Abstract
The impact of RGD integrin binding-peptide concentration and cell phenotype on directing extracellular matrix (ECM) gene expression in vocal fold fibroblasts is little understood. Less is known about cell response to RGD concentration on a biomaterial when fibroblasts are in a scar-like environment compared to a healthy environment. We investigated the effects of varying RGD integrin-binding peptide surface concentration on ECM gene expression of elastin, collagen type 3 alpha 1, decorin, fibronectin, hyaluronan synthase 2, and collagen type 1 alpha 2 in scarred and unscarred immortalized human vocal fold fibroblasts (I-HVFFs). Phenotype and RGD concentration affected ECM gene expression. Phenotype change from healthy to myofibroblast-like resulted in ECM gene up-regulation for all genes tested, except for decorin. Systematically altering RGD concentration affected the expression of elastin and collagen type 3 alpha 1 in a myofibroblast phenotype. Specifically greater up-regulation in gene expression was observed with higher RGD concentrations. This research demonstrates that controlling RGD concentration may influence ECM gene expression levels in fibroblasts. Such knowledge is critical in developing the next generation of bioactive materials that, when implanted into sites of tissue damage and scarring, will direct cells to regenerate healthy tissues with normal ECM ratios and morphologies.
Keywords: RGD, phenotype, fibroblast, extracellular matrix, real-time PCR
1. Introduction
Much work has been done to understand the role of RGD (Arginine-Glycine-Aspartic acid) integrin-binding peptide in cell binding, growth, proliferation, and motility.1–4 This understanding is critical in tissue engineering as investigators seek to incorporate short bioactive sequences, including RGD, into synthetic materials to direct tissue healing and regeneration. With the goal of tissue engineering in mind, the RGD peptide has been incorporated into many different types of materials in order to facilitate the binding to and proliferation of cells on and within normally non-adherent materials.5–7 For example, minimally-adherent hyaluronic acid has been modified with RGD to develop an adherent, bioactive material for correcting vocal fold defects.7 However, simply improving cell adherence does not usually result in the proper full restoration of healthy tissues, vocal folds or otherwise. Healthy tissues are complex heterogeneous structures that require the expression and deposition of ECM components in normal ratios and morphologies in order to maintain their proper function.
Researchers have taken multiple approaches to develop materials that facilitate regeneration. One such method is the use of RGD concentration to modulate the expression of genes critical to tissue regeneration. For example TiO2 nanotube surfaces were modified with varying amounts of RGD and seeded with rat bone marrow stromal cells; this resulted in a dramatic enhancement in the expression of osteogenic genes on nanotube surfaces modified with higher concentrations of RGD versus lower.8 This same trend was seen in a 3D environment when goat bone marrow stromal cells were grown in poly(ethylene glycol) diacrylate hydrogels modified with varying amounts of RGD.9 Again as RGD concentration increased bone-related marker expression also increased.9 Finally, researchers have looked at how cell lines from soft tissue sources behaved when both RGD concentration and integrin type were varied.10 In contrast to the results seen with hard tissue, they found that as the adhesiveness of the surface increased, either due to the increased RGD or the use of a more adherent integrin, a decrease in overall ECM production by the cell lines tested was observed.10 Combined, these results suggest that an understanding of how RGD signal density affects cell behavior is needed for each cell type in order to properly design material cell combinations that promote healthy tissue regeneration.
Although RGD’s impact on gene expression has been studied in relation to bone cell differentiation and impact on overall ECM production of cells from soft-tissue, gaps still exist.8–10 Currently, we have a gross understanding of how RGD concentration impacts overall ECM deposition for some cell types. An improved understanding of how RGD concentration impacts the expression of individual ECM genes is important for designing better biomaterials that facilitate the expression of individual ECM components in healthy ratios. Furthermore, little is understood with regards to how ECM component gene expression is affected by changes in cell phenotype brought on by changes in the environment. This knowledge is especially important to have so one can understand how cells growing on RGD modified materials might behave when implanted into a site of tissue damage and scarring.
