Abstract
Dedifferentiated liposarcomas (DDLPS) are highly resistant to conventional chemo- and radiotherapies, with surgical resection remaining the classic treatment strategy; therefore there is a pressing need for novel anti-DDLPS targeted chemotherapeutics. Hepatocyte growth factor receptor (Met) expression is elevated in DDLPS, but the functional role of Met signaling in this disease is not known. We found that the in vitro stimulation of DDLPS cells with hepatocyte growth factor (HGF) elevated the degree of PI3K/AKT and MAPK pathway signaling, and that pro-tumorigenic phenotypes such as cell proliferation, invasion, and migration, were significantly enhanced. Conversely, Met knockdown using shRNA-mediated interference decreased HGF-induced Met signaling, the invasive and migratory nature of DDLPS cells in vitro, and the tumorigenicity of DDLPS cells in vivo. This data strongly supports the role for Met as a DDLPS therapeutic target. To that end, using EMD1214063, an ATP-competitive kinas inhibitor that targets Met more specifically than other kinases, inhibited Met-dependent signaling, reduced the oncogenecity of DDLPS cells in vitro, and significantly increased the survival of nude mice bearing subcutaneous DDLPS xenografts. These findings support further investigations of HGF-induced Met signaling inhibition in DDLPS, as a potential strategy to enhance clinical outcomes for this disease.
Approximately 15–20% of adult soft tissue sarcomas are adipogenic-lineage liposarcomas1,2, of which the most common histological subtypes include well differentiated (WDLPS) and dedifferentiated (DDLPS) liposarcomas3. DDLPS typically develops within WDLPS as high grade tumor cells with enhanced mitotic activity in demarcated areas lacking characteristic WDLPS lipomatous features4. Although DDLPS can arise in any anatomic site5, it often develops in the retroperitoneum where it is particularly aggressive and associated with a poor prognosis4,6. Surgery is the therapeutic mainstay for localized primary DDLPS; however, the majority of these patients will be diagnosed with and die from locally recurrent or disseminated disease within five years2. DDLPS are highly resistant to conventional cytotoxic or radiation neo-adjuvant therapies; therefore, alternative targeted strategies are needed to improve patient prognosis and outcome7.
Receptor tyrosine kinases (RTKs) are often deregulated in cancer; therefore, inhibiting their catalytic activity has emerged as a focused strategy for targeted therapy8,9. Previous studies have indicated the importance of the Met signaling axis in various sarcoma histologies10–17. Additionally, Peng and colleagues (2011) suggested a potential role for deregulated RTKs in DDLPS. They found that Met, as well as several other RTKs, was overexpressed in DDLPS cell strains when compared with adipocytic lineage control cells, and that Met expression was elevated in two different DDLPS xenograft models18. Met is a RTK that is essential for normal developmental processes, and also has causative oncogenic roles in most cancers when overactive19. Enhanced Met activity may occur in a ligand-dependent or -independent manner, contributing to hallmark malignancy phenotypes9,19,20. The only known activating Met ligand is the hepatocyte growth factor (HGF)21, also known as scatter factor due to its ability to induce epithelial cell scattering22,23. When HGF binds Met in the extracellular Sema24 or IPT3/IPT4 domains25, receptor dimerization26 leads to receptor activation through phosphorylation of Y1234 and Y1235 in the kinase domain27. Once this occurs, signal transduction ensues through C-terminal phosphorylation28 with resultant STAT3, Src, and PI3K/AKT and MAPK pathway activation9,25,29. Met hyperactivation in cancer promotes numerous pathogenic processes, including angiogenesis, proliferation, survival, migration, invasion, and metastasis20,27,29,30. Accordingly, several antagonists of Met signaling have been developed, including anti-HGF antibodies, Met monoclonal antibodies, decoy Met receptors, and specific and dual specificity tyrosine kinase inhibitors, all of which are all in various stages of clinical trials for different cancers9,31,32. Recently, clinical benefits have been reported with Met/HGF axis inhibition for advanced stage or metastatic disease, and for patients whose tumors are resistant to EGFR antagonism32.
Since targeting Met has benefitted patients in the clinical setting9,31,32 and Met overexpression in DDLPS has previously been reported18, we sought to evaluate the functional effects of Met signaling abrogation in DDLPS cells in vitro and in vivo. We show that constitutive and inducible Met activity enhances cell proliferation as well as the migratory and invasive Met-associated phenotypes of DDLPS cells. Furthermore, we found that Met expression and activity was crucial in maintaining not only these Met-specific phenotypes but also DDLPS cell tumorigenicity in pre-clinical animal models.
Materials and Methods
Cell lines and reagents
LPS141, a primary human cell line derived from a high grade retroperotineal DDLPS, was kindly provided by Dr. Jonathan Fletcher (DCFI, Boston, MA). Human DDLPS cell lines Lipo224, Lipo224B, Lipo246, Lipo815, and Lipo863B were established in our laboratory from surgically resected human retroperitoneal DDLPSs and maintained as previously described18. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM 1X) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, or as otherwise described in figure legends. All the established DDLPS cell lines were DNA fingerprinted using short tandem repeat DNA fingerprinting3 (cell line STR results have been previously reported3; STR for Lipo815 is reported in Supplementary Table 1).
Recombinant Human HGF was purchased from R&D Systems, and the Met inhibitors, EMD1214063 and SU11274, were purchased from Chemitek and Selleckchem, respectively. Antibodies used for western blot analyses or for immunohistochemistry were purchased from the following manufacturers: Met, pMet (Tyr1234/Tyr1235), AKT, pAKT (Ser473), ERK, pERK (Y202/T204) (Cell Signaling), CD31 (BD Pharmingen), Ki67 (Thermo/Lab Vision), and β-actin-HRP (Santa Cruz Biotechnology).
MDM2 FISH analyses
FISH for MDM2 in fixed cultured cells were performed as previously described18 (data not shown). Briefly, a BAC labeled probe cocktail for the 12q15 chromosomal region (RP11-185H13, RP11-450G15, RP11-816C9, RP11-630N19, RP11-717F7, RP11-1104N20, and RP11-426B12; (Texas Children’s Hospital, Houston, Tx, USA) was used alongside a specific probe for the centromeric region of chromosome 12 (Abbott Molecular, DesPlaines, IL, USA), and performed as previously described33. No less than 100 nuclei per slide were analyzed for MDM2 and CEP12 signals. MDM2 signal ratio to CEP12 signal ratios were calculated per slide, MDM2 was considered amplified if the MDM2/CEP12 ≥ 2.1 and positive and negative control cell lines were used as appropriate.
