Background: Systemic RNA interference deficiency-1 (SID-1) is a membrane protein required for cellular uptake of RNA in C. elegans.
Results: SID-1 extracellular domain selectively binds long dsRNA.
Conclusion: Binding affinity between SID-1 ECD and dsRNA is related to RNA transport efficiency by SID-1.
Significance: This defines SID-1 ECD as a functional domain for dsRNA recognition.
Keywords: RNA, RNA-binding protein, RNA interference (RNAi), RNA transport, RNA-protein interaction, SID-1, SIDT1, SIDT2
Abstract
During systemic RNA interference (RNAi) in Caenorhabditis elegans, RNA spreads across different cells and tissues in a process that requires the systemic RNA interference deficient-1 (sid-1) gene, which encodes an integral membrane protein. SID-1 acts cell-autonomously and is required for cellular import of interfering RNAs. Heterologous expression of SID-1 in Drosophila Schneider 2 cells enables passive uptake of dsRNA and subsequent soaking RNAi. Previous studies have suggested that SID-1 may serve as an RNA channel, but its precise molecular role remains unclear. To test the hypothesis that SID-1 mediates a direct biochemical recognition of RNA molecule and subsequent permeation, we expressed the extracellular domain (ECD) of SID-1 and purified it to near homogeneity. Recombinant purified SID-1 ECD selectively binds dsRNA but not dsDNA in a length-dependent and sequence-independent manner. Genetic missense mutations in SID-1 ECD causal for deficient systemic RNAi resulted in significant reduction in its affinity for dsRNA. Furthermore, full-length proteins with these mutations decrease SID-1-mediated RNA transport efficiency, providing evidence that dsRNA binding to SID-1 ECD is related to RNA transport. To examine the functional similarity of mammalian homologs of SID-1 (SIDT1 and SIDT2), we expressed and purified mouse SIDT1 and SIDT2 ECDs. We show that they bind long dsRNA in vitro, supportive of dsRNA recognition. In summary, our study illustrates the functional importance of SID-1 ECD as a dsRNA binding domain that contributes to RNA transport.
Introduction
RNA interference (RNAi) in Caenorhabditis elegans is systemic and requires the spread of silencing RNA from an initial site to adjacent tissues and their progeny (1). A genetic screen in C. elegans found a gene termed sid-1 (systemic RNA interference-deficient-1) is required for systemic but not cell-autonomous RNAi (2). sid-1 encodes a novel membrane protein, SID-1, that is widely expressed in non-neuronal cells, especially those exposed to the environment, and localizes to the cell periphery (2). Although C. elegans neuronal cells are normally resistant to systemic RNAi, ectopic expression of SID-1 in these cells sensitize them to systemic RNAi (3). Genetic evidence suggests that SID-1 functions cell-autonomously and is involved in import rather than export of interfering RNAs between cells (2, 4), but the precise molecular function and biochemical activity of SID-1 in RNA transport remains unresolved.
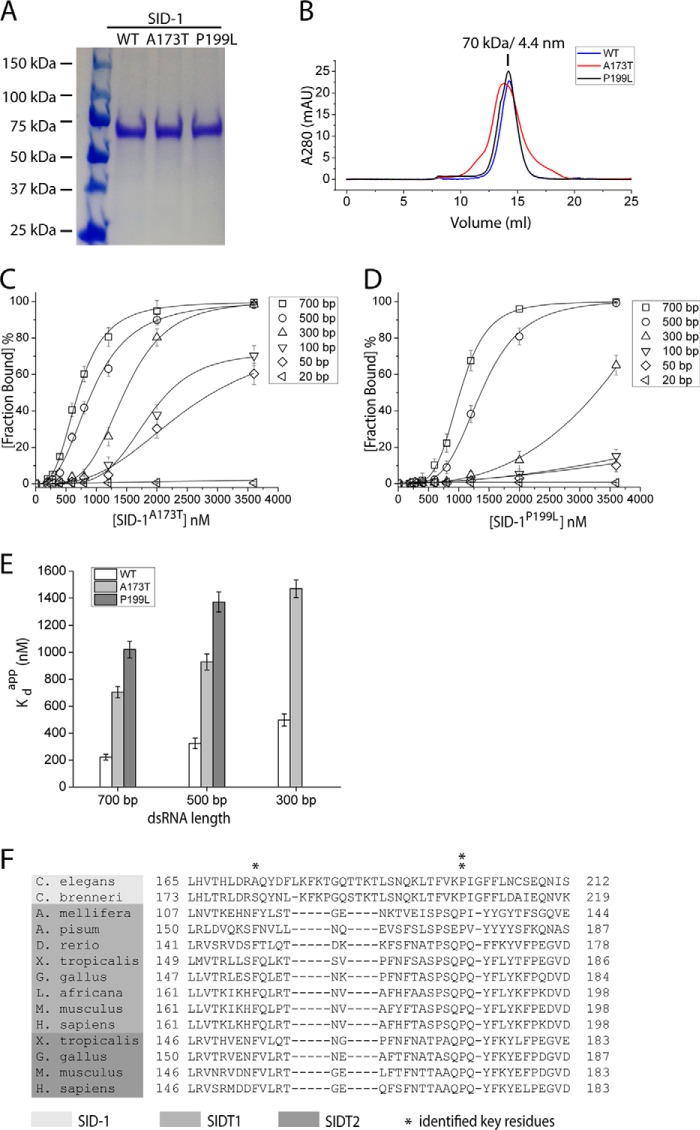
SID-1 is an integral membrane protein that contains a N-terminal extracellular domain (ECD)2 and 11 predicted transmembrane helices (6) (Fig. 1A). Several loss-of-function mutations were identified through a genetic screen in C. elegans (2) and map to both the ECD (A173T and P199L) and the transmembrane regions (P328L, S536I, and R565C), suggesting that both elements are important for SID-1 function.
FIGURE 1.
