Dear Editor
Schizophrenia (SZ) is a devastating psychiatric disorder hypothesized to be a neurodevelopmental condition (1, 2); arising as a consequence of dysregulation of brain development (3, 4). WNT signaling is important for neural patterning, proliferation and migration, and synapse formation (reviewed by (5)); moreover, converging post-mortem (6, 7), rodent (8, 9) and pharmacological (10) evidence suggests that WNT signaling may contribute to SZ (reviewed by (11, 12)). We utilized human induced pluripotent stem cell (hiPSC) derived forebrain patterned neural progenitor cells (NPCs) (13, 14) to investigate canonical WNT activity in a pilot cohort of four SZ patients.
Because all research described herein was performed on deidentified human samples obtained for broadly consented scientific research by either American Type Culture Collection (ATCC) or the Coriell Cell Repository, is was found to be exempt by the Internal Review Committee of the Icahn School of Medicine at Mount Sinai. This work was also reviewed by the Embryonic Stem Cell Research Oversight Committee at the Icahn School of Medicine at Mount Sinai.
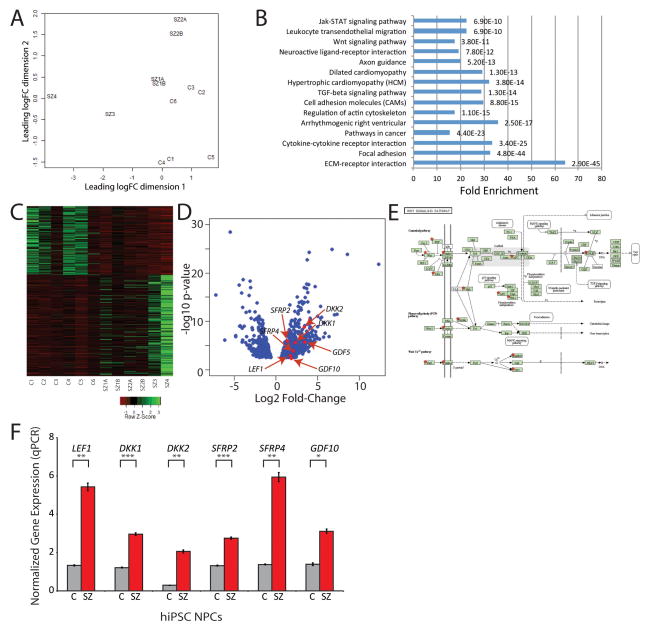
We compared global transcription of forebrain hiPSC NPCs from six control and four SZ patients by RNAseq (Table 1; GSE63738), cultured as described (13, 14). As previously reported, hiPSC forebrain NPCs differentiate to a mixed neuronal population of glutamatergic and GABAergic neurons; there was no difference in the ability of control or SZ hiPSC NPCs to generate βIII-TUBULIN-positive neurons (14) and neither transcriptional nor immunohistochemical characterization revealed any diagnosis-dependent differences in the regional patterning of forebrain NPCs (13). Multi-dimensional scaling (MDS) resolved most SZ and control hiPSC NPC samples (Fig. 1A); 848 genes were significantly differentially expressed (FDR<0.05) (SI Table 1), as illustrated by a heat map (Fig. 1C) and a volcano plot (Fig. 1D). The differentially expressed genes in SZ hiPSC NPCs were significantly 3.6-fold enriched when compared to WNT target genes (p < 10e-20) predicted by standard Classification and Regression Tree (CART) methods (16). The differentially expressed genes (FDR < 0.05) were submitted to DAVID (http://david.abcc.ncifcrf.gov), which identified several significantly enriched pathways (Fig. 1B; Tables 2–3), including the WNT signaling pathway: 17.3-fold enrichment (p < 10e-13; FDR < 10e-11). The perturbed WNT genes are marked by red stars in the WNT signaling pathway diagram (Fig. 1E; Table 2). 6/6 differentially expressed WNT genes identified by RNAseq were confirmed when tested by qPCR (Fig. 1F; Table 2).
Table 1.