Migration and invasion assays
Migration and invasion assays were performed as previously described18. Modified Boyden chamber migration assays were performed using 24-well tissue culture plates that were fitted with polycarbonate filters with 8 mm pores (Becton Dickinson Labware, Franklin Lakes, NJ, USA). Upper chamber compartments were seeded with 5–20×103 cells. Lower chamber compartments contained DMEM 1x that was supplemented with 1% FBS and/50 ng/mL HGF as chemo-attractants where appropriate. After 24 hours, cells on the upper surface of the filters were removed with a cotton swab, and the cells on the lower surface were fixed with 4% formaldehyde and stained with 0.2% crystal violet (Baxter Healthcare, Houston, TX, USA). Migratory activity was calculated by averaging the % area fraction coverage in four fields per well using Image J software. Invasion assays were performed similarly to migration assays using 24-well BioCoat Matrigel invasion chambers (8mm pore size polycarbonate filters) pre-coated with Matrigel were used (Becton Dickinson Labware).
Short tandem repeat DNA fingerprinting
DNA was extracted from all liposarcoma cell lines using the Qiagen DNA Mini-kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). STR DNA fingerprinting was carried out as previously described17.
MTS cell viability assays
Cellular viability was evaluated and calculated as previously described18. Briefly, cells were seeded in 96-well plates at a density of 1500 cells/well in 100 μL of cell culture media. Depending on the experiment (please see specific figure legends), 20 μL of MTS reagent (CellTiter96 Aqueous Non-Radioactive Assay Kit; Promega, Madison, WI, USA) was added per well, incubated for 2 h at 37°C, and absorbance measured at 490 nm. Average absorbance values per treatment were calculated as a percentage of averaged untreated/DMSO absorbance values as appropriate.
In vivo animal experiments
All animal procedures were approved by the UTMDACC Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals”.
For experiments assessing the effect of Met shRNA on tumor growth, 2 × 106 non-targeting (shNT) or shMet cells were injected subcutaneously into the flanks of six week old female hairless SCID mice. Tumor volume was measured twice weekly. Mice were sacrificed 12 days after injection; tumors were resected, weighed, and paraffin embedded for sectioning and staining. For experiments evaluating the effect of EMD1214063 treatment on tumorigenicity of Lipo246 cells in vivo, 2 × 106 cells were injected subcutaneously into the flanks of six week old female hairless SCID mice. Once palpable tumors formed after approximately two weeks, mice were randomized into DMSO control or EMD1214063 (6 mg/kg) twice weekly treatment arms. Mice were sacrificed once tumor burden reached 1.5 cm in height, length, or width, or after two and a half weeks of treatment. Tumors were resected, weighed, and either snap-frozen in liquid nitrogen or paraffin embedded for sectioning and staining once the experiment was terminated.
Quantitative PCR (qPCR)
Extraction of mRNA from parental DDLPS cell lines was conducted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). cDNA synthesis reactions were performed using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Quantitative PCR was performed using the Light Cycler 480 SYBR Green I Master mix and the Light Cycler 480 system (Roche, Indianapolis, IN, USA). Primers used for HGF and β-2-microglobulin have been detailed previously20. Second derivative max values were calculated by Roche’s Light Cycler software. HGF expression levels were normalized to β-2-microglobulin, and calculated using the 2ΔΔCT method.
Western blotting
Western blotting reagents and procedures have been previously described17. Briefly, 20–30 μg of tumor or cell protein lysates were separated by SDS-PAGE electrophoresis and then transferred onto PVDF membranes. Membranes were then blocked with 5% non-fat milk in 1x PBS/T and then probed with commercially available antibodies as previously described17. Horseradish peroxidase-conjugated secondary antibodies were detected using ECL reagent (Amersham Biosciences).
Lentiviral transduction for generation of stable Met knockdown cells
Met shRNA lentiviral particles and transduction procedures were performed as previously described17.
HGF ELISA
Cells were seeded (2.5 × 105 cells/well) in 6-well plates in duplicate wells per experiment and cultured overnight in 10% FBS-containing media. Cells were washed twice with PBS/T and then cultured in 1 mL of 1% FBS-containing media for 48 h. Conditioned media was collected and analyzed for HGF content using the Human HGF DuoSet Kit (R & D Systems, Minneapolis, MN USA) according to the manufacturer’s instructions.
Statistics
Data significance was analyzed using GraphPad Prism 6.01 software. Specific tests of significance are detailed in figure legends. Unpaired, two-tailed t-tests were performed as indicated; P<0.05 =*; P<0.005 = **; and P< 0.0001 = ***.
Results
Met and HGF is expressed in DDLPS cell lines
All DDLPS cell lines were generated from surgical specimens in our laboratory (except for LPS141) and their MDM2 amplification status was confirmed by fluorescence in situ hybridization (FISH; data not shown). Short tandem repeat fingerprinting for most cell lines in these studies has been previously reported3 except for Lipo815 (Table S1). Western blot analysis revealed that the majority of DDLPS cell lines had enhanced Met activity in culture (Figure 1A). To determine whether autocrine HGF expression may be responsible for elevated Met activation in DDLPS cell lines, we evaluated all cell lines of Figure 1A for their relative amounts of autocrine HGF mRNA expression using quantitative RT-PCR (Figure S1A). HGF mRNA expression was highest in Lipo246 and Lipo815, suggesting that the other evaluated cell lines might not rely on autocrine HGF expression for potential receptor activation. Using HGF ELISAs, we showed that HGF was secreted into CM from Lipo246 and Lipo815 cells in an autocrine manner (Figure S1B). These data suggest that autocrine HGF may activate Met in some DDLPS cell lines, but that other mechanisms may contribute to enhanced Met activity in other DDLPS cell lines. In that Met protein expression was high in all of the DDLPS cell lines evaluated and HGF was produced in an autocrine manner in several of these cell lines, the Met:HGF axis could represent an attractive anti-DDLPS therapeutic target.
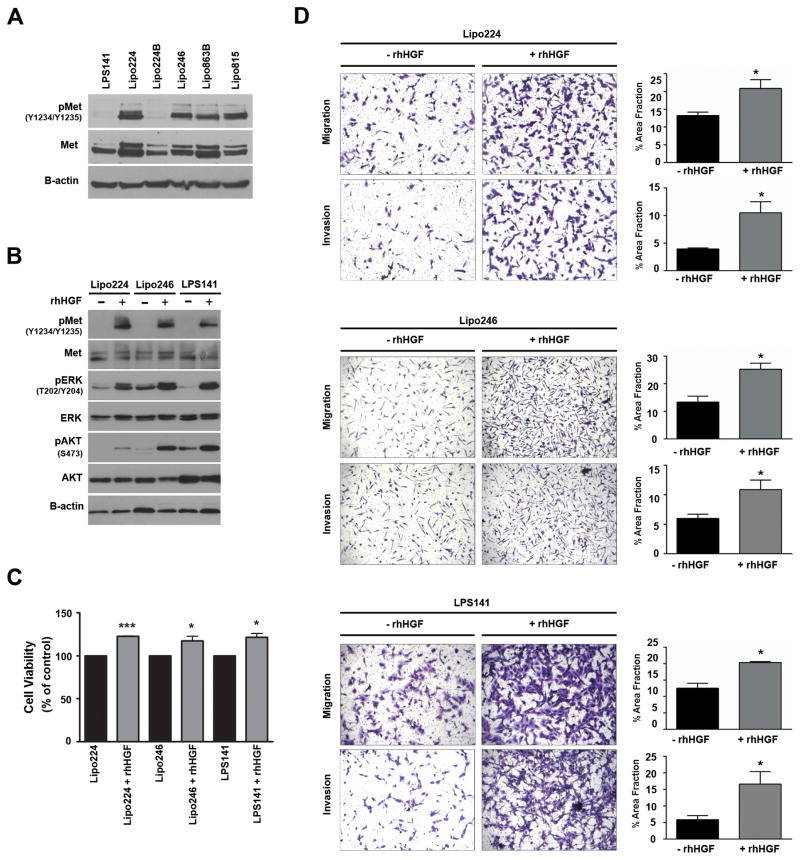
Figure 1. HGF-mediated Met activation enhances oncogenic signaling and phenotypes of DDLPS cells in vitro.