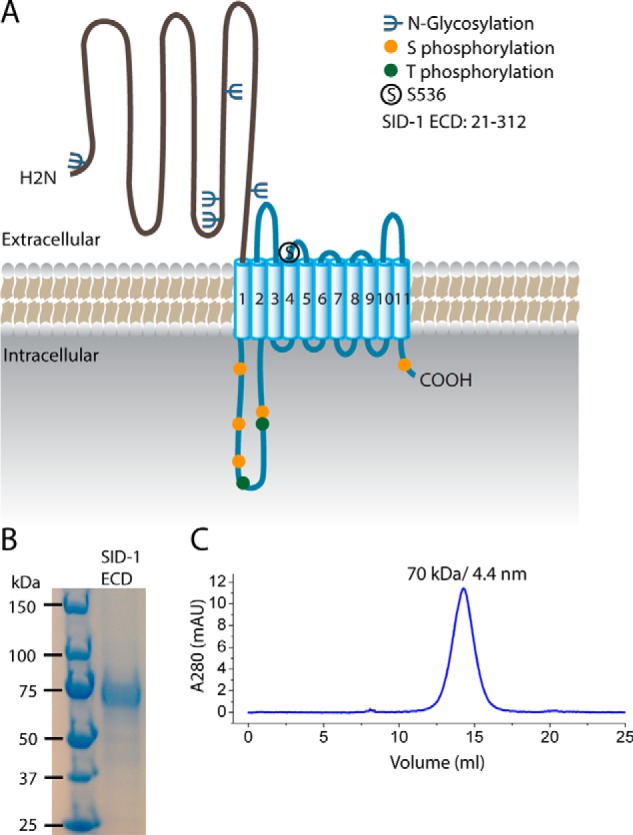
Purification of the SID-1 extracellular domain. A, predicted structural organization and post-translational modifications of SID-1. SID-1 has an extracellular domain (∼300 amino acids) with multiple N-linked glycosylation sites, a multipass transmembrane region carrying a key residue, Ser-536, a cytoplasmic loop having several Ser/Thr phosphorylation sites, and a short cytosolic tail. B, SDS-PAGE analysis and Coomassie Blue staining of purified recombinant SID-1 ECD with hGH-His tag. The disperse band is most likely due to N-linked glycosylation. C, SEC profile of SID-1 ECD using a Superdex 200 column. The main peak corresponds to 70 kDa, as determined by molecular weight standards, and its calculated Stokes radius is 4.4 nm.
Previous studies of the cellular function of SID-1 suggest that SID-1 possesses transport activities for RNA and may function as an RNA channel. First, expression of SID-1 in Drosophila Schneider 2 (S2) cells facilitates soaking RNAi that is more efficient when long dsRNA (50–500 bp) is used rather than siRNA (5). Co-expression of a dominant negative variant SID-1S536I with wild type (WT) SID-1 inhibits soaking RNAi, suggesting that SID-1 may function as an oligomer (6). Second, SID-1 mediates passive cellular uptake of dsRNA in S2 cells. The process is rapid and concentration-dependent (5). Cellular ATP depletion or incubation at 4 °C has little effect on dsRNA uptake by SID-1 (5). Third, exposing SID-1-expressing cells to dsRNA increases membrane conductance, implying that dsRNA transport may involve opening of an RNA channel (7). The change in membrane conductance is SID-1-dependent and does not occur in response to dsDNA (7). Fourth, similar to these results in S2 cells, heterologous expression of SID-1 in Bombyx mori BmN4 cells, Spodoptera frugiperda Sf9 cells, and Mus musculus embryonic stem cells also enables soaking RNAi (8–11). These different lines of evidence argue for a specific biochemical step by which SID-1 recognizes and differentiates RNA substrates for transport. However, no biochemical evidence is available to establish the substrate recognition step.
Sequence similarity has provided evidence for the existence of SID-1 homologues in different species. Except for in nematodes, SID-1 homologs are only sporadically represented in invertebrate genomes but persist in most vertebrates. Most invertebrates, such as S. American, have only one SID-1 homolog, which is probably involved in systemic RNAi (13). Although systemic RNAi is uncommon in vertebrates, most vertebrates have two SID-1 homologs: SIDT1 and SIDT2. Previous studies have suggested that SIDT1 may function similarly to SID-1. For example, expression of human SIDT1 in a pancreatic cell line enhances soaking RNAi by siRNA (12), and overexpression of human SIDT1 in human embryonic kidney cells mediates intercellular transport of small RNA (13). Purified human SIDT1 extracellular domains are glycosylated and form stable tetramers that behave like a compact globular particle (14). Unlike studies of SIDT1, animal models have been used for the study of SIDT2. A fish homolog of SIDT2 is expressed in Siniperca chuatsi and may play a role in exogenous dsRNA uptake and antiviral host defense (15). Thus, one would anticipate that these molecules, similar to SID-1, are likely to possess biochemical recognition for RNA substrates.
Considering that the SID-1 ECD is directly exposed to extracellular RNA, we hypothesized that it may function as a docking site to recognize and recruit RNA at the cell surface. To test this hypothesis, we expressed and purified recombinant SID-1 ECD from mammalian cells. Using an electrophoretic mobility shift assay (EMSA), we quantified the binding affinity of SID-1 ECD to different nucleic acids in vitro. Using an RNA uptake assay in S2 cells ectopically expressing SID-1, we compared RNA transport efficiency between wild-type (WT) and mutant SID-1. Our results support a role for the SID-1 ECD in selectively binding long dsRNA and show that this activity is also required for RNA transport by full-length SID-1.
Experimental Procedures
Molecular Cloning of SID-1 Homologs
cDNAs for SID-1, mouse SIDT1, and mouse SIDT2 were purchased from OriGene. A synthetic SID-1 gene with optimized codon frequency for mammalian expression was ordered from GenScript. Residues included in the ECD are as follows: SID-1(22–312), SIDT1(23–310), and SIDT2(22–292). The SID-1 ECD was cloned into pSGHV0 flanked by BamHI and NotI. pSGHV0 is a mammalian expression plasmid encoding an N-terminal growth hormone and an octahistidine tag (16). SIDT1 ECD and SIDT2 ECD were cloned into this vector in the same fashion. SID-1A173T, SID-1P199L, SIDT1F169T, SIDT1P186L, SIDT2F154T, and SIDT2F171L ECDs were generated using the QuikChange site-directed mutagenesis kit (Agilent). Full-length SID-1 with a C-terminal FLAG tag was cloned into pPacPl flanked by BamHI/NotI or XbaI/NotI. pPacPl is an insect expression vector. Firefly luciferase, Renilla luciferase, SID-1A173T, SID-1P199L, and SID-1S536I were cloned into pPacPl in the same manner.
Expression and Purification of the Extracellular Domain of SID-1 Homologs
CHO-S cells were maintained in suspension culture using serum-free medium supplemented with 1% FBS and 1% l-glutamate. One day before transfection, CHO-S cells were seeded at 1 × 106 cells/ml. Plasmids used for transfection were prepared from Escherichia coli using the PureLink Maxiprep kit (Life Technologies, Inc.). Expression vectors were mixed with polyethyleneimine (PEI) at an optimized ratio of DNA/PEI of 1:3 by weight within one-tenth of the final volume of transfection for a half-hour. The DNA/PEI mixture was then added into the suspension cells. 24 h post-transfection, cells were split by 1:3 using fresh culture medium. 4 days post-transfection, medium was harvested and centrifuged to remove cell debris. The medium was then sterile filtrated and concentrated using a tangential flow filtration system (Centramate, Pall). Various ECDs were isolated by immobilized metal affinity chromatography (IMAC) using nickel-nitrilotriacetic acid-agarose (Qiagen). Medium was loaded onto nickel-nitrilotriacetic acid beads followed by washing using 50 mm phosphate buffer, 250 mm NaCl, 40 mm imidazole. Recombinant protein was eluted using 50 mm phosphate buffer, 250 mm NaCl, 250 mm imidazole. Purified ECD was loaded onto a size exclusion column (Superdex 200 10/300 GL) equilibrated in 50 mm HEPES, pH 7.0, 200 mm KCl, and 1 mm β-mercaptoethanol. Bio-Rad gel filtration standards were used for estimating protein size. The monomeric fractions of ECD were concentrated, checked by Coomassie Brilliant Blue staining, and stored for later use.