Description of known clinical information for the four Coriell SZ patients.
| Coriell ID | Sex | Ethnicity | Age | Age of Onset | Phenotype | Hospitalizations? | Family History |
|---|---|---|---|---|---|---|---|
| GM02038 | M | Causasian | 22y | 6 years | suicide | ? | unknown |
| GM01792 | M | Causasian Jewish/Scandanavian | 26y | unknown | episodes of agitation, delusions of persecution, and fear of assassination; at age four mild features of pervasive developmental disorder | ? | father and sister affected; brother autistic at age four |
| GM01835 | F | Causasian Jewish | 27y | unknown | drug abuse; schizo-affective disorder | Yes | father and brother affected |
| GM02497 | M | Causasian Jewish | 23y | 15 years | paralogical thinking, affective shielding, splitting of affect from content, and suspiciousness | Yes | affected father, anorexic/schizoid sister |
Fig. 1. RNAseq comparisons of control and SZ hiPSC NPCs.
A. Multidimensional scaling (MDS) of RNAseq gene expression of hiPSC NPCs from each of six control and four SZ patients segregates samples along the two leading fold change dimensions. Gene expression analysis was performed on passage-matched hiPSC forebrain NPCs cultured on matrigel. Cells were lysed in RNA BEE (Tel-test, Inc). RNA was chloroform extracted and treated with RQ1 RNAse-free DNAse (Promega). RNAseq samples were prepared using the Illumina HiSeq 2500 RNA kit for 100nt/single end reads, four samples were run per lane. Raw cDNA reads were aligned to the hg19 reference with the spliced gap aligner Spliced Transcripts Alignment to a Reference (STAR) software, with count-based quantitation carried out via the Subread package featureCounts (http://bioconductor.org/packages/release/bioc/html/Rsubread.html) at both the geneic and exonic levels for UCSC and ensemble annotation builds. B. Pathway enrichment analysis based on DAVID. X-axis represents fold enrichment; Y-axis denotes pathways. The FDR are labeled on the right of the bar plot. C. Heat map of control and SZ hiPSC NPCs of 848 unique genes (FDR<0.05). The count data were normalized and modeled as over-dispersed Poisson data using a negative binomial model in the Bioconductor package edgeR (15). Fold changes, p-values and false discovery rates (FDRs) are obtained from the same package for integrative analysis. D. Volcano plots of −log10 p-value versus log2 fold-change mRNA levels for control and SZ hiPSC NPCs. Key canonical WNT signaling genes, including Lymphoid Enhancer-Binding Factor 1 (LEF1), Dickkopf-1 (DKK1), DKK2, Secreted frizzled-related protein-2 (SFRP2), SFRP4, growth differentiation factor 5 (GDF5) and GDF10, are indicated. E. Wnt signaling pathway. The differentially expressed genes by RNAseq are marked by red stars. F. qPCR validation of perturbed WNT gene expression, normalized to the expression of the housekeeping genes GAPDH and ACTIN: LEF1, DKK1, DKK2, SFRP2, SFRP4, and GDF10. Error bars are s.e. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 2.
Selected WNT signalling genes differentially expressed in SZ hiPSC NPCs.
| RNAseq | qPCR | |||||
|---|---|---|---|---|---|---|
| Symbol | Refseq ID | Fold-Change | P-value | FDR | Fold-Change | P-value |
| DKK1 | NM_012242 | 2.32 | 2.23E-06 | 1.43E-04 | 2.43 | 7.78E-04 |
| DKK2 | NM_014421 | 3.35 | 1.45E-09 | 2.38E-07 | 7.11 | 1.08E-03 |
| SFRP2 | NM_003013 | 1.33 | 1.21E-06 | 8.61E-05 | 2.08 | 3.14E-04 |
| SFRP4 | NM_003014 | 2.05 | 1.22E-04 | 3.87E-03 | 4.31 | 3.37E-03 |
| GDF5 | NM_000557 | 3.42 | 1.75E-06 | 1.18E-04 | - | - |
| GDF10 | NM_004962 | 1.82 | 4.16E-03 | 6.58E-02 | 2.24 | 3.77E-02 |
| LEF1 | NM_016269 | 1.68 | 1.19E-03 | 2.51E-02 | 4.09 | 1.73E-03 |
Table 3.