We hypothesized that both cell phenotype and RGD concentration would combine to affect ECM gene expression in vocal fold fibroblasts. We evaluated the effects of RGD surface concentration and cell phenotype on ECM expression by growing adherent immortalized human vocal fold fibroblasts (I-HVFFs) in scar-like/myfibroblastic or healthy environments on NHS-ester polyethylene glycol (PEG) thin film coated glass substrates modified with varying concentrations of RGD integrin-binding peptide. I-HVFFs were chosen because they comprise the major cell type in the vocal folds and are responsible for ECM synthesis.11 Both myofibroblastic and healthy I-HVFF phenotypes were investigated due to their roles in tissue healing and scarring.11 PEG was chosen due to its proven anti-fouling capabilities to limit non-RGD specific attachment of I-HVFFs to the substrate surface.12 Treated I-HVFFs were probed utilizing RT-qPCR to assess the impact of varying RGD concentration and cell phenotype on the expression of six ECM genes including elastin (ELN), decorin (DCN), fibronectin (FN), collagen type 1 alpha 2 (COL1A2), collagen type 3 alpha 2 (COL3A1), and hyaluronan synthase 2 (HAS2). These ECM genes were targeted as they are the primary constituents that dictate the biomechanical properties of vocal fold connective tissue.13–15 Both cell pheonotype and RGD density were shown to influence gene-specific ECM expression. This work is the first step in evaluating if this interplay between RGD and vocal fold fibroblast phenotype could be manipulated in a way to potentially drive proper regeneration of overall damaged or diseased vocal fold tissue. Future work will focus on verifying if our in vitro findings hold up in an in vivo model.
2. Materials & Methods
Peptide Synthesis
Both RGD, an integrin-binding, and RGE (Arginine-Glycine-Glutamic acid), a non-integrin-binding, peptides were synthesized using a 0.4mmol scale Knorr-amide resin (Synbiosci Corp.) and standard FMOC (9-fluorenylmethyloxycarbonyl) chemistry. Once synthesized peptides were capped with an acetyl group, and then cleaved from the resin using 95% trifluoroacetic acid (Sigma-Aldrich), 2.5% water, 1.25% triisopropylsilane (Sigma-Aldrich), and 1.25% ethanedithiol (Sigma-Aldrich). Cleaved peptides were precipitated in cold ether and then recovered utilizing centrifugation. The crude peptides were then further purified using an acetonitrile gradient on an AKTA Explorer FPLC (GE Healthcare) equipped with a 22/250 C18 reversed phase column (Grace Davidson). Molecular weight and purity was confirmed by time of flight MALDI mass spectrometry using a 4800 Plus MALDI TOF/TOF Analyzer (Applied Biosystems).
Substrate Modification
NHS-ester PEG thin film coated glass slides, termed Nexterion H, were purchased from Schott. These substrates once cut to 10x10mm were rinsed with 50mM sodium borate buffer pH 7.5. They were then incubated, according to the literature,12 in varying concentrations of peptide resuspended in 50 mM sodium borate buffer pH 7.5 for ~21 hours with 300 rpm shaking on a plate shaker at room temperature. Post incubation substrates were dried with dry nitrogen gas and used immediately or stored at −20°C.
Cell Culture
I-HVFFs, specifically A8s, were developed and provided to us by Dr. Susan Thibeault at the University of Wisconsin, Madison.16 These I-HVFFs were maintained in an unscarred phenotypic state in normal media composed of Dulbecco’s Modified Eagles Medium (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% MEM non-essential amino acids (Sigma-Aldrich), and 200 ug geneticin (G418; Teknova) per mL of media. I-HVFFs were induced into a myofibroblast phenotypic state by utilizing induction media.17 This media contained all the same aforementioned media components except the 10% FBS was replaced with 10 ng/mL of transforming growth factor – beta 1 (TGF-β1; Biosource) as described by Vyas, et al.17 All cells were used between passage 10–12. Successful transition of fibroblasts to myofibroblasts was confirmed using immunohistochemistry.
Peptide Surface Density
NHS-ester PEG thin film substrates, four for each concentration, were incubated in 0.01, 0.1, and 1.0 mM of biotinylated peptide. Once modified, substrates were blocked with 1% BSA + PBS pH 7.2 solution for 1 hr with shaking at room temperature. All substrates were then probed with a 1:2000 streptavidin-HRP (Life Technologies) solution for 20 min with shaking at room temperature. The substrates were then rinsed three times with 1 mL of 0.05% Tween 20 + PBS pH 7.2 and washed overnight in the same solution. Post washing a development solution (1 to 1 mixture of Color Reagent A Stabilized Peroxide Solution & Color Reagent B Stabilized Chromogen Solution, R&D Systems) was added to the substrates for 20 min. The colorimetric reaction was then neutralized using 2 M sulfuric acid (Mallinckrodt Chemicals). Next 200 uL of colorimetric solution from each substrate was transferred to a 96-well plate and absorbance was read on an M5 spectrophotometer (Molecular Devices) at 450 nm with a correction at 540 nm. Using the same streptavidin-HRP and color solution and incubation times a standard curve was generated to which these absorbances were compared to determine peptide substrate surface density.