Nucleic Acid Synthesis
dsDNA was prepared by PCR and gel purification. Long dsRNA was synthesized by in vitro transcription (MEGAscript T7, Life Technologies) using dsDNA (gel-purified PCR products) as a template. dsRNA was then purified by gel purification. Calf intestinal phosphatase (New England Biolabs) was used for dephosphorylation of 5′ ends of dsRNA. Dephosphorylated RNA was then purified by phenol/chloroform extraction, followed by end-labeling using [γ-32P]ATP (PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). [32P]dsRNA was finally purified by a Quick Spin column twice, and concentrations were measured by a scintillation counter. 20-bp dsRNA and 50-bp dsRNA were synthesized by annealing ordered ssRNA.
Electrophoretic Mobility Shift Assays
Purified ECD was dialyzed into EMSA buffer: 50 mm Tris-HCl, pH 7.0, 200 mm KCl. Protein concentrations were measured by a BCA protein assay kit (Thermo Scientific). Immediately before use, dsRNA was heated to 95 °C and then cooled down at 37 °C for 10 min, followed by folding in 50 mm Tris-HCl, pH 7.0, 200 mm KCl, and 2 mm MgCl2 for 20 min. Protein and dsRNA were then mixed in 20 μl of a buffer containing 50 mm Tris-HCl, pH 7.0, 200 mm KCl, 1 mm MgCl2. After a 30-min incubation at room temperature, samples were loaded into 4% polyacrylamide gel and run at 180 V in TBE (Tris/borate/EDTA) buffer. Then gels were dried onto filter paper using a gel dryer (Bio-Rad) and exposed to a PhosphorImager screen that was then scanned using a Typhoon scanner (GE Healthcare). The fraction bound was quantified using ImageJ. Data were fit to a non-linear curve using the Hill equation to obtain apparent Kd and Hill constant values.
Cell-based RNA Transport Assays
S2 cells were maintained in adherent culture in insect serum-free medium. 24 h before transfection, S2 cells were seeded in 24-well plates at 0.5 × 106 cells/well. Transient transfection was performed using Effectene (Qiagen). The co-transfection ratio of luciferase to SID-1 was optimized to be 1:100. 24 h after transfection, dsRNA against firefly luciferase was added. If necessary, dsRNA was removed by wash with culture medium. 72 h post-transfection, a dual luciferase assay was performed using the Dual-Luciferase® reporter assay kit (Promega). Briefly, lysis buffer was added to 24-well plates, and cell lysate was transferred to a 384-well plate. Firefly luciferase and Renilla luciferase levels were measured separately using a plate reader (POLARstar Omega). Firefly luciferase level was normalized to Renilla luciferase level, which served as an internal control.
Secondary Structure Prediction and Sequence Alignment
Transmembrane helices were predicted by the TMHMM version 2.0 server (17). The signal peptide was predicted by the SignalP version 4.1 server (18). N-Linked glycosylation sites were predicted by the NetNGlyc version 1.0 server. Phosphorylation sites were predicted by the NetPhos version 2.0 server (19). Secondary structure was predicted by Jpred3 (20). Sequence alignment was done using T-coffee (21). A phylogenetic tree was generated by phylowidget (22).
Statistics
Paired or unpaired Student's t test (two-tailed) (Excel, Microsoft) was used to determine statistical significance. A p value of <0.05 was considered statistically significant.
Results
Recombinant SID-1 ECD Selectively Binds dsRNA in Vitro
To characterize the biochemical properties of SID-1 ECD and test its role in dsRNA recognition, we expressed and purified C. elegans SID-1 ECD from mammalian cells. A eukaryotic expression system was used to preserve the potential post-translational modifications on SID-1, which may be essential for its function. Briefly, the coding region for SID-1 ECD (amino acids 22–312; Fig. 1A) was cloned into a mammalian expression vector pSGHV0, which adds a human growth hormone and an octahistidine tag to the SID-1 N terminus to enhance expression and subsequent purification (16). The SID-1 expression construct was codon-optimized for expression in Chinese hamster ovary cells (CHO-S). We purified recombinant SID-1 ECD with high yield (0.5 mg/liter of growth) and purity (Fig. 1B). Purified SID-1 ECD is stable and monomeric as judged by migration on size exclusion chromatography (SEC) (Fig. 1C).
To determine whether SID-1 ECD binds dsRNA in vitro, we performed EMSAs. First, we incubated 32P-5′-end-labeled 700-bp dsRNA (encoding a GFP sequence) with increasing concentrations of SID-1 ECD and separated bound and free forms on native polyacrylamide gels. We observed progressively reduced dsRNA mobility as the protein concentration increased (Fig. 2A). After incubation of dsRNA with SID-1 ECD, we observed an unshifted band corresponding to unbound dsRNA, a smear presumably corresponding to intermediate dsRNA-protein complexes, and a compact shifted band probably corresponding to the saturated dsRNA-protein complexes. Considering that the length of 700 bp dsRNA is about 238 nm and the Stokes radius of SID-1 ECD is about 4.4 nm (judged by SEC), it is likely that one 700-bp piece of dsRNA simultaneously binds multiple SID-1 ECDs in our assays. This is consistent with the notion that we observe both intermediate and saturated dsRNA-protein complexes, which is common for protein binding long nucleic acid (e.g. see Refs. 23–25).
FIGURE 2.