Enrichment analysis for BMP signaling, hedgehog signaling or GPCR signaling pathways.
| Category | Term | Fold Enrichment | FDR |
|---|---|---|---|
| KEGG_PATHWAY | Hedgehog signaling pathway | 18.7 | 0.02 |
| REACTOME_PATHWAY | Signaling by BMP | 0.7 | 1.00 |
| REACTOME_PATHWAY | Signaling by GPCR | 2.1 | 1.00 |
We investigated canonical WNT activity using the well-established T-cell factor (TCF) / Lymphoid enhancer-binding factor (LEF) (TOPFlash) assay, in which transcriptional activation of TCF/LEF binding sites drives expression of a luciferase reporter (17, 18). NPCs were infected 3–7 days prior to analysis with a Lentiviral (LV)-TOPFLASH luciferase reporter, generously provided by Karl Willert (UCSD), as well as a constitutive LV-renilla reporter for normalization. SZ hiPSC NPCs showed increased canonical WNT signaling relative to controls (p<10e-5) (Fig. 2A, Fig. 3), though increased WNT signaling was not necessarily present in every patient and significant outliers often skewed results (Fig. 3). Across six independent experimental replicates, the following fold-changes in canonical WNT signaling we observed: 2.8, 4.2, 3.1, 2.7, 4.7 and 3.3 (Fig. 3). The ultimate effector of canonical WNT signaling is β-CATENIN; Western blot analysis for β-CATENIN protein (1:10,000; Millipore), normalized to β-ACTIN (1:10,000; Ambion), revealed increased β-CATENIN protein levels in SZ hiPSC NPCs (Fig. 2B).
Fig. 2. Perturbed WNT signaling in SZ hiPSC forebrain NPCs.
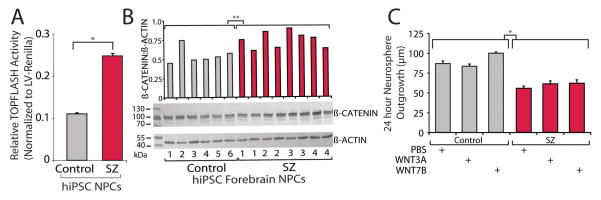
A. Comparison of canonical WNT activity (assayed as LV-TOPFLASH reporter levels relative to LV-renilla florescence) between control and SZ hiPSC forebrain NPCs, averaged by diagnosis. Luciferase levels were determined using the Dual-Glo Luciferase Assay System (Promega), measured on a FlexStation 3 (Molecular Devices) and then normalized to LV-renilla florescence. B. Increased β-CATENIN protein levels in SZ hiPSC forebrain NPCs. Western blot comparison of β-CATENIN and β-ACTIN levels in control and SZ hiPSC NPCs. Western blots were repeated twice using independent protein lysates; Student’s T tests were used to test statistical differences between control and SZ western blot β-CATENIN levels. β-ACTIN was used as a loading control because we have found no evidence, by microarray or Nanostring nCounter gene expression assays or SILAC quantitative protein mass spectrometry, that it is differentially expressed in SZ hiPSC NPCs or neurons (13, 14). C. No effect of WNT on aberrant migration in SZ hiPSC forebrain NPCs. Control and SZ neurosphere outgrowth when cultured with PBS, canonical WNT3A (20 ng/ml) and noncanonical WNT7B (5 ng/ml). Error bars are s.e. *P < 0.05, **P < 0.01.
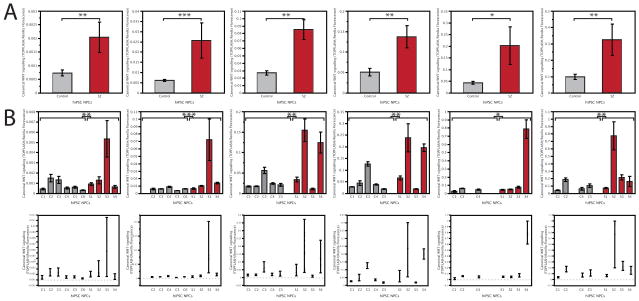
Fig. 3. Experimental variability in assaying WNT signaling in SZ hiPSC forebrain NPCs.