(A) Western blot analysis of activated Met in several DDLPS cell lines (10% FBS media). (B) Western blots monitored HGF-stimulated Met, and downstream Erk1/2 and AKT activation in DDLPS cells. Lipo224, Lipo246, and LPS141 cells were serum-starved overnight and stimulated with rhHGF (50 ng/mL) for 20 minutes. (C) MTS assays measured cell proliferation of 24 hr serum-starved DDLPS cells over a 48 hr period with or without rhHGF (50 ng/mL) stimulation (n=3 ± SEM; t-test: *=P<0.05, ***=P<0.0001; samples were analyzed at least in duplicate per experiment). (D) Modified Boyden chamber assays measured the invasion and migration capabilities of DDLPS cells with or without the use of rhHGF (50 ng/mL) as a chemoattractant. (Images are representative, and graphs represent n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; samples were analyzed at least in duplicate per experiment).
Met activation enhances oncogenic phenotypes and signaling in DDLPS cells
To determine whether stimulation of DDLPS cells with recombinant human HGF (rhHGF) could activate (or further activate) the Met receptor, we treated serum-starved DDLPS cells with rhHGF for 15 minutes and examined Met activation and Met:HGF axis canonical signaling pathways by western blot analyses (Figure 1B). We found that phosphorylation of Met at Y1234/Y1235 increased with HGF stimulation in the DDLPS cell lines evaluated and that downstream oncogenic signaling through the mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) pathways were acutely activated. It is known that HGF-mediated Met activation stimulates MAPK and PI3K pathway signaling, which are necessary for cell proliferation and invasion and migration, respectively20. To assess DDLPS cell proliferation rates when stimulated with HGF in vitro, we exposed serum-starved DDLPS cells to either rhHGF or serum-free media for 48 h, demonstrating that proliferation was significantly enhanced in DDLPS cell lines when stimulated with rhHGF (Figure 1C).
The Met:HGF axis is critical for invasion and migration in many malignancies30; however, very little is known about the role of this axis on these phenotypes in DDLPS. Therefore, we evaluated HGF-mediated changes to these processes in DDLPS cell lines in vitro. Stimulation of serum-starved DDLPS cells with rhHGF significantly increased the invasive and migratory characteristics of DDLPS cells in vitro (Figure 1D). These findings suggest that the HGF paracrine stimulation of DDLPS cells enhances their malignant phenotype.
Met knockdown suppresses AKT signaling, proliferation, invasion, and the migration of DDLPS cells in vitro
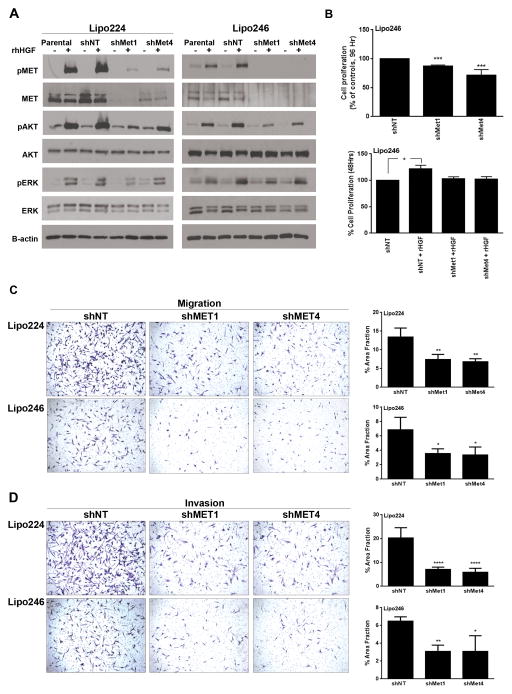
Since DDLPS cells require Met activation for cell proliferation, migration, and invasion (Figure 1), we hypothesized that Met knockdown, even when HGF-stimulated, would reduce the aggressiveness of DDLPS cells in vitro. Using shRNA-mediated interference we knocked Met expression down stably in Lipo246 and Lipo224 DDLPS cells, and evaluated downstream signaling consequences of Met knockdown cells when stimulated with rhHGF compared to control cells (Figure 2A). For both Lipo224 and Lipo246 cells, AKT activity was reduced in shMet cells even with rhHGF stimulation when compared to control cells; however, interestingly, ERK activity was unaffected. Next, we evaluated the proliferative capacity of Lipo224 and Lipo246 cells with Met knockdown in comparison with parental sh-non-targeting (shNT) control cells (Figure 2B; upper panel). We found that both Met knockdown polyclonal populations (shMet1 and shMet4) exhibited reduced capabilities to proliferate in full serum after 96 hours. However, when these cell lines were serum-starved for 48 hours, their capacity to proliferate resembled those of shNT cells even when stimulated with rhHGF (Figure 2B; lower panel). In both Lipo246 and Lipo224 cells, migration (Figure 2C) and invasion (Figure 2D) was significantly abrogated in the two polyclonal populations of Met knockdown cells compared to controls. These results suggest that Met expression is crucial for the migratory and invasive properties of DDLPS cells in vitro.
Figure 2. Met knockdown decreases PI3K and MAPK pathway signaling, and the proliferative, invasive, and migratory phenotypes of DDLPS cells in vitro.
(A) Left panel: Western blot analyses of downstream signaling in Met knockdown Lipo224 cells that were serum-starved and then stimulated with 50 ng/mL rhHGF for 15 minutes (− represents no rhHGF stimulation; + represents cells that were stimulated with rhHGF). Right panel: Western analysis was performed as in the left panel, however this panel evaluated Lipo246 cells with Met knockdown and rhHGF stimulation. (B) Upper Panel: MTS assays measured the proliferative capabilities of shNT, shMet1, and shMet4 polyclonal populations after 96 hours in culture in Lipo246 Met knockdown cells (10%-FBS supplemented media; n=3 experiments ± SEM; t-test: *=P<0.05, ***=P<0.0001; samples were analyzed at least in duplicate per experiment). Lower Panel: 50 ng/mL rhHGF stimulated Lipo246 DDLPS cells with Met knockdown were serum-starved for 24 h prior to stimulation with 50 ng/mL rhHGF for 48 h. MTS assays measured cell proliferation (n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; samples were analyzed at least in duplicate per experiment). Lipo224 and Lipo246 shMet cells exhibit a statistically significant decrease in migration (C) and invasion (D) when compared with shNT cells (rhHGF was used as a chemoattractant; n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; samples were analyzed at least in duplicate per experiment).