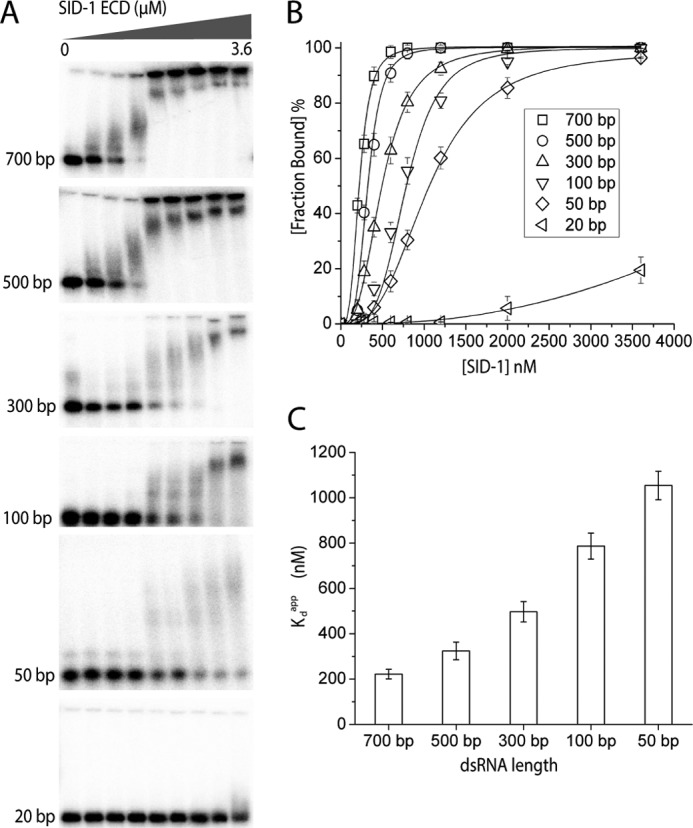
Recombinant SID-1 ECD binds dsRNA in vitro in a length-dependent manner. A, EMSA using recombinant SID-1 ECD and 32P-labeled dsRNA of different lengths (encoding GFP sequences). Increasing concentrations of SID-1 ECD (0, 200, 280, 400, 600, 800, 1200, 2000, and 3600 nm) and a fixed concentration of dsRNA (0.1 nm) were used for all EMSAs. B, quantified binding curves of EMSA results in A. The fraction bound was calculated by Sbound/(Sbound + Sfree). Sbound is determined by the 32P signal for all shifted dsRNA, including both saturated and intermediate dsRNA-protein complex. Lines are fit using the Hill equation (number of independent experiments (n) = 3; ±S.D. (error bars)). Here the fraction bound is defined as 1/(1 + (Kdapp/[SID-1])Hill constant. C, plot of calculated Kdapp against dsRNA length for SID-1 ECD (n = 3; ±S.D.).
To determine whether SID-1 ECD binding dsRNA depends on RNA length, we performed EMSAs using dsRNA of various lengths (Fig. 2A). Our results demonstrate that SID-1 ECD binds longer RNA more robustly but less effectively to shorter dsRNAs. SID-1 ECD is unable to bind 20-bp dsRNA. This result is consistent with previous studies showing that the minimal length of dsRNA required for efficient RNA uptake by SID-1 is 50 bp (5, 6). We quantified these EMSA results and fitted the binding data using the Hill equation (Fig. 2B). Steep sigmoid binding curves were seen for long dsRNAs, suggesting that binding is cooperative. The apparent dissociation constant (Kdapp) and Hill coefficient were calculated for each binding assay. The SID-1 ECD binds long dsRNA with modest Kdapp values ranging from 200 to 1000 nm (Table 1). The Hill coefficients are larger than 1, suggesting that binding is positively cooperative (Table 2). By plotting Kdapp against dsRNA length, we find that SID-1 ECD binds preferentially to longer dsRNA (Fig. 2C). The Hill coefficients also decrease as Kdapp increases, implying that stronger binding affinity and positively cooperative binding could be related.
TABLE 1.
Calculated Kdapp (nm) for different SID ectodomains and dsRNA with different lengths
Kdapp values (nm) ± S.D. were calculated from multiple experiments (n ≥ 3). For low affinity binding, the absolute value of Kdapp could not be determined but is greater than value stated.
|
Kdapp |
||||||
|---|---|---|---|---|---|---|
| 700 bp | 500 bp | 300 bp | 100 bp | 50 bp | 20 bp | |
| nm | ||||||
| SID-1 | 222 ± 15 | 324 ± 39 | 497 ± 45 | 787 ± 57 | 1054 ± 63 | >3600 |
| SID-1A173T | 704 ± 43 | 928 ± 59 | 1470 ± 65 | >2000 | >2000 | >3600 |
| SID-1P199L | 1021 ± 62 | 1373 ± 73 | >2000 | >3600 | >3600 | >3600 |
| SIDT1 | 1094 ± 59 | >2000 | >2000 | >3600 | >3600 | >3600 |
| SIDT2 | 851 ± 47 | 1058 ± 63 | 1394 ± 112 | >3600 | >3600 | >3600 |
TABLE 2.
Calculated Hill coefficient (nH) from in vitro binding assays using different SID ectodomains and dsRNA with different lengths
Hill coefficients (nH) were calculated from multiple experiments (n ≥ 3). ND, n could not be determined.
| Hill coefficient |
||||||
|---|---|---|---|---|---|---|
| 700 bp | 500 bp | 300 bp | 100 bp | 50 bp | 20 bp | |
| SID-1 | 4.0 ± 0.3 | 3.8 ± 0.5 | 3.5 ± 0.2 | 3.3 ± 0.3 | 2.9 ± 0.1 | N.D. |
| SID-1A173T | 3.1 ± 0.1 | 2.9 ± 0.2 | 2.7 ± 0.5 | ND | ND | ND |
| SID-1P199L | 3.3 ± 0.2 | 3.4 ± 0.4 | ND | ND | ND | ND |
| SIDT1 | 3.2 ± 0.4 | ND | ND | ND | ND | ND |
| SIDT2 | 3.1 ± 0.3 | 3.0 ± 0.4 | 2.6 ± 0.2 | ND | ND | ND |
To examine any nucleotide sequence specificity of SID-1 ECD binding dsRNA, we assessed whether SID-1 ECD binding dsRNA depends on specific RNA sequence and whether the SID-1 ECD binds dsDNA. First we generated 32P-labeled 500-bp dsRNA composed of different sequences (i.e. encoding GFP, luciferase, and mCherry) and performed EMSAs with the ECD of SID-1 (Fig. 3A). After fitting the binding curves (Fig. 3D) and calculating Kdapp values (Fig. 3E), we found no significant difference in binding affinity for these various RNA sequences. We then performed EMSAs using either 500-bp dsDNA (Fig. 3B) or 500-bp dsRNA encoding the same GFP sequences. SID-1 ECD binds dsDNA but requires higher protein concentrations compared with binding dsRNA (Fig. 3D). The calculated Kdapp for 500-bp dsDNA is 1.45 ± 0.08 μm (n = 3), which is 5-fold higher than the Kdapp for 500 bp dsRNA (Fig. 3E). dsRNA differs from dsDNA by a 2′-hydroxyl group on the ribose and by using the base uridine rather than thymidine. It is possible that SID-1 ECD distinguishes dsRNA from dsDNA by recognizing the base uridine. To test this possibility, we transcribed a 500-bp dsRNA using aminoallyl-UTP instead of UTP and then assessed its binding to SID-1 ECD using EMSA. SID-1 ECD binds dsRNA containing aminoallyl-uridine (Fig. 3C), and there is no significant difference in calculated Kdapp between dsRNA containing aminoallyl-uridine and dsRNA containing uridine (Fig. 3, D and E). This suggests that the difference in affinities between dsRNA and dsDNA does not arise from the differences in the nucleotide bases alone. To further investigate the discrimination between dsRNA and dsDNA, we performed competition binding assays using unlabeled nucleic acids. We mixed 32P-end-labeled 500-bp dsRNA (0.1 nm) with SID-1 ECD (2 μm), which is a condition in which protein-dsRNA complexes are formed. We then added different amounts of unlabeled 500-bp dsRNA as competitor. As the unlabeled competitor concentration increases, 32P-end-labeled 500-bp dsRNA shifts from the protein-bound form to the free form (Fig. 3F). Calculated inhibition constant IC50 (the concentration of competitor at the half-maximal inhibition) using unlabeled dsRNA is 10 ± 2 nm. We also performed similar competition binding assays using unlabeled 500-bp dsDNA as competitor (Fig. 3G), and the calculated IC50 is 400 ± 80 nm. These results further indicate that SID-1 ECD preferentially binds dsRNA relatively to dsDNA. Therefore, our EMSA results show that recombinant SID-1 ECD binds dsRNA in vitro. Binding is length-dependent, has no sequence preference, and is selective for dsRNA over dsDNA.
