformation is unclear, but statin-mediated inhibition of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) – a critical enzyme in cholesterol biosynthesis – has been reported to prevent32 and treat33 brain tumors. Therefore, we reasoned that the downregulation of cholesterol biosynthesis in ependymoma cells might render them susceptible to further pathway suppression. In keeping with this notion, six different statins markedly and selectively impaired the proliferation of mouse cerebral ependymoma cells relative to parental cNSCsnull and a variety of other brain tumor and control cell lines (Fig. 7b).
Figure 7.
Suppression of cholesterol biosynthesis in cerebral ependymomas. (a) Heatmap (right) reporting the mRNA expression of critical cholesterol biosynthesis enzymes (left) in parental cNSCsnull and daughter Rab3a, Bcl7c and Znf668 driven ependymomas. (b) Dose response-growth curves of activity of six different statins against the indicated cells in vitro. (BJ=normal human foreskin; Hepg2=hepatocellular carcinoma).
DISCUSSION
We report the identification of eight new ependymoma oncogenes and 10 new ependymoma TSGs from among 123 genes located within 28 recurrent CNAs. These genes emerged from screens of oncogenes and TSGs that converged on just a handful of cell functions; providing new animal models of ependymoma and insights into disease biology and treatment.
Aberrant vesicle trafficking is thought to transform cells by disrupting the recycling of growth factor receptors, prolonging cell signaling, and abolishing cell polarity28, 29. For example, disruption of Rab25 that regulates various aspects of vesicle trafficking promotes intestinal tumorigenesis in mice34 and the endosome regulators Rbsn, Vps45, Avl, and Rab5 serve as TSGs in Drosophila35. Dysregulation of vesicle trafficking has also been observed in human cancer; although most studies report altered expression rather than mutation of vesicle trafficking genes, making it difficult to determine if it is a cause or consequence of human malignancy28, 29. Our data provide evidence that ependymoma CNAs amplify pro-exocytotic oncogenes and delete pro-endocytotic TSGs, driving mis-trafficking of growth factor receptors in cNSCs. In this regard, we validated RAB3A as a new brain tumor oncogene. Although RAB3A is a known regulator of late stage vesicle trafficking in neuronal cells12, 13, subsequent modulation of growth factor receptor signaling has not been reported. We also identified seven CNA-associated TSGs that populate key points in the receptor endocytosis pathway: ULK2 (deleted from 17p11.2) positively regulates receptor endocytosis in neural cells20 while SGSM3 (22q13.1) binds RAB proteins in endosomes that orchestrate endocytosis21. Receptors within endosomes are then recycled to the trans-Golgi network by the retromer – a heteropentameric protein complex that includes SNX6 (14q13.1). SNX6 organizes the retromer within the endosomal network by mediating direct interactions with the dynactin subunit p150glued22. p150glued binds a further dynactin subunit, ACTR1A23 (10q24.32). Recycling of receptor endosomes is further regulated by AKAP10 (17p11.2) that binds RAB4/11 labeled vesicles24, PRKRA2A (3p21.31) and TOR1A/B (9q34.11)25. Our observation that gain of RAB3A, or deletion of SNX6 or ULK2, prolongs EGFR and FGFR signaling in cNSCs, supports the notion that CNAs promote the trafficking of oncogenic receptors to the surface of ependymoma cells.
We also identified new ependymoma oncogenes (BCL7C and ZNF668) and TSGs (TET1, CDK19, STAG1 and DNA2) that regulate chromatin modification and remodeling, or telomere maintenance. These data are particularly interesting since aggressive forms of ependymoma possess aberrant chromatin states7 and dysfunctional telomeres36: BCL7C is a close relative of the lymphoma translocation partner BCL7A37 and a subunit of the nucleosome remodeling SWI/SNF complex11; ZNF668 links chromatin relaxation state with the DNA damage response10; TET1 is a known TSG that converts 5-methylcytosine into 5-hydroxymethylcytosine18; and CDK19 represses C/EP2β target gene transcription27. With regard to telomere function, STAG1 encodes a component of the cohesion complex that is essential for telomere cohesion26, while DNA2 recognizes and cleaves telomeric G4, protecting cells from chromosome segregation errors16.