A. Six experimental replicates comparing canonical WNT activity (assayed as LV-TOPFLASH reporter levels relative to LV-renilla florescence) between control and SZ hiPSC forebrain NPCs, averaged by diagnosis. With increasing passage, NPC lines can show reduced ability to differentiate to neurons or undergo spontaneous transformation to a highly proliferative cell with rounded morphology that cannot undergo neural differentiation at all; when either event occurred, that NPC line was dropped from subsequent experiments, for this reason, not all NPC lines were analysed in independent experiments. B. Six experimental replicates comparing canonical WNT activity (assayed as LV-TOPFLASH reporter levels relative to LV-renilla florescence) between control and SZ hiPSC forebrain NPCs, averaged by individual. (Top row: Mean +/− s.e. Bottom row: Variability chart showing individual data points). For phenotypic analysis, statistical analysis was performed using JMP (Carey, NC). Box-Cox transformation of raw data was performed to correct non-normal distribution of the data and means were compared within diagnosis by Oneway analysis using both Student’s T test and Tukey Kramer HSD. A nested analysis of values for individual patients was performed using standard least squares analysis comparing means for all pairs using Student’s T test for specific pairs and Tukey Kramer HSD for multiple comparisons. Error bars are s.e. *P < 0.05, **P < 0.01, ***P < 0.001.
WNT signaling has been implicated in neural migration (19). Following 48 hours of culture with either canonical (20ng/ml WNT3A) or non-canonical (5ng/ml WNT7B) WNT signals, neither control nor SZ hiPSC forebrain NPCs showed significant changes in radial migration (98 total SZ neurospheres were analyzed relative to 56 total control neurospheres) (Fig. 2C); increased canonical WNT signaling was not sufficient to recapitulate SZ aberrant migration in control hiPSC derived neurospheres (14).
Consistent with evidence suggesting that the WNT pathway could be aberrant in SZ (20), we demonstrate that SZ hiPSC forebrain NPCs derived from four patients have perturbations in WNT signaling, but caution that i) due to our small sample size, these phenotypes may not generalize across all SZ patients and ii) there was substantial variation in the specific SZ hiPSC NPC lines with increased WNT signaling between experimental replicates. SZ hiPSC NPCs with elevated canonical WNT signaling showed significantly increased experimental variation, suggesting that this phenotype might be more accurately reflect an increased variability in WNT signaling, perhaps due to increased susceptibility to an extrinsic factor, rather than implying a cell-autonomous difference in canonical WNT signaling.
A question of immediate interest is whether WNT signaling is also perturbed in SZ hiPSC neurons, and if so, in which neuronal cell types this is most evident. WNT signaling has been implicated in neural patterning, proliferation, differentiation, migration and activity-dependent synaptic modulation (12, 19, 21–25). Given that WNT signaling is typically believed to increase neurogenesis (26), and that we and others have reported reduced neuronal connectivity in SZ hiPSC neurons (14, 27), we note that the (increased) direction of change in WNT signaling observed in our hiPSC NPCs is potentially surprising, though it may reflect an attempt at compensation for neural defects in other pathway(s). Perturbations in canonical WNT signaling in SZ hiPSC NPCs foretells a practical confound for future hiPSC-based studies of SZ because aberrant canonical WNT signaling might affect the specification of SZ hiPSCs to certain neural fates. During neuronal differentiation, active WNT signaling is required for the specification of hippocampal (28) and midbrain dopaminergic fate (29, 30), while repression of WNT signaling is required for cortical interneuron (31, 32) and striatum (33, 34) neuronal patterning; two recent publications have reported differing abilities of SZ hiPSCs to differentiate into dopaminergic neurons (27, 35).