Peptide Functionality
NHS-ester PEG thin film substrates modified with 0.1, 0.01, or 0.001 mM of RGD or RGE, three substrates for each concentration, were incubated with 20,000 I-HVFFs/mL in normal unscarred media for 4 hrs at 37°C at 5% CO2. Post-incubation all substrates were briefly rinsed in PBS pH 7.4 to dislodge any non-adhered I-HVFFs. Adhered I-HVFF nuclei were then stained with Hoescht dye. Ten random images of each substrate’s surface were taken using a 10x objective on a Nikon Eclipse TE2000-S fluorescent microscope & digital camera. Finally, the number of I-HVFFs on the different substrates were counted manually by two different individuals via visual inspection of each image.
Unscar vs Scar RT-qPCR
Six plain glass substrates, not coated with a PEG thin film or peptide, were seeded with 20,000 I-HVFFs/mL and three were grown in normal media for ~9 days in the case of the unscarred control substrates. While the other three were grown for ~4 days (Chosen as it provided the I-HVFFs time to establish themselves on the substrates.) in normal media and ~5 days (Chosen as any longer and the I-HVFFs would have become confluent on the substrates.) in induction media in the case of scarred experimental substrates. Post growth, RNA was isolated from each sample utilizing the Nucleospin total RNA isolation kit (Clontech). Purified RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo). Reverse transcription of the pure RNA was achieved using a high-capacity cDNA reverse transcription kit (Life Technologies). Finally, real-time or quantitative polymerase chain reaction (qPCR) was conducted utilizing an Applied Biosystems 7500 real-time PCR machine. Briefly, qPCR reactions were set-up utilizing the TaqMan gene expression master mix (Life Technologies), a probe (Life Technologies) to detect the scar-like/myfibroblastic phenotype [Including human alpha-smooth muscle actin (alpha-SMA) {Hs00426835_g1}], six ECM gene-specific probes (Life Technologies) [Including human ELN {Hs00355783_m1}, DCN {Hs00754870_s1}, FN {Hs00365052_m1}, COL1A2 {Hs00164099_m1}, COL3A1 {Hs00943809_m1}, and HAS2 {Hs00193435_m1}], and template cDNA from the different samples. qPCR reaction conditions were as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec, and 60°C for 1 min. Human β-actin (Life Technologies) {Hs99999903_m1} was used as the endogenous control. The average cycle threshold (Ct) of each sample was then used for the calculation of expression levels using the relative comparative Ct method.
Varying RGD RT-qPCR
NHS-ester PEG thin film substrates were modified as described above with 0.01, 0.1, or 1 mM of RGD peptide, three substrates per concentration. Peptide-conjugated film coated substrates were seeded and grown as outlined in the previous section. RNA purification and RT-qPCR were also conducted in the same way as outlined in the previous section.
Statistical Analysis
Data were summarized as means ± SD. A one-way ANOVA with Tukey post hoc testing was utilized for statistical analysis. Non-matching symbols represent statistical significance. The alpha level was set to 0.05. All statistical analysis were computed using Minitab statistical software.
3. Results
Peptide surface density was determined to be dependent on the concentration of the peptide solution in which the NHS-ester PEG thin film substrates were incubated. The 0.01 mM peptide solution generated a lower peptide surface density of 0.8 ± 0.3 pg per mm2, 0.1 mM peptide solution generated a peptide surface density of 7.9 ± 0.6 pg per mm2, and 1 mM peptide solution generated the highest peptide surface density at 10.3 ± 0.8 pg per mm2 (Figure 1A). Note due to the low number of cells adherent to the 0.001 mM RGD surfaces (Figure 1B), peptide surface density studies did not include this surface concentration, and instead included 0.01 to 1.0 mM peptide concentrations.
Figure 1.