One surprising result of our study was the observation that ependymomas driven by each validated-oncogene downregulate cholesterol biosynthesis, rendering them highly sensitive to further pathway suppression. This phenomenon might also involve mis-trafficking of oncogenic receptors. Endocytosed receptors face two, alternative fates: recycling back to the cell membrane (clatherin-mediated endocytosis, CME), or degradation (non-clatherin mediated endocytosis, NCE). NCE is acutely dependent upon cholesterol and disruption of cholesterol supplies pushes EGFR toward CME, recycling receptors to the cell membrane and prolonging EGFR signaling38. Thus, downregulation of cholesterol synthesis in transforming cNSCs might promote oncogenic EGFR signaling but render these cells sensitive to further pathway inhibition.
As well as providing important new insights into brain oncogenesis and treatment, the results of our screens also pose some important questions: why did some of the most enriched candidate oncogenes emerging from our cNSC pool screen (e.g., ARRDC2 and RNASE2H) fail to drive tumors when transduced alone into cNSCs, and why did pooled tumors, but not those driven by single oncogenes, include PNET-like tumors? Further work will be required to answer these questions, but one possibility is that certain oncogenes only drive transformation in the context of clonal heterogeneity, perhaps requiring paracrine factors from surrounding, but unrelated tumor cells. Indeed, pool-derived tumors that enriched ARRDC2 and RNASE2H also enriched the validated oncogenes ZNF668 and PRDX2, respectively. Clonal heterogeneity might also explain the variety of histologies that we observed among pooled tumors. But it is also important to consider if ex vivo manipulation and implantation of NSCs that is used to generate our models also impacts tumor morphology. Finally, an important caveat of our oncogene screening approach was the use of a single TSG background (Ink4a/Arf null). CDKN2A/B is deleted in ~50% of human cerebral ependymomas4, but is rarely deleted from hindbrain or spinal tumors. Thus, pairing oncogene candidates with alternative TSGs might validate additional oncogenes.
In summary, we describe a novel and comprehensive cross-species approach to identify oncogenes and TSGs within recurrent tumor CNAs. The approach described here could be applied to other cancers, enabling the identification of critical oncogenic drivers in tumors that lack recurrent alterations of single genes.
ONLINE METHODS
Human Genomic Analysis
Our original set of 85 candidate oncogenes and 39 candidate TSGs were identified and reported previously by integrating the results of Affymetrix GeneChip Human Mapping 500K and U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA) of human ependymomas4. 98% (n=83/85) of candidate oncogenes screened in the current study were selected from an original list of 85 candidates reported in reference 4, Supplementary Table 5a column ‘subgroup specific expression’ that included genes that displayed copy number-driven expression within a specific genomic subgroup of ependymoma, rather than generally across the disease. Note that LOC90835 and POL3S have since been renamed C16orf93 and PRSS53, respectively. The cDNA for BTBD14B reported as a candidate oncogene in Ref. 4 was not commercially available when we initiated our study and LOC730744 has since been withdrawn from the RefSeq database. These two genes were therefore replaced by EXO1 that was highly amplified in cerebral ependymoma (confirmed by fluorescence in situ hybridization, data not shown) and FAM53A that is present on the 4p16.3 ependymoma amplicon.
The 39 candidate TSGs were all drawn from 130 candidates identified as deleted in Ref. 4 Supplementary Table 5b. These 130 TSG candidates we triaged for screening based on evidence of subtype-specific copy number-driven expression (as for oncogenes), but also on evidence of candidate involvement at least two or more of the following processes: neural development, cancer (general), or brain tumors (see Supplementary Table 2 for more details). 77% (n=30/39) candidate TSGs selected for screening displayed both copy number-driven decreased expression in ependymoma and evidence of involvement in two or more of the processes defined above. Four additional candidate TSGs we selected for screening since they were located within the one of the most recurrent, minimal regions of deletion in ependymoma (chromosome 22q13.1) and five candidates were screened because of significant evidence implicating them in neural development, cancer (general), or brain tumors (see Supplementary Table 2 for details).