FIGURE 3.
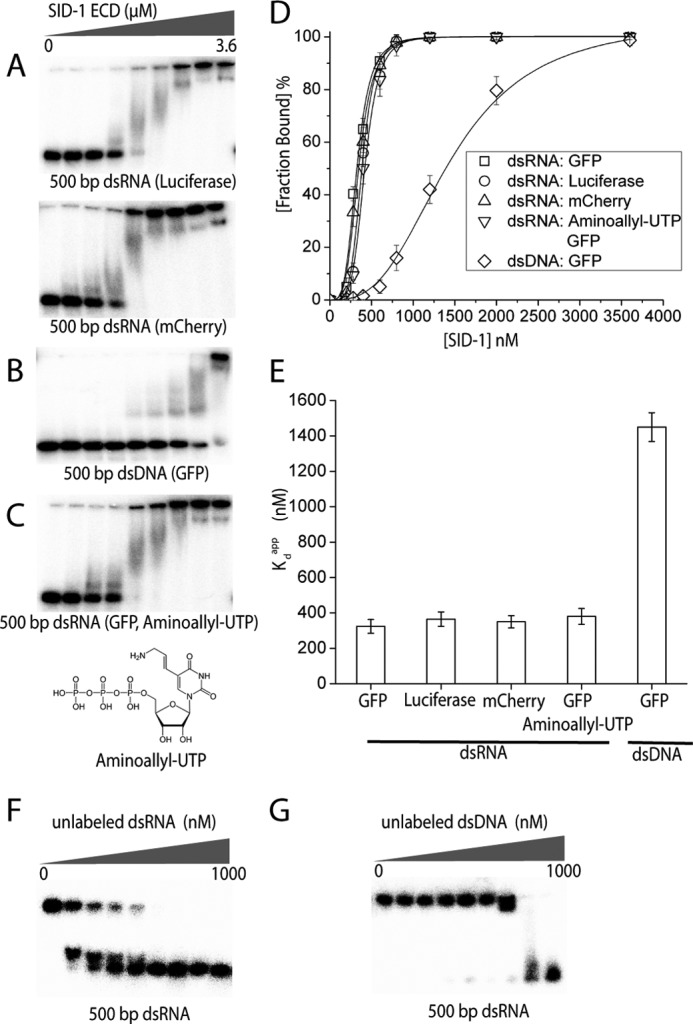
SID-1 ECD has no sequence preference but distinguishes dsRNA from dsDNA. A–C, EMSAs using recombinant SID-1 ECD (0, 200, 280, 400, 600, 800, 1200, 2000, and 3600 nm) and various nucleic acids (0.1 nm). A, SID-1 ECD binds 500-bp dsRNA encoding different sequences (firefly luciferase and mCherry). B, SID-1 ECD binds 500-bp dsDNA (encoding GFP sequence) but requires higher protein concentrations. C, SID-1 ECD binds 500-bp dsRNA containing aminoallyl-uridine, showing a similar binding pattern as WT dsRNA. The chemical structure of aminoallyl-UTP is shown. D, quantified binding curves of EMSA results in A. E, a plot of calculated Kdapp against various nucleic acids used in A (n = 3; ±S.D. (error bars)). F and G, competition binding assay using 32P-end-labeled 500-bp dsRNA (0.1 nm) with SID-1 ECD (2 μm) and different amounts of unlabeled 500 bp dsRNA (F) or dsDNA (G) as competitor (0, 5, 15, 30, 60, 125, 250, 500, and 1000 nm).
Inhibition of Binding between SID-1 ECD and dsRNA Decreases dsRNA Transport Efficiency by Full-length SID-1
If the binding between dsRNA and SID-1 ECD is essential for RNA transport by SID-1, one expects that a correlation between the binding affinity and RNA transport efficiency. To test this hypothesis, we focused on key residues in the SID-1 ECD. A previous genetic screen had identified several loss-of-function mutations in SID-1, two of which are missense mutations in the ECD: A173T and P199L (2). To test whether these mutations change the dsRNA binding properties of SID-1 ECD, we purified SID-1A173T ECD and SID-1P199L ECD from CHO-S cells (Fig. 4A). Both variants are secreted as stable soluble proteins and are monodisperse as judged by SEC (Fig. 4B). By EMSA, recombinant SID-1A173T ECD and SID-1P199L ECD no longer bind dsRNA shorter than 100 bp. When tested for dsRNA longer than 300 bp, higher protein concentrations are necessary for detection of the binding. Thus, the mutant variants retain a preference for longer dsRNA (Fig. 4, C and D). Kdapp values were calculated for comparison (Table 1). After plotting Kdapp against dsRNA length, we found that Kdapp decreases as dsRNA length increases, which is consistent between both WT and mutated ECDs (Fig. 4E). However, at fixed dsRNA length, the Kdapp for mutant variants is 3–5-fold higher than the Kdapp for WT ECD. Sequence alignment using various SID-1 homologs revealed that both residues are in a conserved region of SID-1 ECD (Fig. 4F). Ala-173 is conserved in all invertebrate SID-1 homologs, and Pro-199 is broadly conserved across both vertebrate and invertebrate SID-1 homologs. The sequence conservation therefore also argues for a potentially shared function of RNA binding.
FIGURE 4.
Mutating key residues in SID-1 ECD (A173T or P199L) reduces binding affinity between SID-1 ECD and dsRNA. A, SDS-PAGE analysis and Coomassie Blue staining of purified recombinant SID-1 ECD (WT, A173T, and P199L). B, SEC profile of WT and mutant SID-1 ECD using a Superdex 200 column. C, quantification of EMSAs using recombinant SID-1A173T ECD (0, 200, 280, 400, 600, 800, 1200, 2000, and 3600 nm) and different lengths of dsRNA (0.1 nm; encoding GFP sequence). D, quantification of EMSAs using recombinant SID-1P199L ECD (0, 200, 280, 400, 600, 800, 1200, 2000, and 3600 nm) and different lengths of dsRNA (0.1 nm; encoding GFP sequence). E, a plot of calculated Kdapp for SID-1/SID-1A173T/SID-1P199L ECD against dsRNA lengths (n = 3; ± S.D. (error bars)). F, alignment of partial ECD sequences using various SID-1 homologs. Two identified conserved key residues are shown by asterisks. mAU, milliabsorbance units.