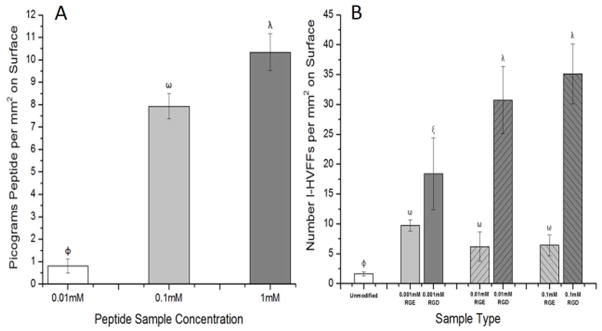
(A)Peptide substrate surface density was found to be proportional to the peptide concentration of the solution NHS-ester PEG thin film substrates were incubated in. (B)Peptide and NHS-ester PEG thin film functionality confirmed. High concentrations of RGD, 0.1mM, bind more I-HVFFs then lower concentrations. PEG thin film has anti-fouling capabilities with unmodified control binding the lowest number of I-HVFFs per mm2. Non-matching symbols represent statistical significance (p< 0.05).
Peptide grafted NHS-ester PEG thin film promoted fibroblast adhesion with films containing a higher peptide density supporting greater numbers of adherent cells. Substrates modified with 0.1 mM RGD had an average of ~35 I-HVFFs per mm2 adhered while 0.001 mM RGD had only an average of ~18 I-HVFFs per mm2 (Figure 1B). Furthermore, for each peptide concentration tested more I-HVFFs per mm2 adhered to the integrin-binding peptide, RGD, then the non-integrin binding control, RGE, peptide (Figure 1B). Finally, the fewest number of I-HVFFs, at an average of ~2 I-HVFFs per mm2, bound to unmodified control NHS-ester PEG thin film substrates (Figure 1B) confirming the relative inability of PEG to support cell adhesion. These data overall are consistent with literature reports stating that the PEG and the RGE control peptide do not support cell adhesion while the RGD peptide supports adhesion.12,18
RT-qPCR results confirmed that I-HVFFs grown in a healthy environment, without TGF-β1, were not myofibroblasts as determined by the lack of α-SMA expression (Figure 2). Fibroblasts grown with TGF-β1, became myofibroblastic, as determined by the expression of α-SMA (Figure 2).17 α-SMA immunohistochemistry (Figure S1) also showed this to be the case and that increasing RGD concentration from 0.01 mM to 1 mM resulted in greater spreading of the scarred I-HVFFs.18 We also observed that changes in phenotype from healthy to myofibroblastic resulted in significant changes in all six ECM genes tested. With an increase in gene expression of ELN by 14.8 ± 0.8, COL3A1 by 1.6 ± 0.1, FN by 2.3 ± 0.2, and COL1A2 by 1.8 ± 0.2 (Figure 2). Simultaneously, decreases in gene expression were observed for DCN by 0.4 ± 0.1 and HAS2 by 0.5 ± 0.1 (Figure 2). Such changes in phenotype have also been shown to impact many other types of genes in the vocal folds with relation to cell adhesion and other ECM components.19
Figure 2.
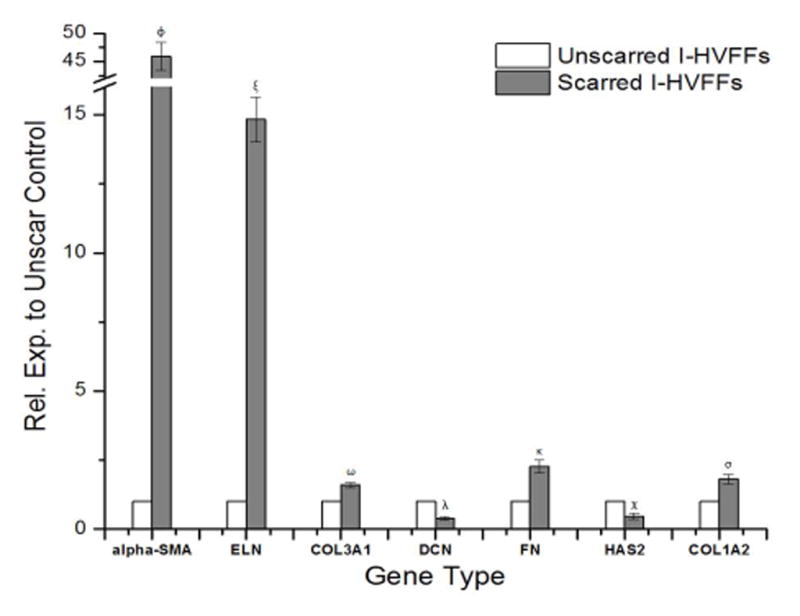
Varying cell phenotype from unscarred to scarred/myofibroblastic causes significant changes in gene expression of alpha smooth muscle actin (alpha-SMA) and all six ECM genes tested including elastin (ELN), collagen 3 alpha 1 (COL3A1), hyaluronan synthase 2 (HAS2), decorin (DCN), fibronectin (FN), collagen 1 alpha 2 (COL1A2). Non-matching symbols represent statistical significance (p< 0.05).