Two methods were used to assess the relative gain or loss of candidate oncogenes and TSGs in human cancers. We used the cBioPortal (see URLs) web resource to obtain allelic gain or loss data from 15,752 samples of 17 different types of human cancer that are approved for use from the TCGA with cBioPortal. For each tumor type the ratio of gain to loss of candidates was recorded. We used the COSMIC database (see URLs) to calculate the net gain or loss of each candidate oncogene and TSG in 3,116 samples of nine different human cancers. In both analyses the Mann-Whitney test was used to assess the significance of the difference in relative gain or loss of candidate oncogenes and TSGs.
Generation of candidate oncogene retroviruses
Retroviral plasmids containing the open reading frames of each candidate oncogene were generated by sub-cloning into the pCX4-IRES-Red vector. cDNA clones for each candidate were obtained from a variety of sources including the American Type Culture Collection, Invitrogen, OpenBiosystems, Addgene, GeneCopoeia or Origene or were generated in house by reverse transcription and polymerase chain reaction. The sequence of all cloned inserts was verified by Sanger sequencing. Retroviral plasmids were then transfected together with pMD-old-gag-pol and CAG4-Eco plasmids into 293T cells. After 20 hours the medium was exchanged for serum-free neurobasal medium (Invitrogen) containing 2 mM L-glutamine, N2 supplement (Invitrogen), B27 supplement (Invitrogen), 20 ng/ml hrEGF (Invitrogen), 20 ng/ml hrbFGF (Invitrogen ) and 50 µg/ml BSA. Every subsequent 8–10 hours virus containing medium was harvested for up to 3 days. Virus containing medium was centrifuged, filtered (0.45 µm filter, Millipore) and concentrated using Centricon plus 70-Millipore. Viral titers were calculated by assessing transduction efficiency at different dilutions in NIH3T3 cells.
Preparation of candidate oncogene transduced NSCs pools
All mouse studies were performed according to protocols approved by the St. Jude Children’s Research Hospital Animal Care and Usage Committee. Enhanced Green Fluorescence Protein expressing (eGFP+) Neural stem cells were isolated from the embryonic (E14.5) cerebrum (cNSCsnull) and hindbrain (hNSCsnull) and adult spine (sNSCsnull) of Blbp-eGFP Ink4a/Arf−/− mice exactly as described previously4. Briefly, forebrain, hindbrain and spine macrodisected from embryonic and adult mice, were minced and digested separately with collagenase type IV and hyaluronidase and isolated cells cultured in Neurobasal medium4. We have shown previously that the cells cultured in this manner are enriched (~100%) for self-renewing eGFP+ NSCs (demonstrated previously using serial potency assays in Ref. 4). NSCsnull were then transduced with a Luciferase-IRES-Yellow Fluorescence Protein (YFP) retrovirus to allow bioluminescence imaging of cells in vivo. eGFP+/YFP+ cNSCsnull were then fluorescence activated cell sorted and separated into 84 fractions. Although cell sorting does not completely resolve eGFP+/YFP+ and eGFP+/YFP− cNSCsnull, our lenitviral transductions are highly efficient, leading to consistent transduction of >80% of eGFP+ cells (ref. 4). Each fraction was transduced with one of 84 retroviruses; each encoding a different candidate oncogene upstream of an IRES-red fluorescence protein (RFP) reporter. eGFP+/YFP+/RFP+ cNSCsnull were then sorted from each fraction and randomly mixed together in equal proportions, creating 11 cell pools, each comprising mixtures of cells that each carried one of up to eight different candidate oncogenes (Fig. 2a,b). Thus, cell pools comprised mixtures of NSCs that each carried a single candidate oncogene rather than cNSCs carrying multiple candidates (see Fig. 2a). This pool design served as the basis of our in vivo tumorigenesis screen that was predicated on the on the notion that cNSCs within pools that carried oncogenic candidates would outgrow those cNSCs in pools that were transduced with non-oncogenic candidates.