Met is required for tumorigenicity and the aggressiveness of Lipo246 DDLPS cells in vivo
To characterize the biological implications of stable Met knockdown in Lipo246 DDLPS cells, we injected parental shNT and Met silenced cells (shMet1 and shMet417) subcutaneously into nude mice. Tumors in mice injected with Lipo246 shMet cells exhibited abrogated tumor growth as well as decreased cumulative growth rates compared to mice injected with Lipo246 shNT cells (Figure 3A); furthermore, the overall tumor volume was less in mice injected with the shMet cells (Figure 3B). Met protein expression remained suppressed in Lipo246 shMet expressing xenografts compared with Lipo246 shNT xenografts andLipo246 parental cells at conclusion of the in vivo experiment (Figure 3C).
Figure 3. Met knockdown decreases tumorigenicity of DDLPS cells in vivo.
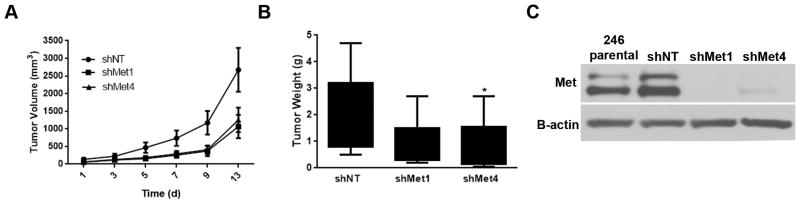
(A) Lipo246 cells expressing a non-targeting construct (shNT), or two different shMet targeting constructs were injected subcutaneously, and the tumor volume measured over time. Met KD xenografts had an overall slower growth rate and a decreased tumor weight (B) at termination as compared to control shNT tumor bearing control mice. (Mean tumor volume at 12 days for each group of mice was recorded ± SEM; t-test: *=P<0.05) (C) Western blot confirmed Met KD in xenograft tumors.
Taken together these data suggest that Met expression is crucial to maintain the tumorigenicity and aggressiveness of Lipo246 cells in vivo. However, these results also suggest that other mechanisms may compensate for Met loss over time, thereby maintaining tumorigenic potential.
The in vitro anti-DDLPS effects of the Met tyrosine kinase inhibitor EMD1214063
Initially, Met inhibition in Lipo246 was tested by using SU11274, a Met inhibitor that has received considerable research attention19,34–36. Western blot analyses of Lipo246 cells that had been serum-starved for 24 hr then treated with rhHGF and increasing concentrations of SU11274 showed that Met activity was reduced in a dose-dependent manner by SU11274 (Figure S2A). In addition, we found that the activity of AKT was reduced with nanomolar concentrations of SU11274, but the inhibition of ERK signaling required greater amounts of SU11274 to produce the same effect. To determine whether Met activity inhibition may play a role in the proliferation, migratory capacity, and invasiveness of Lipo246 cells, we treated Lipo246 cells with SU11274. Cells were treated with either increasing concentrations of SU11274 (to determine relative effects on cell viability; Figure S2B), or with 1 μM SU11274 to test its effects on cell migration and invasion (Figure S2C). We found that SU11274 reduced the proliferative and aggressive capacities of Lipo246 cells in vitro.
A novel Met tyrosine kinase inhibitor, EMD1214063, has recently been approved for use in a Phase 1 human clinical trial for advanced solid tumors refractory to standard care or without other potentially beneficial therapeutic options (http://clinicaltrials.gov; Identifier: NCT01014936). EMD1214063 is a potent and highly selective therapeutic compound with a wide therapeutic index, it can target HGF-dependent and HGF–independent tumor xenografts37. Therefore, we sought to evaluate its anti-DDLPS efficacy compared to SU11274.
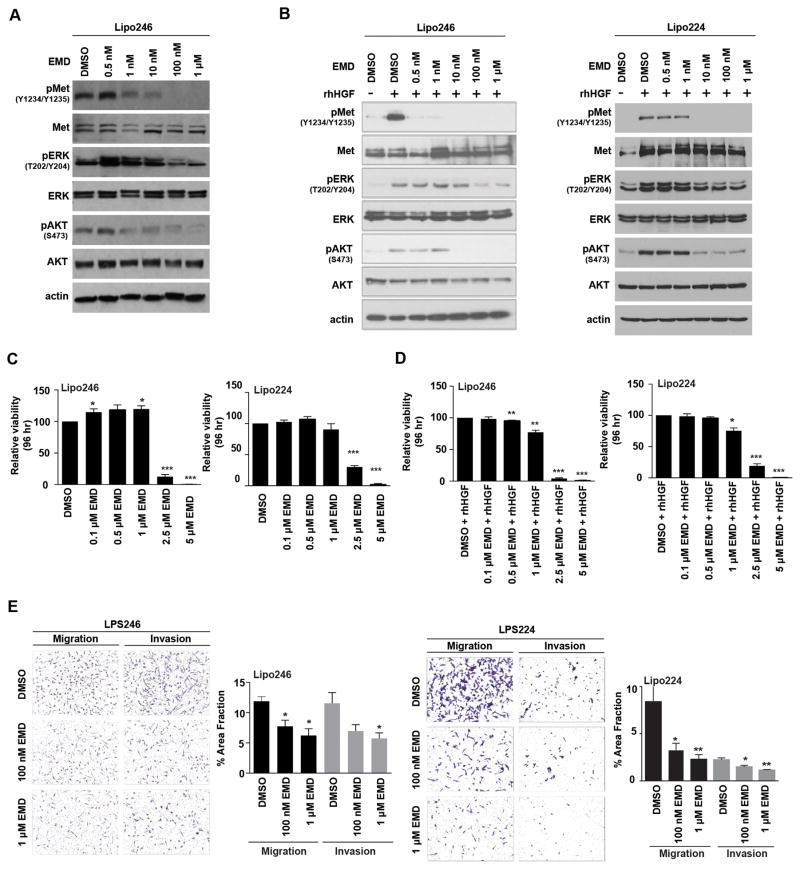
Met is constitutively active in Lipo246 cells (Figure 1A), possibly in an autocrine manner involving HGF (Figure S1), so we used this cell line to test whether EMD1214063 was capable of suppressing Met-dependent signaling (Figure 4A). Similarly to Lipo246 treatment with SU11274 (Figure S2A), we found that EMD1214063 inhibited AKT phosphorylation (S473) at a low nanomolar concentration (1 nM), whereas inhibition of ERK activity required 100-fold higher EMD1214063 concentrations (Figure 4A). Consequently, EMD1214063 inhibited both PI3K and MAPK signaling in Met-dependent Lipo246 cells.