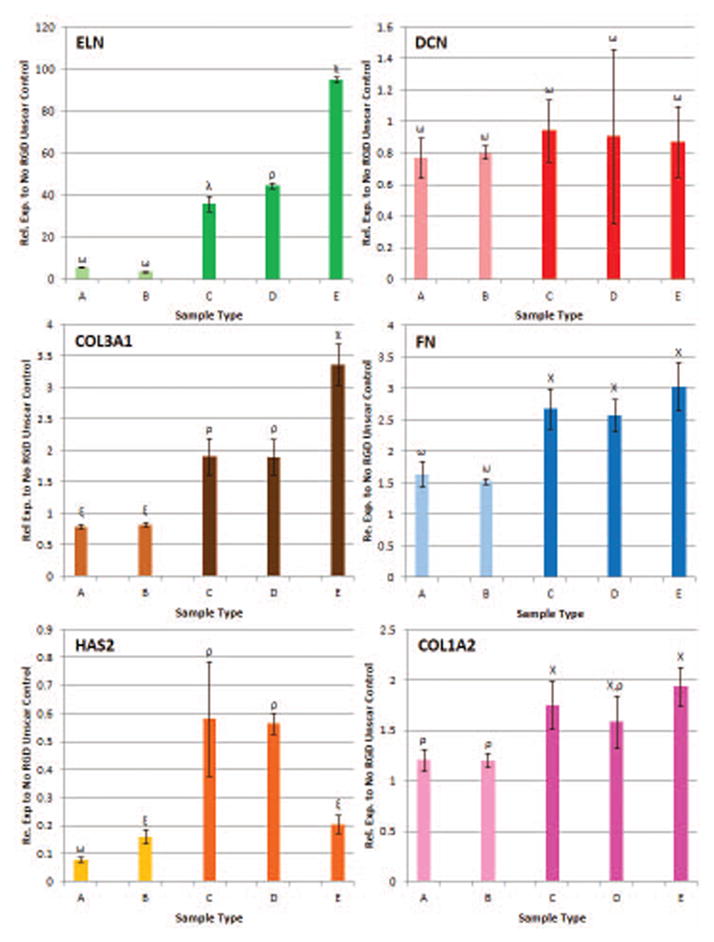
Variations in RGD peptide concentration impacted ECM gene expression. With unscarred I-HVFFs being unaffected, except for HAS2, by changes in RGD peptide concentration (Figure 3). While scarred I-HVFFs were affected, with ELN gene expression being up-regulated as the concentration of RGD increased (Figure 3). Similar results, albeit of smaller magnitude were obtained for COL3A1 gene expression (Figure 3). HAS2 expression was also impacted with a down regulation in its expression at the highest RGD concentration in the myofibroblast phenotype (Figure 3). DCN, FN, & COL1A2 remained unchanged in myofibroblastic I-HVFFs with respect to changes in RGD concentration (Figure 3). Note due to the low number of cells adherent to the 0.001 mM RGD surfaces (Figure 1B), RT-qPCR studies did not include this surface concentration, and instead included 0.01 to 1.0 mM peptide concentrations.
Figure 3.
Myofibroblastic phenotype and varying RGD surface densities have a significant impact on the gene expression of ELN, COL3A1, and HAS2. While the same variations have no significant impact on DCN, FN, & COL1A2. A = 0.01mM RGD Unscar/Healthy, B = 1mM RGD Unscar/Healthy, C = 0.01mM RGD Myofibroblastic, D = 0.1mM RGD Myofibroblastic, E = 1mM RGD Myofibroblastic. Non-matching symbols represent statistical significance (p< 0.05).