Generation of candidate tumor suppressor gene (TSG) shRNA lentiviral pools
Three 19mer shRNAs targeting each candidate TSG were designed using the shRNA sequence prediction algorithm from Dharmacon/thermo Scientific (see URLs). shRNAs were cloned into the pFUGWH1-CFP vector and transformed into bacteria as one ligation product. The transformed bacteria were then spread on an LB/AMP plate and individual colonies were screened for unique shRNA constructs by sequencing. All 117 constructs were sequence verified. shRNAs were then pooled as described in the main text as concentration normalized DNA mixtures. This equimolar DNA mix was then used to transform bacteria and the mixed population bacteria culture was expanded, harvested and subjected to endotoxin-free DNA maxiprep. Prior to DNA maxiprep, a sample of this bacteria was streaked onto an LB/AMP plate and individual colonies screened for unique shRNA constructs to verify that pool complexity was maintained. The DNA mix was fugene (Roche) transfected into 293FT cells (Invitrogen) along with lentiviral packaging plasmids for viral production. Resulting virus titre was determined by cyan fluorescence protein (CFP) expression in transduced E14.5 cNSCnull. Finally, ependymoma prone cNSCsnull cells transduced with EPHB2 and luciferase were mass transduced with pools of shRNA lentivirus at a multiplicity of infection of 1 or 5. Transduced cNSCsnull were then expanded through one passage. A sample of cells was then taken for deep sequencing (see below) and 1.5×106 of the remaining transduced cells suspended in 5 µl of matrigel were implanted into the cerebrum of CD1 nude mice exactly as described below. Tumor growth was monitored by bioluminescence xenogen imaging.
Orthotopic cerebral, cerebellar and spinal NSC implants
Appropriately transduced NSCs were resuspended in matrigel at a concentration of 1.5×106 cells/5µL and injected using a Hamilton Syringe into the cerebral cortex or cerebellum of CD-1 nu/nu immunocompromised mice with the support of a small animal stereotactic frame as described4. Mice were monitored daily for neurological symptoms and imaged biweekly using bioluminescence (see below). Symptomatic mice were euthanized, the brain removed and subject to direct fluorescence microscopy to detect the presence of GFP+/RFP+ tumors. Tumors were macro-dissected and subject to three separate preparations: (i) Portions were flash frozen tissue in liquid nitrogen for RNA and DNA analysis. (ii) Adjacent portions were formalin fixed and embedded in paraffin wax. Wax-embedded tissues were sectioned at 5µm thicknesses and mounted on SuperfrostPlus microscope slides (Fisher Scientific). Hematoxylin/eosin (H&E; Fisher Scientific) stains of de-waxed and rehydrated sections were performed to a standard protocol. Standard immunohistochemical methods were used to detect proteins in tissue sections. (iii) Suspensions of live cells were prepared from remaining tumor fragments and cultured in Neurobasal medium.
For spinal implants, mice were anesthetized and treated with metacam (1–2mg/kg) immediately prior to surgery to reduce tissue inflammation. The skin over the thoracic spine was cleansed with iodine, covered with a sterile drape and a dorsal vertical incision of ~1.5 cm made in the skin over vertebrae T8 toT12. The spinal column at T9 was firmly secured with the forceps and a laminectomy performed. 1.5×106 transduced sNSCsnull suspended in 2.5 µL matrigel were injected at a 70° angle using a Hamilton syringe into the gray matter proximal to the central canal. Immediately after injection, sutures were applied to the superficial fascia on both sides of the incision and closed with 3–6 staples. Post-operative analgesia was given as Buprenorphine/Buprenex 0.05–0.1 mg/kg every 6 to 12 hours and metacam 1–2 mg/kg dosage for 4–5 days.
Xenogen imaging and tumor growth velocity calculations
Bioluminescence imaging was performed using a Xenogen IVIS-200 imaging system (Caliper Life Sciences, Hopkinton, MA). For each imaging session a total of 150 mg/kg of Luciferin was injected into the peritoneal cavity of mice. Animals were anesthetized using 2–3% isoflurane and imaged 5 minutes after Luciferin injection to ensure consistent photon flux. Post imaging, animals were placed under observation on a heating blanket. Imaging data were analyzed using Living Image software 4.0 (Caliper biosciences). Linear mixed effects models with random intercepts and slopes were used to estimate the tumor growth velocity based on natural log transformed bioluminescence data using the lme package from TIBCO Spotfire S+® 8.2 for Windows.