Figure 4. Decreased oncogenic signaling and aggressiveness of DDLPS cells with Met inhibition in vitro.
(A) Western blot analysis monitored decreasing Met, AKT, and Erk1/2 activity levels in Lipo246 cells with increasing concentrations of EMD1214063 (4 h; 10%-FBS containing media). (B) Left Panel; Lipo246 cells. Right Panel; Lipo224 cells: Serum-starved cells were pretreated with increasing concentrations of EMD1214063 followed by rhHGF stimulation for 20 min. Western blot analyses revealed that Met, AKT, and ERK1/2 signaling decreased in a dose-dependent manner. (C) Left Panel: Lipo246; Right panel: Lipo224. MTS assays measured proliferation rates as a consequence of increasing EMD1214063 treatment (n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; samples were analyzed at least in duplicate per experiment). (D) Serum starved Lipo246 (Left Panel) and Lipo224 (Right Panel) were simultaneously treated with increasing concentrations of EMD1214063 and rhHGF (50 ng/mL) for 96 h. MTS assays revealed that proliferation rates decreased with increasing concentrations of EMD1214063 (n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; samples were analyzed at least in duplicate per experiment). (E) Left Panel: Lipo246 cells were either treated with DMSO, 100 nM, or 1 μM EMD1214063 for 24 h, and migration and invasion results recorded (3 images per well; at least 2 replicates per experiment; n=3 experiments ± SEM; t-test: *=P<0.05). Right Panel: Lipo224 cells were also either treated with DMSO, 100 nM, or 1 μM EMD1214063 for 24 h (3 images per well; at least 2 replicates per experiment; n=3 experiments ± SEM; t-test: *=P<0.05, **=P<0.005).
To assess whether EMD1214063 could inhibit exogenous HGF-induced Met phosphorylation, we pre-treated serum-starved Lipo246 (Figure 4B; left panel) and Lipo224 (Figure 4B; right panel) DDLPS cells with increasing concentrations of EMD1214063 for 4 hr followed by rhHGF stimulation. We found that EMD1214063 inhibited Met (Y1234/Y1235), AKT (S473), and ERK (T202/Y204) activity in a dose-dependent manner even when stimulated with rhHGF. To evaluate whether HGF-induced Met phosphorylation could be inhibited by EMD1214063 in another DDLPS cell line, we next repeated the above experiment in LPS141 cells (Figure S3A) using 1 μM EMD1214063 with or without rhHGF stimulation. Met activation and the activities of key signaling components of the PI3K and MAPK pathways (pAKT S473 and pERK Y204/T202, respectively) decreased in HGF-stimulated EMD1214063-treated cell lines, whereas increased AKT and ERK activities occurred when the cell lines were HGF-stimulated without Met kinase inhibition (Figure S3A).
Most likely as a consequence of MAPK pathway inhibition in Lipo246 (Figure 4B; left panel) and Lipo224 cells when treated with EMD1214063 (Figure 4B; right panel), we found that cell proliferation was also inhibited with increasing doses of EMD1214063 after 96 hr in culture (Figure 4C). However, when examining the proliferative capacity of LPS141 which does not have constitutive Met activity (Figure 1A), we found that a higher dose of EMD1214063 was necessary to suppress cell proliferation (Figure S3B). We found that when serum-starved Lipo246 (Figure 4D; upper panel) and Lipo224 (Figure 4D; lower panel) DDLPS cells were treated concurrently with EMD1214063 and rhHGF for 96 hours, lower doses of this small molecule kinase inhibitor were needed to reduce cell proliferation. Interestingly, cell proliferation of the HGF-inducible LPS141 cell line was similarly affected compared to Lipo224 and Lipo246 when stimulated with HGF and treated with EMD1214063 (Figure S3C). Following a 4 hr pretreatment of Lipo224, Lipo246 (Figure 4E), and LPS141 (Figure S3D) with either DMSO, or 100 nM or 1 μM EMD1214063, cells were plated in chambers using rhHGF as a chemo-attractant with migration and invasion determined after 24 hr. In all cell lines examined, we found that both migration and invasion were significantly inhibited in a dose-dependent manner in EMD1214063-treated cells as compared to control cells.
Met tyrosine kinase inhibition with EMD1214063 suppresses the tumorigenicity of DDLPS xenografts
Next, we evaluated the potential tumor inhibitory effects of EMD1214063 in vivo using the Lipo246 xenograft mouse model18. Lipo246 cells were injected subcutaneously in the flanks of NOD/SCID mice, and tumor growth was measured six times over a period of 17 days in mice receiving the vehicle control or in those dosed twice weekly with 6 mg/kg EMD1214063. Results demonstrated that xenograft volume (Figure 5A) and weight (Figure 5B) decreased significantly in EMD1214063-treated versus vehicle control-treated mice. To ensure that Met activity was indeed affected by EMD1214063 in vivo, vehicle control and EMD1214063 treated xenografts were evaluated for Met activity by western analysis (Figure 5C). We found that Met kinase activity was inhibited with EMD1214063 treatment as compared to the vehicle control-treated xenografts. These data suggest that targeting the Met signaling axis in DDLPS may be an important anti-DDLPS therapeutic strategy.
Figure 5. Met kinase inhibition decreases the tumorigenicity of Lipo246 xenografts in vivo.

(A) EMD1214063 treatment of Lipo246 xenografts decreased tumor volume and (B) tumor weight significantly within 17 days. (C) Western blots monitored whether EMD1214063 inhibited Met Y1234/Y1235 phosphorylation in EMD1214063 treated xenografts compared with vehicle control treated xenografts.
Discussion
DDLPS is an aggressive malignancy characterized by a high recurrence rate (>80%), disease progression, and an overall grim five-year disease-specific survival rate (40%)38. Despite aggressive surgery and systemic chemotherapy, little improvement has been made in DDLPS outcome, suggesting the need for therapeutic strategies targeting specific molecular abnormalities. Aberrantly activated Met signaling has been implicated in multiple human cancers; however, the role of Met in DDLPS yet to be elucidated. Evidence suggesting the critical role of Met in sarcomagenesis was proposed by data from a large panel of human sarcoma samples39; compared to controls, high expression of Met as well as elevated levels of HGF have been reported in multiple STS histologies compared to fibroblast control cells39. Consequently, and as per previously published data18, we evaluated the expression status of Met and assessed the clinical relevance of Met overexpression as a therapeutic target for DDLPS.
Using a panel of DDLPS cell lines, we observed that the Met protein and its activating ligand, HGF, is highly expressed; we investigated the specific cell-directed effects of Met inhibition. Utilizing an shRNA MET-targeted model system, it has been observed that aberrant activation of the Met signaling pathway promotes tumor progression by stimulating cell proliferation. Consistent with this observation, our results demonstrated activation of Met with rHGF promoted cell growth (Figure 1A); when Met was silenced, proliferation was inhibited (Figure 2B), an effect specific to HGF/Met activation in that cells were serum starved prior to rhHGF treatment, thereby blocking access to many nutrients that aid in cell proliferation.