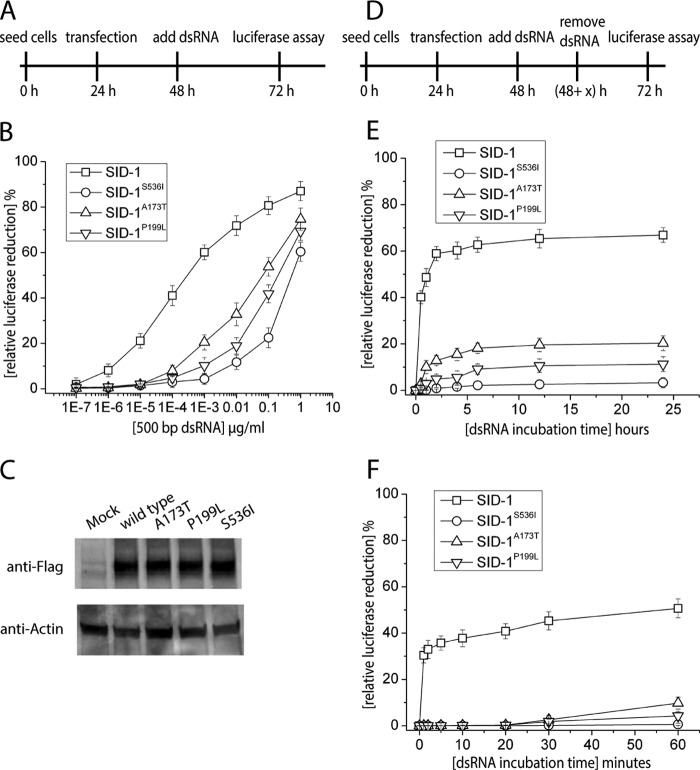
To test whether reduced binding affinity of SID-1 ECD results in a decrease in RNA transport efficiency, we performed an RNA transport assay in Drosophila S2 cells. Briefly, full-length SID-1 and luciferase were co-expressed in S2 cells, which were subsequently incubated with media containing dsRNA targeting luciferase. If SID-1 transports the dsRNA into S2 cells, expression of luciferase will be silenced. Thus, RNA transport is assessed by monitoring a relative reduction in luciferase activity. RNA transport by SID-1 is both concentration- and time-dependent. First, we fixed the dsRNA incubation time at 24 h and analyzed the RNA transport efficiency of SID-1, SID-1A173T, and SID-1P199L at various RNA concentrations (Fig. 5A). SID-1S536I carries a previously identified dominant negative mutation in the transmembrane region, causing it to be deficient in dsRNA transport (6). Our results show that WT and mutant variants exhibited different RNA transport capabilities (Fig. 5B). Relative luciferase reduction increases as dsRNA concentration increases. However, compared with WT SID-1, the dose-response curves for SID-1A173T and SID-1P199L are shifted to the right, indicating that to achieve the same level of silencing, the mutant variants require higher concentrations of dsRNA. WT SID-1 requires about 0.1 ng/ml dsRNA to reach 50% silencing, whereas SID-1A173T and SID-1P199L require about 100 ng/ml dsRNA to reach the same level of silencing. The differential efficiency cannot be readily explained by expression variation because the different SID-1 variants have a similar expression level in S2 cells, as judged by Western blot (Fig. 5C), supporting that the difference in silencing arises from function (i.e. the difference in affinity for the substrate). Our results suggest that the A173T and P199L mutations decrease SID-1 RNA transport efficiency, which is consistent with EMSA results suggesting that they reduce RNA binding affinity.
FIGURE 5.
Mutating key residues in SID-1 ECD (A173T or P199L) decreases RNA transport efficiency by full-length SID-1. A, a time line for a dsRNA uptake assay in S2 cells. To study dose-dependent RNA transport by SID-1, different concentrations of 500-bp dsRNA (encoding firefly luciferase sequence) were added at 48 h. RNA treatment time was fixed at 24 h. B, quantification of concentration-dependent dsRNA uptake by different SID-1 variants (n = 3; ±S.D. (error bars)). The percentage of relative luciferase reduction is calculated by 1 − L/Lfree. Lfree is the control luciferase level without adding dsRNA. C, expression of different FLAG-tagged SID-1 variants was detected by Western blot using anti-FLAG antibody. Endogenous actin was detected by anti-actin antibody as loading control. D, a modified time line for dsRNA uptake assay in S2 cells. To study time-dependent RNA transport by SID-1, a fixed concentration of dsRNA (1 ng/ml) was added at 48 h. After various dsRNA treatment times, dsRNA was removed by washing with culture medium. E, quantification of time-dependent dsRNA uptake by different SID-1 variants (n = 3; ±S.D.). dsRNA treatment times tested were 0, 0.5, 1, 2, 4, 6, 12, and 24 h. F, kinetic study of dsRNA uptake by SID-1 variants using shorter dsRNA treatment times (0, 1, 2, 5, 10, 20, 30, and 60 min).
To better understand the kinetics of dsRNA uptake by SID-1, we varied dsRNA incubation time using a fixed concentration of dsRNA (1 ng/ml). Briefly, SID-1 variants and luciferase were expressed in S2 cells. dsRNA:Luci was applied for various lengths of time and then removed (Fig. 5D). At 24 h after initial RNA exposure, relative luciferase reduction was quantified. For cells expressing WT SID-1, 50% luciferase reduction was observed with 1 h of RNA incubation, and maximum luciferase reduction (60%) was reached with 2 h of RNA treatment (Fig. 5E). For SID-1 variants, 5–10% luciferase reduction was observed with 1 h of RNA incubation, and maximum luciferase reduction (10–20%) was reached with 6 h of RNA treatment (Fig. 5E). We then assayed shorter RNA incubation times and found that for cells expressing WT SID-1, 1 min of RNA exposure is enough to confer 30% luciferase reduction. In contrast, cells expressing SID-1A173T or SID-1P199L required at least 1 h of RNA exposure to reach detectable luciferase reduction (5–10%) (Fig. 5F). These results suggest that at a fixed dsRNA concentration, SID-1A173T and SID-1P199L have slower RNA transport rates than WT SID-1. In summary, the A173T or P199L mutation reduce the binding affinity between SID-1 ECD and dsRNA, which may explain how these mutants decrease RNA transport efficiency by SID-1.