DNA and RNA isolation
Tumor samples or pre-injection NSCs were snap frozen and stored at −80° until analysis. Samples were lysed with buffer containing Proteinase K (20 mg/ml), homogenized on dry ice and incubated at 37°C overnight. DNA was extracted using Manual Phase Lock Gel™ (PLG) - 5 PRIME and phenol chloroform iso-amyl alcohol, and precipitated by adding cold isopropanol and centrifugation. The DNA pellet was washed in 70% ethanol and suspended in DNA/RNA free water. Total RNA was isolated from tumors by addition of RNA-STAT-60 (Tel Tech, Inc.) and homogenized on dry ice followed by 75% ethanol wash, drying and resuspended in DEPC treated water. The Aurium Total RNA Mini Kit (Bio-RAD) was used to isolate RNA from NSC cells.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed using a MyiQ™ Single-Color Real-Time PCR Detection System (Bio-Rad) and 20µl reaction mixtures that included iQ™ SYBR® green supermix (Bio-Rad), template DNA or cDNA, and specific primers for the corresponding template. Relative gene quantification was calculated according to the comparative Ct method: 2−(ΔΔCt) ΔΔCt = ΔCt tumor - ΔCt preinjection pool with ΔCt corrected for GAPDH level..
Identification of shRNA species in tumors
Genomic DNA was extracted from each tumor as described above and then amplified using one of six pairs of primers recognizing unique sequences engineered to flank the lentiviral vector insert. Amplification primers were designed in a staggered pattern on either side of the cloning sites to permit identification of unique tumor-derived PCR products after pooling. Sequencing libraries were generated from pooled equimolar amounts of PCR products using the Illumina TruSeq DNA Sample Prep Kit v2 protocol. Each library was indexed using one of 12 available sequence barcodes. Twelve libraries containing PCR products from up to 72 unique tumors were sequenced in a single lane of an Illumina GAIIx flow cell using paired-end 100 bp sequencing chemistry or in a single MiSeq flow cell using paired-end 150 bp sequencing chemistry. An average of 436,000 reads per tumor were generated on the GAIIx flow cell, while an average of 135,000 reads per tumor were generated on the MiSeq flow cell. Sequences derived from each tumor were identified by first demultiplexing sequence reads according to library index and then by demultiplexing according to unique vector sequences adjacent to the staggered PCR primers. After quality trimming, the reads containing tumor-derived shRNA sequences were aligned against the shRNA sequences originally constructed in the pFUGWH1 expression vector using CLC Genomic Workbench software v5.1 (CLC Bio, Denmark). shRNA sequence reads were then counted and binned according to the sequences represented within each tumor pool.
Gene expression microarray and agreement of differential expression (AGDEX) and gene set enrichment analyses
RNA was isolated from mouse tumor samples and cells as described above. Gene expression profiles were generated using and HT MG-430PM microarrays (Affymetrix, Santa Clara, USA). Gene expression data were summarized and normalized using the RMA method as implemented in Partek Genomics Suite 6.6 (St. Louis MO, USA). The data was then imported into Spotfire Decision Site (Palo Alto CA, USA) and z-scores calculated by computing the mean and standard deviation across subjects within each probeset. Differences in gene expression between defined groups were calculated using a series of Welch t-tests with a Bonferroni threshold at 0.05 percent. The AGDEX procedure was performed exactly as described previously4 and gene set enrichment analyses performed using the online analytical resource from the Broad Institute (see URLs).