Activation of Met signaling initiates downstream signaling pathways that promote migration and invasion (i.e., PI3K/AKT and ERK signaling)40. Multiple reports have demonstrated the importance of these Met-activated pathways as contributing to tumor growth, tumor cell migration, and invasion41–44. Our data demonstrate that silencing Met in DDLPS cells inhibited cellular proliferation as well as perturbed motility and invasiveness, perhaps due to the decreased activation of these downstream signaling pathways following Met activation.
DDLPS tumors are very locally recurrent and readily invade surrounding organs4,45. Our data demonstrated that Met expression and activation is critical for DDLPS cells to migrate and invade surrounding matrices as observed in cell culture-based assays. Xenograft models incorporating shMET constructs demonstrated inhibitory effects on tumor growth mediated by Met knockdown. Additionally, shMet xenograft models showed decreased Met-mediated tumorigenicity, a finding consistent with the findings in other cancer model systems.
Due to the anti-DDLPS impact of Met silencing, we investigated the pre-clinical therapeutic effects of Met inhibition. Small molecule Met inhibitors bind to the ATP-binding pocket in the kinase domain, thereby preventing activation of downstream signaling pathways46. In this study we demonstrated that the Met tyrosine kinase inhibitor, EMD1214063, is an effective anti-DDLPS therapeutic agent through inhibition of its activation, thereby preventing downstream signaling activation that underlies motility, invasion, viability, and tumorigenicity.
Aberrant Met signaling may be due to receptor overexpression or increased ligand autocrine/paracrine expression47. We detected variable Met expression and activity in DDLPS; however, there was no direct correlation of Met activity with autocrine HGF levels. Therefore, the efficacy of EMD1214063 in DDLPS cells suggests that basal Met expression may be sufficient for cell viability. Constitutive activation of Met was observed in Lipo224 despite negligible HGF levels; perhaps this activation is functioning in a ligand-independent manner, whereas the high level of HGF detected in Lipo246 cells may be a contributing factor for the observed constitutively activated Met expression.
The significant interface between tumor cells and the surrounding microenvironment may contribute to receptor-ligand activation of Met. Consequently, HGF is not recognized by human Met, and suggests that the EMD1214063 inhibited activation of Met in Lipo246 xenograft models is Met-specific. Although EMD1214063 is a potent Met inhibitor and is more than 200-fold more selective for Met than other RTKs, its therapeutic efficacy in DDLPS still may be a result of co-targeting other highly expressed RTKs (i.e., EGFR, AXL)48; a possibility we are currently exploring.
In conclusion, our data demonstrate that dysregulation of the Met signaling pathway contributes to the DDLPS malignant phenotype; inhibition of Met activation in vitro decreased tumor cell proliferation, migration and invasion. Consistent with these in vitro results, Met silencing and EMD1214063 treatment led to abrogated tumor growth in xenograft models. Molecular-driven strategies to personalize clinical approaches to DDLPS have yet to be exploited. Our data suggests that EMD1214063 may be a novel and therapeutically effective strategy for DDLPS patients, and therefore merits further laboratory and clinical research validation.
Supplementary Material
(A) qRT-PCR measured autocrine HGF mRNA levels in DDLPS cell lines cultured in media containing 10% FBS (n=3 ± SEM; samples were analyzed at least in duplicate per experiment). (B) ELISAs demonstrated that Lipo246 and Lipo815 DDLPS cell lines secrete HGF in an autocrine manner (n=2 ± SEM; t-test: *=P<0.05, **=P<0.005; at least duplicate samples per experiment).
(A) Serum-starved Lipo246 cells were pre-treated with increasing concentrations of SU11274 for 4 h, and then rhHGF stimulated for 20 min. Western blot analyses evaluated Met, AKT, and ERK suppression as a consequence of SU11274 treatment. (B) Cell proliferation of Lipo246 cells were measured via MTS assays following SU11274 treatment for 48 h (10% FBS containing media; n=3 ± SEM; t-test: *=P<0.05, ***=P<0.0001; at least duplicate samples per experiment). (C) Lipo246 cells exhibit statistically significantly decreased migratory and invasive characteristics when treated with 1 μM SU11274 for 24 h (representative images of n=3 experiments; at least duplicate samples per experiment).
(A) Western blots measured decreased Met, ERK, and AKT activity in serum-starved LPS141 cells that were pretreated for 4 h with 1 μM EMD1214063 prior to HGF stimulation (50 ng/mL) for 20 min. (B) LPS141 cells, cultured in 10% FBS-containing media, were treated with increasing concentrations of EMD1214063 for 96 h prior to examination of proliferation rates via MTS assays (n=3; at least duplicate samples per experiment). (C) LPS141 cells that were deprived of serum for 24 h were pretreated with increasing concentrations of EMD1214063 for 4 h. MTS assays measured decreased cell proliferation following 96 h of rhHGF stimulation (n=3 ± SEM; t-test: *=P<0.05, ***=P<0.0001; at least duplicate samples per experiment). (D) Migration and invasion assays evaluated LPS141 cells that had been pretreated with either DMSO, or 100 nM EMD1214063, or 1 μM EMD1214063 for 4 h after a 24 h period (n=3 ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; at least duplicate samples per experiment).
Short tandem repeat DNA fingerprinting of Lipo815. Lipo815 was profiled using short tandem repeat DNA fingerprinting technology, such that the future use of this cell line can be authenticated.
Acknowledgments
Grant Numbers and Sources of Support: These studies were supported in part by grants from the National Cancer Institute of the National Institutes of Health: U54CA168512 (R.E.P).
The authors thank Dr. Jonathan Fletcher (Brigham and Women’s, Boston, MA, USA) for kindly providing us with the LPS141 cell line. We would like to thank the Lobo and Margolis families for their philanthropic support of our liposarcoma studies. We greatly appreciate the support of the Amschwand Foundation (to D. Lev). This manuscript was supported by the SARC sarcoma SPORE (to R.E. Pollock).
Abbreviations
- DDLPS
dedifferentiated liposarcoma
- FISH
fluorescence in situ hybridization
- HGF
hepatocyte growth factor
- Met
hepatocyte growth factor receptor
- rhHGF
recombinant human HGF
- RTK
receptor tyrosine kinase
- WDLPS
well-differentiated liposarcoma
Footnotes
Supplementary information is available at Laboratory Investigation’s website.
Disclosure/Duality of Interest:
The authors declare no conflict of interest.