Recent rodent-(36), hiPSC-(13, 27, 37, 38) and olfactory neural stem cell-(39) based studies of SZ have reported increased oxidative stress and reactive oxygen species. There is a well-documented cross-talk between redox and WNT/β-catenin signaling (40–44); for example, treatment of cells with a low dose of H2O2, induces a rapid stabilization of β-catenin (43), while down-regulation of canonical WNT signaling can decrease oxidative stress (45). If increased oxidative stress does indeed contribute to perturbed canonical WNT signaling in SZ hiPSC NPCs, small variations in tissue culture induced oxidative stress between experimental replicates may be one source of the large experimental variation observed between SZ patients. Future studies, across larger patient cohorts, will be necessary to determine whether aberrant canonical WNT signaling is a causal molecular factor contributing to aberrant neural patterning and neuronal maturation in SZ, or simply a non-cell autonomous consequence of increased oxidative stress (46).
Supplementary Material
Acknowledgments
We would like to thank the laboratory of Fred H. Gage for supporting the early experiments of this work. Kristen Brennand is a New York Stem Cell Foundation - Robertson Investigator. The Brennand Laboratory is supported by a Brain and Behavior Young Investigator Grant, National Institute of Health (NIH) grant R01 MH101454 and the New York Stem Cell Foundation. This work is also partially supported by NIH grant R01 MH097276 (G.F.).
ABBREVIATIONS
- SZ
schizophrenia
- hiPSC
human induced pluripotent stem cell
- NPC
neural progenitor cell
- FDR
false discovery rate
Footnotes
AUTHOR CONTRIBUTIONS
A.T., A.S., and N.T. performed and analyzed the experiments. S.Z. and G.F. completed the RNAseq analysis. K.J.B. designed the experiments and wrote the manuscript.
As per our agreement with Coriell Cell Repository, some hiPSC lines generated from control and SZ fibroblasts will be available from Coriell.
The authors have declared that no competing interests exist.
References
- 1.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 2.Marin O, Baker J, Puelles L, Rubenstein JL. Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development. 2002;129:761–773. doi: 10.1242/dev.129.3.761. [DOI] [PubMed] [Google Scholar]
- 3.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarskog LF, Miyamoto S, Lieberman JA. Schizophrenia: new pathological insights and therapies. Annu Rev Med. 2007;58:49–61. doi: 10.1146/annurev.med.58.060904.084114. [DOI] [PubMed] [Google Scholar]
- 5.Gage FH. Molecular and cellular mechanisms contributing to the regulation, proliferation and differentiation of neural stem cells in the adult dentate gyrus. The Keio journal of medicine. 2010;59:79–83. doi: 10.2302/kjm.59.79. [DOI] [PubMed] [Google Scholar]
- 6.Pietersen CY, Mauney SA, Kim SS, Passeri E, Lim MP, Rooney RJ, et al. Molecular profiles of parvalbumin-immunoreactive neurons in the superior temporal cortex in schizophrenia. J Neurogenet. 2014;28:70–85. doi: 10.3109/01677063.2013.878339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter D, Kerwin R, al-Sarraji S, Brion JP, Chadwich A, Lovestone S, et al. Abnormalities of Wnt signalling in schizophrenia--evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–1383. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- 8.Durak O, de Anda FC, Singh KK, Leussis MP, Petryshen TL, Sklar P, et al. Ankyrin-G regulates neurogenesis and Wnt signaling by altering the subcellular localization of beta-catenin. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, et al. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutton LP, Rushlow WJ. Regulation of Akt and Wnt signaling by the group II metabotropic glutamate receptor antagonist LY341495 and agonist LY379268. J Neurochem. 2011;117:973–983. doi: 10.1111/j.1471-4159.2011.07268.x. [DOI] [PubMed] [Google Scholar]