Drug treatment studies
Suspensions of the indicated single cells were seeded in 30 µl of Neurobasal medium in each well of 384-well plates (Corning) using an automated plate filler (Wellmate, Matrix). The RTBDN-driven model of ependymoma was used for these studies since this tumor mode most closely recapitulated the morphology of ependymoma in the cerebrum (Fig. 4b). After 24 hours, 60 nL of solution containing appropriate compounds were pin transferred into the 384-well plates to provide the desired and indicated final drug concentration. Each plate also included DMSO only negative controls and cycloheximide single point (0.5µM) and dose-response (0.5 µM-0.01 nM) positive controls. Cell number was determined in each well using the Cell Titer Glo reagent (Promega) and read in an automated Envision plate reader (Perkin-Elmer) after 72 hours incubation. Luminescence data were normalized by log10 transformation and the percentage inhibition = 100 × (sample result – negative control mean)/(positive control mean – negative control mean) calculated. All studies were repeated in triplicate. All data processing and visualization was performed using custom programs written in the Pipeline Pilot platform (Accelrys, v.7.0.1) and the R program.
Western blotting
5×106 NSCs transduced with the indicated retrovirus or lentivirus were washed and seeded in non-growth factor containing neurobasal medium in 10 cm dishes. Sixteen hours later, human EGF and human FGF-2 (Miltenti Biotech) was added to each dish and cells were harvested and washed in cold PBS after the indicated incubation time. For analyses of cell signaling, NSCs were then pelleted and then lysed in m-PER mammalian protein extraction reagent plus Halt protease and phosphatase inhibitors (Thermo Scientific) on ice for 30 minutes. Following centrifugation for 30 minutes, supernatant protein was quantitated with the Qubit protein assay kit and the Qubit Fluorometer 2.0(Invitrogen). Protein lysates were electrophoresed on Novex Nu Page 4–12% BisTris Gel in MOPS SDS running buffer. Transfer was performed with the IBlot® gel transfer device (Invitrogen). The blot was then processed using the SnapId 2.0 system (EMD Millipore Corporation) with blocking, primary and secondary antibody incubation and the Novex® ECL Chemiluminescent substrate for detection (Invitrogen). Primary antibodies included: FGFR (D8E4) XP®Rabbit mAb, Phospho-FGF Receptor (Tyr653/654), total EGFR, Phospho-EGFR (Tyr1173), Phospho-S6 Ribosomal Protein (Ser235/236), total S6, Phospho-Erk1/2 (Thr202/Tyr204), total Erk1/2, all from Cell Signaling Technologies; RAB3A Antibody ab3335 (Abcam inc); SNX6-76 (Sigma-Aldrich); and Tubulin (B-5-1-2, Santa Cruz SC23948).
Endocytosis and exocytosis assays
Cells transduced with the indicated test or control retrovirus or lentivirus were plated at the density of 100,000 cells per well on the Labtek-Tek II chambered #1.5 German cover glass system in medium without antibiotics. After 24 hours cells were placed on ice for 10 minutes and then washed with chilled live cell imaging solution (LCIS) containing 20 mM Glucose and 1% BSA. Cells were then incubated with fluorescein transferrin conjugate (cat number :T2871) or pHrodo® Red transferrin conjugate. Cells were then washed with LCIS and analyzed. Cells incubated with the pHrodo® Red transferrin conjugate (cat number: P3576) were imaged using Nikon C1Si confocal laser fluorescence microscope with a 60× 1.45 NAO TIRF objective; image processing was performed using Fiji software. Cells incubated with the fluorescein transferrin conjugate were imaged using a Nikon C2 confocal microscope with a plan fluor 40×1.3 NAO objective and processing of images was performed with NIS Elements software.
Exocytosis assays were performed using the Neon transfection system at a cell density at 100,000 per well and 0.5µg VAMP7-phlorin construct (generous gift from Frank B. Gertler, MIT and Stephanie Gupton University of North Carolina). The 10µl Neon tip was used with resuspension buffer R and optimized parameters that included: pulse voltage 1200, pulse width 20 and pulse number 2. Cells were plated on the Labtek-Tek II chambered #1.5 German cover glass system in medium without antibiotics. After 24 hrs, images were collected with a Zeiss LSM 780 NLO confocal microscope using a LDC- Apochromat 40xNA 1.1W objective or with a Nikon C2 confocal microscope using a plan fluor 40× NA 1.3 oil objective and all images were analyzed and quantified using Fiji software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (R01CA129541, P01CA96832 and P30CA021765, R.J.G), the Collaborative Ependymoma Research Network (CERN), and by the American Lebanese Syrian Associated Charities (ALSAC). We are grateful to Frank B. Gertler, MIT, and Stephanie Gupton University of North Carolina for the generous gift of the VAMP7-phlorin construct and the staff of the Hartwell Center for Bioinformatics and Biotechnology, Small Animal Imaging Center, Animal Resources Center, Cell and Tissue Imaging Center and Flow Cytometry & Cell Sorting Shared Resource at St Jude Children’s Research Hospital for technical assistance.