4. Discussion
Understanding the interplay between cell phenotype and the environment in which a cell is grown. Will prove invaluable in designing biomaterials that can facilitate the proper regeneration of damaged or diseased tissues. Here we begin to explore the complex interplay between cell phenotype and environment by growing I-HVFFs in myofibroblast-inducing or healthy environments on NHS-ester PEG thin film coated glass substrates modified with varying concentrations of RGD-binding peptides. We observed that changes in phenotype alone caused significant changes in gene expression of all six ECM genes tested while changes in RGD surface density impacted some of the ECM genes tested but not all.
It is of note that I-HVFFs for the most part only became responsive to varying RGD surface densities when they were induced into a myofibroblastic phenotype. This is an interesting finding, as it shows that myofibroblasts are much more sensitive to changes in RGD density then healthy fibroblasts. This is consistent with their normal role in the body which is to help drive wound healing of damaged tissues.20 It also suggests that being highly responsive to the surrounding environment facilitates rapid wound repair. Unfortunately, it also means that myofibroblasts have a higher probability of failing to sense when to quit. As there is often an increase in ECM during wound healing and thus potentially an increase in integrin-ligand interactions. This then helps drive a feedback loop that exists between TGF-β1 activity and the perceived stiffness of the surrounding environment,21–24 where the greater the perceived stiffness of the surrounding tissue by myofibroblasts the more TGF-β1 is activated. This then results in the production of more ECM, which leads to more RGD production that allows the cells to perceive the increase in stiffness due to the increased production of ECM, which then drives the production of yet more active TGF-β1 and the loop continues. It is possible then that here the myofibroblastic I-HVFFs growing on higher densities of RGD better perceive the glass substrate stiffness. This may in turn stimulate more endogenous TGF-β1 production, which would drive an increase in ECM gene expression. In comparison, the myofibroblastic I-HVFFs grown on the lower densities of RGD did not behave similarly due to the poor perception of the stiffness of the glass substrate.
The reasons for why only some ECM genes responded in myofibroblastic I-HVFFs to changes in RGD density while others did not is unclear. It could be that only some ECM genes respond to mechanotransduction mechanisms related to stiffness of the perceived environment as is the case with TGF-β1.21–24 Here ELN, COL3A1, and HAS2 gene expression appeared to be the most sensitive to changes in perceived substrate stiffness in myfibroblastic I-HVFFs while the other three ECM genes tested were not. More investigation is needed to determine if a response to stiffness through increased RGD density is indeed a driving factor.
Cells, like I-HVFFs, have evolved to respond to all aspects of their environments. Our data shows this in that not only do I-HVFFs respond to changes in soluble biochemical cues, like TGF-β1, but they also respond to what they are grown on through matrix biochemical cues and mechanotransduction.8–10,25,26 Here we see that as matrix biochemical signals change with the increase or decrease in RGD peptide there are changes in gene expression levels by I-HVFFs. This suggests that researchers not only need to control soluble cues, like TGF-β1, but also need to consider the bigger picture with regards to matrix biochemical cues. This understanding of the importance of the matrix in delivering biochemical cues and or modulating biochemical cues from soluble factors is in its infancy as researchers work to understand this highly complex interplay between the two.8–10,25,26
In this study biotinylated peptide was used to determine the concentration of RGD peptide grafted to the PEG surfaces. While it is possible that the biotin tag influences conjugation to the surface, it is unlikely as the biotin is conjugated to the amine-terminus of the peptide. A second limitation of the study is that serum levels were not consistent between the TGF-β1 treated I-HVFFs and the healthy I-HVFF growth environments. It is possible then that the differences observed in gene expression could be attributed to this serum concentration difference. Retaining serum in the TGF-β1 treatments would add tremendous complexity to results interpretation due to the relative unknown composition of serum; thus it is common practice to remove serum in these types of studies. Despite this concern we remain confident that serum changes contribute minimally and that the major changes were seen either with changing RGD surface concentration (Since serum concentration remained constant as RGD surface concentrations changed.), or are consistent with the known effects of TGF-β1 on fibroblasts.17,27 As the in vitro system is further optimized studies will be done to attempt to further tease out any potential contribution that the absence of serum may have. A third limitation is the difficulty in elucidating how much the induced TGF-β1 myofibroblast-like phenotype versus the eventual stable myofibrolast phenotype had in driving ECM gene expression changes in concert with the changes in integrin binding to variations in RGD surface substrate density. This elucidation is made even more complex as the literature has shown that some cell lines once induced into a stable myofibroblastic phenotype never revert back while others will.28–30 Answering this question is important, however it falls outside the scope of this manuscript, and will therefore be addressed in future work. A fourth limitation is that although the glass substrates provide a straight-forward in vitro model for studying this interaction between RGD and cell phenotype, the design is limited, in that the stiffness of this material significantly differs then that which is seen in either healthy or scarred vocal fold tissue. Future studies will utilize materials that more accurately mimic the rigidity of healthy or scarred vocal fold tissue.