References
- 1.Lahat G, Zhu Q-S, Huang K-L, Wang S, Bolshakov S, Liu J, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. In: Bauer JA, editor. PLoS One. 4. Vol. 5. Public Library of Science; 2010. Jan, p. e10105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Taylor BS, DeCarolis PL, Angeles CV, Brenet F, Schultz N, Antonescu CR, et al. Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Discov. 2011 Dec;1(7):587–97. doi: 10.1158/2159-8290.CD-11-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, et al. MiR-155 is a liposarcoma oncogene that targets casein kinase-1α and enhances β-catenin signaling. Cancer Res. 2012 Apr 1;72(7):1751–62. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman A, Lazar AJ, Pollock RE, Lev D. Drug Resist Updat. 1. Vol. 14. Elsevier Ltd; 2011. Feb, New frontiers in the treatment of liposarcoma, a therapeutically resistant malignant cohort; pp. 52–66. [DOI] [PubMed] [Google Scholar]
- 5.Singer S, Socci ND, Ambrosini G, Sambol E, Decarolis P, Wu Y, et al. Gene expression profiling of liposarcoma identifies distinct biological types/subtypes and potential therapeutic targets in well-differentiated and dedifferentiated liposarcoma. Cancer Res. 2007 Jul 15;67(14):6626–36. doi: 10.1158/0008-5472.CAN-07-0584. [DOI] [PubMed] [Google Scholar]
- 6.Lahat G, Anaya DA, Wang X, Tuvin D, Lev D, Pollock RE. Resectable well-differentiated versus dedifferentiated liposarcomas: two different diseases possibly requiring different treatment approaches. Ann Surg Oncol. 2008 Jun;15(6):1585–93. doi: 10.1245/s10434-007-9805-x. [DOI] [PubMed] [Google Scholar]
- 7.Crago AM, Singer S. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol. 2011 Jul;23(4):373–8. doi: 10.1097/CCO.0b013e32834796e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arena S, Pisacane A, Mazzone M, Comoglio PM, Bardelli A. Genetic targeting of the kinase activity of the Met receptor in cancer cells. Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11412–7. doi: 10.1073/pnas.0703205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012 Jun;9(6):314–26. doi: 10.1038/nrclinonc.2012.71. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. [DOI] [PubMed] [Google Scholar]
- 10.Cortner J, Vande Woude GF, Rong S. The Met-HGF/SF autocrine signaling mechanism is involved in sarcomagenesis. EXS. 1995 Jan;74:89–121. doi: 10.1007/978-3-0348-9070-0_6. [DOI] [PubMed] [Google Scholar]
- 11.Ferracini R, Olivero M, Di Renzo MF, Martano M, De Giovanni C, Nanni P, et al. Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene. 1996 Apr 18;12(8):1697–705. [PubMed] [Google Scholar]
- 12.Ferracini R, Di Renzo MF, Scotlandi K, Baldini N, Olivero M, Lollini P, et al. The Met/HGF receptor is over-expressed in human osteosarcomas and is activated by either a paracrine or an autocrine circuit. Oncogene. 1995 Feb 16;10(4):739–49. [PubMed] [Google Scholar]
- 13.Fukuda T, Ichimura E, Shinozaki T, Sano T, Kashiwabara K, Oyama T, et al. Coexpression of HGF and c-Met/HGF receptor in human bone and soft tissue tumors. Pathol Int. 1998 Oct;48(10):757–62. doi: 10.1111/j.1440-1827.1998.tb03834.x. [DOI] [PubMed] [Google Scholar]
- 14.Patanè S, Avnet S, Coltella N, Costa B, Sponza S, Olivero M, et al. MET overexpression turns human primary osteoblasts into osteosarcomas. Cancer Res. 2006 May 1;66(9):4750–7. doi: 10.1158/0008-5472.CAN-05-4422. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Farenholtz KE, Yang Y, Guessous F, Dipierro CG, Calvert VS, et al. Hepatocyte growth factor sensitizes brain tumors to c-MET kinase inhibition. Clin Cancer Res. 2013 Mar 15;19(6):1433–44. doi: 10.1158/1078-0432.CCR-12-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006 May 1;66(9):4742–9. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 17.Torres KE, Zhu Q-S, Bill K, Lopez G, Ghadimi MP, Xie X, et al. Activated MET is a molecular prognosticator and potential therapeutic target for malignant peripheral nerve sheath tumors. Clin Cancer Res. 2011 Jun 15;17(12):3943–55. doi: 10.1158/1078-0432.CCR-11-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng T, Zhang P, Liu J, Nguyen T, Bolshakov S, Belousov R, et al. An experimental model for the study of well-differentiated and dedifferentiated liposarcoma; deregulation of targetable tyrosine kinase receptors. Lab Invest. 2011 Mar;91(3):392–403. doi: 10.1038/labinvest.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnett J, Chumbalkar V, Vaillant B, Gururaj AE, Hill KS, Latha K, et al. Regulation of HGF expression by ΔEGFR-mediated c-Met activation in glioblastoma cells. Neoplasia. 2013 Jan;15(1):73–84. doi: 10.1593/neo.121536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migliore C, Giordano S. Molecular cancer therapy: can our expectation be MET? Eur J Cancer. 2008 Mar;44(5):641–51. doi: 10.1016/j.ejca.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Goldoni S, Humphries A, Nyström A, Sattar S, Owens RT, McQuillan DJ, et al. Decorin is a novel antagonistic ligand of the Met receptor. J Cell Biol. 2009 May 18;185(4):743–54. doi: 10.1083/jcb.200901129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maulik G, Madhiwala P, Brooks S, Ma PC, Kijima T, Tibaldi EV, et al. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. J Cell Mol Med. 6(4):539–53. doi: 10.1111/j.1582-4934.2002.tb00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, et al. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009 May;11(5):448–58. doi: 10.1593/neo.09230. 2 p following 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gherardi E, Youles ME, Miguel RN, Blundell TL, Iamele L, Gough J, et al. Functional map and domain structure of MET, the product of the c-met protooncogene and receptor for hepatocyte growth factor/scatter factor. Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12039–44. doi: 10.1073/pnas.2034936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010 Dec;11(12):834–48. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 26.Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of Met is necessary for receptor dimerization and activation. Cancer Cell. 2004 Jul;6(1):75–84. doi: 10.1016/j.ccr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Bardelli A, Pugliese L, Comoglio PM. “Invasive-growth” signaling by the Met/HGF receptor: the hereditary renal carcinoma connection. Biochim Biophys Acta. 1997 Dec 9;1333(3):M41–51. doi: 10.1016/s0304-419x(97)00026-7. [DOI] [PubMed] [Google Scholar]
- 28.Peruzzi B, Bottaro DP. Targeting the c-Met signaling pathway in cancer. Clin Cancer Res. 2006 Jun 15;12(12):3657–60. doi: 10.1158/1078-0432.CCR-06-0818. [DOI] [PubMed] [Google Scholar]