Mammalian SIDT1 and SIDT2 ECD Bind dsRNA with Lower Affinity than SID-1 ECD
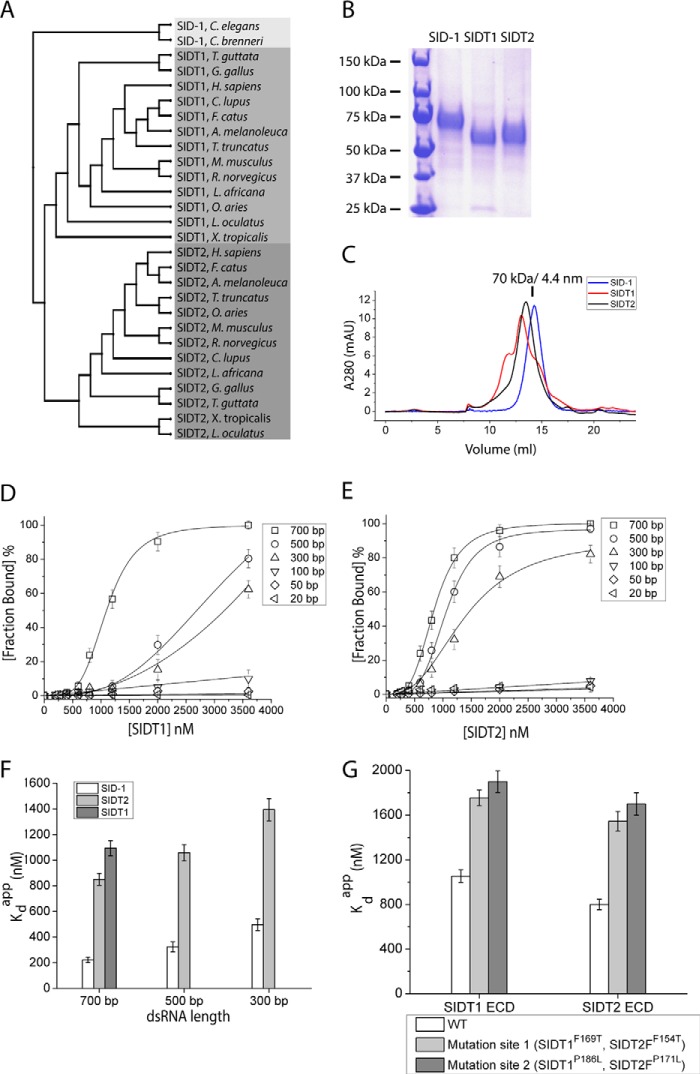
SID-1 homologs are conserved from worms to mammals. A phylogenetic tree made using the sequences of various SID-1 homologs shows that the SID-1 family can be divided into three classes (SID-1, SIDT1, and SIDT2) (Fig. 6A). Like C. elegans SID-1, mammalian SIDT1 and SIDT2 have a large extracellular domain and multiple transmembrane helices. Similar structural organization suggests that they may have a shared functionality with SID-1. To determine whether SIDT1/SIDT2 ECDs bind nucleic acid, we purified mouse SIDT1 and SIDT2 ECDs from CHO-S cells. Both mSIDT1 ECD and mSIDT2 ECD are secreted as stable soluble proteins (Fig. 6B). SIDT1 ECD exhibited a multidisperse elution profile on SEC (Fig. 6C), suggesting that it may form multimers. SIDT2 ECD migrates as a monodisperse protein on SEC (Fig. 6C). EMSAs were performed using recombinant SIDT1 ECD or SIDT2 ECD and dsRNA of various lengths. Both SIDT1 and SIDT2 ECDs bind 500- or 700-bp dsRNA but require higher protein concentrations compared with SID-1 ECD (Fig. 6, D and E). SIDT1 ECD does not bind dsRNA shorter than 300 bp, whereas SIDT2 ECD does not bind dsRNA shorter than 100 bp. Kdapp values were calculated (Table 1). After plotting Kdapp against dsRNA length, we found that SIDT2 ECD retains length preference for dsRNA similar to SID-1 ECD (Fig. 6F). However, at fixed dsRNA lengths, the Kdapp value for SIDT2 ECD is 3–4-fold higher than the Kdapp value for SID-1 ECD. Most Kdapp values for SIDT1 ECD were not readily measurable or were weaker than 2 μm, because saturated binding could not be reached. To further investigate whether mutation of the corresponding residues Ala-173 and Pro-199 in SIDT1 and SIDT2 ECD diminishes their dsRNA-binding capability, we purified extracellular domains containing SIDT1F169T, SIDT1P186L, SIDT2F154T, and SIDT2P171L, which were used to perform binding assays with 32P-end-labeled 700-bp dsRNA. Both site 1 mutation and site 2 mutation decreased binding affinity between SIDT1, SIDT2 ECD, and 700-bp dsRNA (Fig. 6G).
FIGURE 6.
SIDT1/SIDT2 ECD binds long dsRNA with lower affinity compared with SID-1 ECD. A, a phylogenetic tree of various SID-1 homologs. Based on primary sequence similarity, SID-1 homologs can be divided into three classes (SID-1, SIDT1, and SIDT2) shown in different colors. B, SDS-PAGE analysis and Coomassie Blue staining of purified recombinant SID-1/SIDT1/SIDT2 ECD. C, SEC profile of SID-1/SIDT1/SIDT2 ECD using a Superdex 200 column. D and E, quantification of EMSAs using recombinant SIDT1 ECD (D) or SIDT2 ECD (E) and different lengths of dsRNA. Protein concentrations used were 0, 200, 280, 400, 600, 800, 1200, 2000, and 3600 nm. dsRNA (encoding GFP sequence) concentration used was 0.1 nm. F, A plot of calculated Kdapp for SID-1/SIDT1/SIDT2 ECD against dsRNA length (n = 3; ±S.D. (error bars)). G, a plot of calculated Kdapp for mutant SIDT1/SIDT2 ECD with 700-bp dsRNA (n = 3; ±S.D.). mAU, milliabsorbance units.
Discussion
SID-1-dependent RNA uptake has been demonstrated in various in vitro and in vivo systems, but the mechanism of this uptake, specifically the substrate-SID-1 recognition, remains unclear. Our study shows that the SID-1 ECD selectively binds dsRNA and that disruption of dsRNA binding by the ECD diminishes dsRNA uptake. Hence, the evidence provides a direct biochemical link from binding to transmembrane RNA transport.
First, we show that soluble SID-1 ECD can be expressed and secreted as stable protein that binds dsRNA in vitro. Multiple β strands and α helices are predicted in SID-1 ECD, but no known discernable dsRNA binding domain or motif can be identified by sequence alone. SID-1 ECD contains predicted immunoglobulin-like β-sandwich folds, which may mediate the cooperative interaction with long dsRNA (26). Interestingly, the predicted isoelectric point of SID-1 ECD is 8.8; therefore, at physiological pH, SID-1 ECD should carry an overall positive charge capable of complementing negatively charged RNA. We further show that the SID-1 ECD binds preferentially longer dsRNA compared with shorter dsRNA, has no sequence preference, and does not specifically recognize uracil relative to thymine bases. However, SID-1 ECD does distinguish dsRNA from dsDNA. We suggest that differences in dsRNA and dsDNA structure, such as differences in groove size, may contribute to how SID-1 ECD preferentially interacts with dsRNA. Our binding results are consistent with previous characterizations of SID-1 function. Previous reports showed that SID-1-mediated soaking RNAi in S2 cells has a dsRNA length-dependent potency (5), SID-1-mediated neuronal RNAi in C. elegans utilizes dsRNA of various sequences (3), and dsRNA but not dsDNA induces membrane conductance changes in SID-1-expressing S2 cells (7). Our studies suggest that SID-1 ECD may facilitate the first step of RNA recognition and contributes to subsequent RNA transport.