Footnotes
URLS
Catalog of Somatic Mutations in Cancer (COSMIC) (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). Dharmacon siRNA design center (http://www.dharmacon.com/designcenter/DesignCenterPage.aspx). Gene set enrichment algorithm (http://www.broadinstitute.org/gsea/index.jsp). cBioportal algorithm (http://cbioportal.org)
ACCESSION CODES
All previously unpublished microarray data presented in this study are available through the Gene Expression Omnibus: GSE67497.
AUTHOR CONTRIBUTIONS
K.M.M. and D.S.C. conducted the great majority of the experiments and contributed to their design. E.W., N.B., J.D., C.E., B.N., R.T., M.C., and T.A.K. conducted mouse experimental work. G.N., S.O., Y-D.W., and D.F. conducted microarray and deep sequencing studies. K.W. assisted with human genomic analyses. K.G. and D.W.E. conducted all neuropathological reviews. A.O.T. conducted all statistical analyses. R.J.G. conceived, designed and oversaw the study. All authors contributed to the writing.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 2.Nobori T, et al. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 3.Zender L, et al. An Oncogenomics-Based In Vivo RNAi Screen Identifies Tumor Suppressors in Liver Cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson RA, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker M, et al. C11orf95-RELA fusions drive oncogenic NF-kB signaling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson Jennifer M, et al. An Integrated In Vitro and In Vivo High-Throughput Screen Identifies Treatment Leads for Ependymoma. Cancer Cell. 2011;20:384–399. doi: 10.1016/j.ccr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506 doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. 2013:pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chautard E, Ouédraogo Z, Biau J, Verrelle P. Role of Akt in human malignant glioma: from oncogenesis to tumor aggressiveness. Journal of Neuro-Oncology. 2014;117:205–215. doi: 10.1007/s11060-014-1382-9. [DOI] [PubMed] [Google Scholar]
- 10.Hu R, Wang E, Peng G, Dai H, Lin S-Y. Zinc finger protein 668 interacts with Tip60 to promote H2AX acetylation after DNA damage. Cell Cycle. 2013;12:2033–2041. doi: 10.4161/cc.25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller M, Pym Edward CG, Tong A, Davis Graeme W. Rab3-GAP Controls the Progression of Synaptic Homeostasis at a Late Stage of Vesicle Release. Neuron. 2011;69:749–762. doi: 10.1016/j.neuron.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian M, Xu CS, Montpetit R, Kramer RH. Rab3A Mediates Vesicle Delivery at Photoreceptor Ribbon Synapses. The Journal of Neuroscience. 2012;32:6931–6936. doi: 10.1523/JNEUROSCI.0265-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, et al. Peroxiredoxin 2 is upregulated in colorectal cancer and contributes to colorectal cancer cells’ survival by protecting cells from oxidative stress. Molecular and Cellular Biochemistry. 2014;387:261–270. doi: 10.1007/s11010-013-1891-4. [DOI] [PubMed] [Google Scholar]
- 15.Donson AM, et al. Immune Gene and Cell Enrichment Is Associated with a Good Prognosis in Ependymoma. The Journal of Immunology. 2009;183:7428–7440. doi: 10.4049/jimmunol.0902811. [DOI] [PubMed] [Google Scholar]
- 16.Lin W, et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. The EMBO Journal. 2013;32:1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anazawa Y, Arakawa H, Nakagawa H, Nakamura Y. Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene. 2004;23:7621–7627. doi: 10.1038/sj.onc.1207270. [DOI] [PubMed] [Google Scholar]