In summary, the results demonstrate that RGD concentration and cell phenotype impact expression of the ECM genes investigated here. This suggests that controlling RGD concentration with respect to both cell phenotype and the environment in which the material is to be used may improve material bioactivity and thereby improve the intended tissue regeneration when the material is implanted within the vocal fold or other tissue type. Future work will focus on ascertaining if these changes in ECM gene expression also result in changes in protein expression.
Supplementary Material
Acknowledgments
This research was supported by funding from NIDCD/NIH R01DC005788. The authors would like to thank Dr. Susan Thibeault at the University of Wisconsin, Madison & her funding source NIDCD/NIH R21DC008428 for providing the I-HVFFs. We acknowledge the contributions of Jewel Pothen in synthesizing the peptides.
References
- 1.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 2.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 3.Massia SP, Hubbell JA. An RGD Spacing of 440nm is sufficient for integrin alpha-v-beta-3-mediated fibroblast spreading and 140nm for focal contact and stress fiber formation. J Cell Biol. 1991;114:1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes RO. Integrins: Versatility, Modulation, and Signaling in Cell Adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 5.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto K, Matsuura T, Hosokawa R, Akagawa Y. RGD peptides regulate the specific adhesion scheme of osteoblasts to hydroxyapatite but not to titanium. J Dent Res. 1998;77:481–487. doi: 10.1177/00220345980770030701. [DOI] [PubMed] [Google Scholar]
- 7.Shu XZ, Ghosh K, Liu Y, Palumbo FS, Luo Y, Clark RA, Prestwich GD. Attachment and spreading of fibroblasts on an RGD peptide-modified injectable hyaluronan hydrogel. J Biomed Mater Res A. 2004;68A:365–375. doi: 10.1002/jbm.a.20002. [DOI] [PubMed] [Google Scholar]
- 8.Cao X, Yu WQ, Qiu J, Zhao YF, Zhang YL, Zhang FQ. RGD peptide immobilized on TiO2 nanotubes for increased bone marrow stromal cells adhesion and osteogenic gene expression. J Mater Sci: Mater Med. 2012;23:527–536. doi: 10.1007/s10856-011-4479-0. [DOI] [PubMed] [Google Scholar]
- 9.Yang F, Williams CG, Wang DA, Lee H, Manson PN, Elisseeff J. The effect of incorporating RGD adhesive peptide in polyethylene glycol diacrylate hydrogel on osteogenesis of bone marrow stromal cells. Biomaterials. 2005;26:5991–5998. doi: 10.1016/j.biomaterials.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Mann BK, Tsai AT, Scott-Burden T, West JL. Modification of surfaces with cell adhesion peptides alters extracellular matrix deposition. Biomaterials. 1999;20:2281–2286. doi: 10.1016/s0142-9612(99)00158-1. [DOI] [PubMed] [Google Scholar]
- 11.Kutty JK, Webb K. Tissue Engineering Therapies for the Vocal Fold Lamina Propria. Tissue Engineering Part B-Reviews. 2009;15:249–262. doi: 10.1089/ten.TEB.2008.0588. [DOI] [PubMed] [Google Scholar]
- 12.Harbers GM, Emoto K, Greef C, Metzger SW, Woodward HN, Mascali JJ, Grainger DW, Lochhead MJ. Functionalized poly(ethylene glycol)-based bioassay surface chemistry that facilitates bio-immobilization and inhibits nonspecific protein, bacterial, and mammalian cell adhesion. Chem Mater. 2007;19:4405–4414. doi: 10.1021/cm070509u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn MS, Kobler JB, Starcher BC, Zeitels SM, Langer R. Quantitative and comparative studies of the vocal fold extracellular matrix I: elastic fibers and hyaluronic acid. Ann Otol Rhinol Laryngol. 2006;115:156–164. doi: 10.1177/000348940611500213. [DOI] [PubMed] [Google Scholar]