- 29.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003 Dec;4(12):915–25. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 30.Tolbert WD, Daugherty-Holtrop J, Gherardi E, Vande Woude G, Xu HE. Structural basis for agonism and antagonism of hepatocyte growth factor. Proc Natl Acad Sci U S A. 2010 Jul 27;107(30):13264–9. doi: 10.1073/pnas.1005183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009 Apr 1;15(7):2207–14. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- 32.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012 Feb;12(2):89–103. doi: 10.1038/nrc3205. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. [DOI] [PubMed] [Google Scholar]
- 33.Weaver J, Downs-Kelly E, Goldblum JR, Turner S, Kulkarni S, Tubbs RR, et al. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod Pathol. 2008 Aug;21(8):943–9. doi: 10.1038/modpathol.2008.84. [DOI] [PubMed] [Google Scholar]
- 34.Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011 Aug 18;30(33):3625–35. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moschetta M, Basile A, Ferrucci A, Frassanito MA, Rao L, Ria R, et al. Novel targeting of phospho-cMET overcomes drug resistance and induces antitumor activity in multiple myeloma. Clin Cancer Res. 2013 Aug 15;19(16):4371–82. doi: 10.1158/1078-0432.CCR-13-0039. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011 Jun 14;108(24):9951–6. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bladt F, Faden B, Friese-Hamim M, Knuehl C, Wilm C, Fittschen C, et al. EMD 1214063 and EMD 1204831 constitute a new class of potent and highly selective c-Met inhibitors. Clin Cancer Res. 2013 Jun 1;19(11):2941–51. doi: 10.1158/1078-0432.CCR-12-3247. [DOI] [PubMed] [Google Scholar]
- 38.Fletcher CDM, Unni KK, Mertens F. In: World Health Organization Classification of Tumours Pathology and Genetics of Tumours of Soft Tissue and Bone. 4. Fletcher CDM, Unni KK, Mertens F, editors. Lyon, France: IARC Press; 2013. [Google Scholar]
- 39.Rong S, Jeffers M, Resau JH, Oskarsson M, Vande Woude GF. Met Expression and Sarcoma Tumorigenicity. Cancer Res. 1993;53:5355–60. [PubMed] [Google Scholar]
- 40.Scagliotti GV, Novello S, von Pawel J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat Rev. 2013 Nov;39(7):793–801. doi: 10.1016/j.ctrv.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Que W, Chen J. Knockdown of c-Met inhibits cell proliferation and invasion and increases chemosensitivity to doxorubicin in human multiple myeloma U266 cells in vitro. Mol Med Rep. 2011;4(2):343–9. doi: 10.3892/mmr.2011.426. [DOI] [PubMed] [Google Scholar]
- 42.Previdi S, Abbadessa G, Dalò F, France DS, Broggini M. Breast cancer-derived bone metastasis can be effectively reduced through specific c-MET inhibitor tivantinib (ARQ 197) and shRNA c-MET knockdown. Mol Cancer Ther. 2012 Jan;11(1):214–23. doi: 10.1158/1535-7163.MCT-11-0277. [DOI] [PubMed] [Google Scholar]
- 43.Scorsone K, Zhang L, Woodfield SE, Hicks J, Zage PE. The novel kinase inhibitor EMD1214063 is effective against neuroblastoma. Invest New Drugs. 2014 May 16; doi: 10.1007/s10637-014-0107-4. [DOI] [PubMed] [Google Scholar]
- 44.Humbert M, Medová M, Aebersold DM, Blaukat A, Bladt F, Fey MF, et al. Protective autophagy is involved in resistance towards MET inhibitors in human gastric adenocarcinoma cells. Biochem Biophys Res Commun. 2013 Feb 8;431(2):264–9. doi: 10.1016/j.bbrc.2012.12.120. [DOI] [PubMed] [Google Scholar]
- 45.Ghadimi MP, Al-Zaid T, Madewell J, Peng T, Colombo C, Hoffman A, et al. Diagnosis, management, and outcome of patients with dedifferentiated liposarcoma systemic metastasis. Ann Surg Oncol. 2011 Dec;18(13):3762–70. doi: 10.1245/s10434-011-1794-0. [DOI] [PubMed] [Google Scholar]
- 46.Cui JJ. Targeting Receptor Tyrosine Kinase MET in Cancer: Small Molecule Inhibitors and Clinical Progress. J Med Chem. American Chemical Society. 2013 Dec 18; doi: 10.1021/jm401427c. [DOI] [PubMed] [Google Scholar]
- 47.Organ SL, Tsao M-S. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011 Nov;3(1 Suppl):S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comoglio PM, Giordano S, Trusolino L. Nat Rev Drug Discov. 6. Vol. 7. Nature Publishing Group; 2008. Jun, Drug development of MET inhibitors: targeting oncogene addiction and expedience; pp. 504–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) qRT-PCR measured autocrine HGF mRNA levels in DDLPS cell lines cultured in media containing 10% FBS (n=3 ± SEM; samples were analyzed at least in duplicate per experiment). (B) ELISAs demonstrated that Lipo246 and Lipo815 DDLPS cell lines secrete HGF in an autocrine manner (n=2 ± SEM; t-test: *=P<0.05, **=P<0.005; at least duplicate samples per experiment).
(A) Serum-starved Lipo246 cells were pre-treated with increasing concentrations of SU11274 for 4 h, and then rhHGF stimulated for 20 min. Western blot analyses evaluated Met, AKT, and ERK suppression as a consequence of SU11274 treatment. (B) Cell proliferation of Lipo246 cells were measured via MTS assays following SU11274 treatment for 48 h (10% FBS containing media; n=3 ± SEM; t-test: *=P<0.05, ***=P<0.0001; at least duplicate samples per experiment). (C) Lipo246 cells exhibit statistically significantly decreased migratory and invasive characteristics when treated with 1 μM SU11274 for 24 h (representative images of n=3 experiments; at least duplicate samples per experiment).
(A) Western blots measured decreased Met, ERK, and AKT activity in serum-starved LPS141 cells that were pretreated for 4 h with 1 μM EMD1214063 prior to HGF stimulation (50 ng/mL) for 20 min. (B) LPS141 cells, cultured in 10% FBS-containing media, were treated with increasing concentrations of EMD1214063 for 96 h prior to examination of proliferation rates via MTS assays (n=3; at least duplicate samples per experiment). (C) LPS141 cells that were deprived of serum for 24 h were pretreated with increasing concentrations of EMD1214063 for 4 h. MTS assays measured decreased cell proliferation following 96 h of rhHGF stimulation (n=3 ± SEM; t-test: *=P<0.05, ***=P<0.0001; at least duplicate samples per experiment). (D) Migration and invasion assays evaluated LPS141 cells that had been pretreated with either DMSO, or 100 nM EMD1214063, or 1 μM EMD1214063 for 4 h after a 24 h period (n=3 ± SEM; t-test: *=P<0.05, **=P<0.005, ***=P<0.0001; at least duplicate samples per experiment).
Short tandem repeat DNA fingerprinting of Lipo815. Lipo815 was profiled using short tandem repeat DNA fingerprinting technology, such that the future use of this cell line can be authenticated.