- 18.Hsu C-H, et al. TET1 Suppresses Cancer Invasion by Activating the Tissue Inhibitors of Metalloproteinases. Cell Reports. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Poage GM, et al. Identification of an Epigenetic Profile Classifier That Is Associated with Survival in Head and Neck Cancer. Cancer Research. 2012;72:2728–2737. doi: 10.1158/0008-5472.CAN-11-4121-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proceedings of the National Academy of Sciences. 2007;104:5842–5847. doi: 10.1073/pnas.0701402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Sasaki T, Minoshima S, Shimizu N. Identification of three novel proteins (SGSM1, 2, 3) which modulate small G protein (RAP and RAB)-mediated signaling pathway. Genomics. 2007;90:249–260. doi: 10.1016/j.ygeno.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Wassmer T, et al. The Retromer Coat Complex Coordinates Endosomal Sorting and Dynein-Mediated Transport, with Carrier Recognition by the trans-Golgi Network. Developmental Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur ELF. Ordered Recruitment of Dynactin to the Microtubule Plus-End is Required for Efficient Initiation of Retrograde Axonal Transport. The Journal of Neuroscience. 2013;33:13190–13203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggers CT, Schafer JC, Goldenring JR, Taylor SS. D-AKAP2 Interacts with Rab4 and Rab11 through Its RGS Domains and Regulates Transferrin Receptor Recycling. Journal of Biological Chemistry. 2009;284:32869–32880. doi: 10.1074/jbc.M109.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojtal KA, de Vries E, Hoekstra D, van IJzendoorn SCD. Efficient Trafficking of MDR1/P-Glycoprotein to Apical Canalicular Plasma Membranes in HepG2 Cells Requires PKA-RIIα Anchoring and Glucosylceramide. Molecular Biology of the Cell. 2006;17:3638–3650. doi: 10.1091/mbc.E06-03-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. The EMBO Journal. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsui T, et al. Mediator Complex Recruits Epigenetic Regulators via Its Two Cyclin-dependent Kinase Subunits to Repress Transcription of Immune Response Genes. Journal of Biological Chemistry. 2013;288:20955–20965. doi: 10.1074/jbc.M113.486746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldenring JR. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat Rev Cancer. 2013;13:813–820. doi: 10.1038/nrc3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 30.Sie M, et al. Growth-Factor-Driven Rescue to Receptor Tyrosine Kinase (RTK) Inhibitors through Akt and Erk Phosphorylation in Pediatric Low Grade Astrocytoma and Ependymoma. PLoS One. 2015;10:e0122555. doi: 10.1371/journal.pone.0122555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikkema AH, et al. Kinome Profiling in Pediatric Brain Tumors as a New Approach for Target Discovery. Cancer Research. 2009;69:5987–5995. doi: 10.1158/0008-5472.CAN-08-3660. [DOI] [PubMed] [Google Scholar]
- 32.Gaist D, et al. Use of statins and risk of glioma: a nationwide case-control study in Denmark. Br J Cancer. 2013;108:715–720. doi: 10.1038/bjc.2012.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaist D, Hallas J, Friis S, Hansen S, Sørensen HT. Statin use and survival following glioblastoma multiforme. Cancer Epidemiology. 2014;38:722–727. doi: 10.1016/j.canep.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Nam KT, et al. Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. The Journal of Clinical Investigation. 2010;120:840–849. doi: 10.1172/JCI40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrison HA, et al. Regulation of Early Endosomal Entry by the Drosophila Tumor Suppressors Rabenosyn and Vps45. Molecular Biology of the Cell. 2008;19:4167–4176. doi: 10.1091/mbc.E08-07-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabori U, et al. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol. 2006;24:1522–1528. doi: 10.1200/JCO.2005.04.2127. [DOI] [PubMed] [Google Scholar]
- 37.Zani VJ, et al. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood. 1996;87:3124–3134. [PubMed] [Google Scholar]
- 38.Sigismund S, et al. Clathrin-Mediated Internalization Is Essential for Sustained EGFR Signaling but Dispensable for Degradation. Developmental Cell. 2008;15:209–219. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.