- 11.Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am J Psychiatry. 2010;167:388–396. doi: 10.1176/appi.ajp.2009.08121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodar C, Assar R, Colombres M, Aravena A, Pavez L, Gonzalez M, et al. Genome-wide identification of new Wnt/beta-catenin target genes in the human genome using CART method. BMC Genomics. 2010;11:348. doi: 10.1186/1471-2164-11-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 18.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 19.De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- 20.Kalkman HO. Altered growth factor signaling pathways as the basis of aberrant stem cell maturation in schizophrenia. Pharmacology & therapeutics. 2009;121:115–122. doi: 10.1016/j.pharmthera.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Woodhead GJ, Mutch CA, Olson EC, Chenn A. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J Neurosci. 2006;26:12620–12630. doi: 10.1523/JNEUROSCI.3180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verani R, Cappuccio I, Spinsanti P, Gradini R, Caruso A, Magnotti MC, et al. Expression of the Wnt inhibitor Dickkopf-1 is required for the induction of neural markers in mouse embryonic stem cells differentiating in response to retinoic acid. J Neurochem. 2007;100:242–250. doi: 10.1111/j.1471-4159.2006.04207.x. [DOI] [PubMed] [Google Scholar]
- 23.Patil ST, Zhang L, Martenyi F, Lowe SL, Jackson KA, Andreev BV, et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: a randomized Phase 2 clinical trial. Nat Med. 2007;13:1102–1107. doi: 10.1038/nm1632. [DOI] [PubMed] [Google Scholar]
- 24.Patterson SL, Pittenger C, Morozov A, Martin KC, Scanlin H, Drake C, et al. Some forms of cAMP-mediated long-lasting potentiation are associated with release of BDNF and nuclear translocation of phospho-MAP kinase. Neuron. 2001;32:123–140. doi: 10.1016/s0896-6273(01)00443-3. [DOI] [PubMed] [Google Scholar]
- 25.Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- 26.Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 27.Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- 28.Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. 2014;2:295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hook V, Brennand K, Kim Y, Toneff T, Funkelstein L, Ziegler M, et al. Human iPSC Neurons Display Activity-Dependent Neurotransmitter Secretion: Aberrant Catecholamine Levels in Schizophrenia Neurons. Stem Cell Reports. 2014:3. doi: 10.1016/j.stemcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atkin TA, MacAskill AF, Brandon NJ, Kittler JT. Disrupted in Schizophrenia-1 regulates intracellular trafficking of mitochondria in neurons. Mol Psychiatry. 2011;16:122–124. 121. doi: 10.1038/mp.2010.110. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto-Torii K, Torii M, Fujimoto M, Nakai A, El Fatimy R, Mezger V, et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron. 2014;82:560–572. doi: 10.1016/j.neuron.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen BD, Maciel RD, Galina A, da Silveira MS, Souza CD, Drummond H, et al. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2011 doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- 39.Kano S, Colantuoni C, Han F, Zhou Z, Yuan Q, Wilson A, et al. Genome-wide profiling of multiple histone methylations in olfactory cells: further implications for cellular susceptibility to oxidative stress in schizophrenia. Mol Psychiatry. 2013;18:740–742. doi: 10.1038/mp.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 41.Tao GZ, Lehwald N, Jang KY, Baek J, Xu B, Omary MB, et al. Wnt/beta-catenin signaling protects mouse liver against oxidative stress-induced apoptosis through the inhibition of forkhead transcription factor FoxO3. J Biol Chem. 2013;288:17214–17224. doi: 10.1074/jbc.M112.445965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, et al. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol. 2010;30:2636–2650. doi: 10.1128/MCB.01194-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol. 2006;8:501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- 44.Kajla S, Mondol AS, Nagasawa A, Zhang Y, Kato M, Matsuno K, et al. A crucial role for Nox 1 in redox-dependent regulation of Wnt-beta-catenin signaling. FASEB J. 2012;26:2049–2059. doi: 10.1096/fj.11-196360. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Sun Y, Wang F, Wang Z, Peng Y, Li R. Downregulating the canonical Wnt/beta-catenin signaling pathway attenuates the susceptibility to autism-like phenotypes by decreasing oxidative stress. Neurochem Res. 2012;37:1409–1419. doi: 10.1007/s11064-012-0724-2. [DOI] [PubMed] [Google Scholar]
- 46.Emiliani FE, Sedlak TW, Sawa A. Oxidative stress and schizophrenia: recent breakthroughs from an old story. Curr Opin Psychiatry. 2014;27:185–190. doi: 10.1097/YCO.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.