- 14.Hahn MS, Kobler JB, Zeitels SM, Langer R. Quantitative and comparative studies of the vocal fold extracellular matrix II: collagen. Ann Otol Rhinol Laryngol. 2006;115:225–232. doi: 10.1177/000348940611500311. [DOI] [PubMed] [Google Scholar]
- 15.Hahn MS, Kobler JB, Zeitels SM, Langer R. Midmembranous vocal fold lamina propria proteoglycans across selected species. Ann Otol Rhinol Laryngol. 2005;114:451–462. doi: 10.1177/000348940511400607. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Thibeault SL. Novel isolation and biochemical characterization of immortalized fibroblasts for tissue engineering vocal fold lamina propria. Tissue Eng Part C Methods. 2009;15:201–212. doi: 10.1089/ten.tec.2008.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vyas B, Ishikawa K, Duflo S, Chen X, Thibeault SL. Inhibitory effects of hepatocyte growth factor and interleukin-6 on transforming growth factor-beta 1 mediated vocal fold fibroblast-myofibroblast differentiation. Ann Otol Rhinol Laryngol. 2010;119:350–357. doi: 10.1177/000348941011900513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peppas NA, Hilt JZ, Thomas JB. Nanotechnology in Therapeutics: Current Technology and Applications. Wymondham: Horizon Scientific Press; 2007. [Google Scholar]
- 19.Jette ME, Hayer SD, Thibeault SL. Characterization of human vocal fold fibroblasts derived from chronic scar. Laryngoscope. 2013;123:738–745. doi: 10.1002/lary.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- 21.Wells RG. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J Clin Gastroenterol. 2005;39:S158–S161. doi: 10.1097/01.mcg.0000155516.02468.0f. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Dranoff JA, Chan EP, Uemura M, Sevigny J, Wells RG. Transforming growth factor-beta and substrate stiffness regulate portal fibroblast activation in culture. Hepatology. 2007;46:1246–1256. doi: 10.1002/hep.21792. [DOI] [PubMed] [Google Scholar]
- 23.Hinz B. Tissue stiffness, latent TGF-beta1 activation, and mechanical signal transduction: implications for the pathogenesis and treatment of fibrosis. Curr Rheumatol Rep. 2009;11:120–126. doi: 10.1007/s11926-009-0017-1. [DOI] [PubMed] [Google Scholar]
- 24.Chia HN, Vigen M, Kasko AM. Effect of substrate stiffness on pulmonary fibroblast activation by TGF-β. Acta Biomaterialia. 2012;8:2602–2611. doi: 10.1016/j.actbio.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Faramarz E, Bae H, Manoucheri S, Cha JM, Khademhosseini A. Engineering approaches toward deconstructing and controlling the stem cell environment. Ann Biomed Eng. 2012;40:1301–1315. doi: 10.1007/s10439-011-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters NJ, Gentleman E. Evolving insights in cell-matrix interactions: Elucidating how non-soluble properties of the extracellular niche direct stem cell fate. Acta Biomaterialia. 2015;11:3–16. doi: 10.1016/j.actbio.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branski RC, Barbieri SS, Weksler BB, Saltman B, Krishna P, Kraus DH, Broadbelt NV, Chen J, Poppas DP, Felsen D. Effects of transforming growth factor-beta1 on human vocal fold fibroblasts. Ann Otol Rhinol Laryngol. 2009;118:218–226. doi: 10.1177/000348940911800310. [DOI] [PubMed] [Google Scholar]
- 28.Yang AH, Chen JY, Lin JK. Myofibroblastic conversion of mesothelial cells. Kidney International. 2003;63:1530–1539. doi: 10.1046/j.1523-1755.2003.00861.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Haeger SM, Kloxin AM, Leinwand LA, Anseth KS. Redirecting valvular myofibroblasts into dormant fibroblasts through light-mediated reduction in substrate modulus. PLos One. 2012;7:e39969. doi: 10.1371/journal.pone.0039969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisseleva T, Cong M, Paik YH, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. PNAS. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.