A reason that SID-1 preferentially binds and transports dsRNA may be that long dsRNA often arises from exogenous genetic information, such as dsRNA viruses, and poses a threat to the cell that needs to be nullified. The RNAi machinery in C. elegans has been shown to contribute to antiviral immunity (27); thus, SID-1 may play a role in the recognition of exogenous viral long dsRNA. Consistent with this role, SID-1 does not display any apparent substrate sequence preferences, which would allow it to recognize a variety of invasive genetic targets. Binding parameters for interactions between nucleic acid binding proteins and oligonucleotides of defined length have been modeled, and the observed binding constants are a linear function of oligonucleotide chain length (28). Indeed, the Kdapp for SID-1 ECD that we determined is nearly a linear function with respect to dsRNA length (Fig. 2C). The observed steep transition from free dsRNA to bound dsRNA suggests a cooperative binding mechanism between the SID-1 ECD and long dsRNA. Cooperative binding has been seen for various proteins that preferentially bind long dsRNA or dsDNA (29, 30), and our work is thus consistent with this broadly observed phenomenon. In many cases, cooperativity decreases as dsRNA length decreases, so that cooperativity could be the underlying mechanism for length discrimination.
We also show that dsRNA binding by SID-1 ECD is related to RNA transport through SID-1. SID-1A173T ECD and SID-1P199L ECD, which carry mutations in conserved residues, have reduced affinity with dsRNA compared with SID-1WT ECD. More importantly, cells expressing SID-1A173T and SID-1P199L require higher dsRNA concentrations and longer incubation times to achieve RNA uptake levels similar to those of cells expressing SID-1WT. These results suggest that defective dsRNA binding to SID-1 ECD results in decreased dsRNA transport through SID-1. There are at least two potential ways that the SID-1 ECD could contribute to RNA transport. First, considering that long dsRNA is a large hydrophilic charged molecule, the SID-1 ECD may function as a docking site that recruits exogenous dsRNA through electrostatic interactions and anchors it to the cell surface. Second, a constitutively open RNA channel would be toxic to cells because the size of such a channel's pore region could be large enough to pass various small metabolites and ions. SID-1 ECD could function as the gate of the channel's pore region and open only when dsRNA is bound. To further understand the potential selecting and/or gating mechanism of SID-1, future study of coordination between the SID-1 ECD and transmembrane regions will be necessary.
Although the physiological function of most vertebrate SID-1 homologs is unknown, we show that purified mouse SIDT1 ECD and SIDT2 ECD bind long dsRNA in vitro in a length-dependent manner, albeit with reduced affinity compared with C. elegans SID-1 ECD. We failed to express full-length SIDT1 or SIDT2 in S2 cells, so we could not compare their RNA transport efficiency with that of SID-1. Although SIDT1 and SIDT2 can be expressed in mammalian cells, the long dsRNA transport assay proved unsuccessful due to high endogenous SIDT1/SIDT2 expression and long dsRNA-induced innate immune responses in mammalian cells. Although mammals have sophisticated immune systems and long dsRNA induces innate immune responses in mammalian cells, recent studies show that long dsRNA-mediated RNA interference exists and functions as an antiviral mechanism in mammals (31, 32). M. musculus and Rattus norvegicus endogenously express SIDT2, which is an integral membrane protein that is primarily localized in the lysosome (33). Lysosomal SIDT2 may recognize dsRNA that localizes in the endocytic pathway. Future study of SIDT1 and SIDT2 function will benefit from genetic studies using animal models.
In summary, our studies define an important molecular function for the SID-1 ECD by showing that it selectively binds dsRNA and contributes to dsRNA transport by SID-1. This provides a foundation to begin dissecting the remaining biochemical steps by which an RNA molecule travels across plasma membrane.
Author Contributions
W. L., K. K., D. L., and M. L. designed the research; W. L. performed and analyzed the experiments; K. K. and D. L. provided technical assistance and contributed to the preparation of the figures; and W. L., K. K., D. L., and M. L. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank members of the Min Li and Daniel Leahy laboratories and Yun Qing for valuable discussions and comments on the manuscript. We thank Professor Rachel Green for technical support with EMSAs.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078579 and MH084691 (to M. L.), R01GM099321 (to D. L.), and F32 GM100608 (to K. S. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- ECD
- extracellular domain
- S2
- Schneider 2
- SID-1
- systemic RNA interference deficiency-1
- SIDT1 and SIDT2
- SID1 transmembrane family member 1 and 2, respectively.
References
- 1. Hunter C. P., Winston W. M., Molodowitch C., Feinberg E. H., Shih J., Sutherlin M., Wright A. J., Fitzgerald M. C. (2006) Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 71, 95–100 [DOI] [PubMed] [Google Scholar]
- 2. Winston W. M., Molodowitch C., Hunter C. P. (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 [DOI] [PubMed] [Google Scholar]
- 3. Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M. (2010) Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods 7, 554–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jose A. M., Smith J. J., Hunter C. P. (2009) Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. U.S.A. 106, 2283–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Feinberg E. H., Hunter C. P. (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 [DOI] [PubMed] [Google Scholar]
- 6. Shih J. D., Fitzgerald M. C., Sutherlin M., Hunter C. P. (2009) The SID-1 double-stranded RNA transporter is not selective for dsRNA length. RNA 15, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shih J. D., Hunter C. P. (2011) SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu J., Nagata Y., Mon H., Li Z., Zhu L., Iiyama K., Kusakabe T., Lee J. M. (2013) Soaking RNAi-mediated modification of Sf9 cells for baculovirus expression system by ectopic expression of Caenorhabditis elegans SID-1. Appl. Microbiol. Biotechnol. 97, 5921–5931 [DOI] [PubMed] [Google Scholar]
- 9. Mon H., Kobayashi I., Ohkubo S., Tomita S., Lee J., Sezutsu H., Tamura T., Kusakabe T. (2012) Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol. 9, 40–46 [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi I., Tsukioka H., Kômoto N., Uchino K., Sezutsu H., Tamura T., Kusakabe T., Tomita S. (2012) SID-1 protein of Caenorhabditis elegans mediates uptake of dsRNA into Bombyx cells. Insect Biochem. Mol. Biol. 42, 148–154 [DOI] [PubMed] [Google Scholar]
- 11. Tsang S. Y., Moore J. C., Huizen R. V., Chan C. W., Li R. A. (2007) Ectopic expression of systemic RNA interference defective protein in embryonic stem cells. Biochem. Biophys. Res. Commun. 357, 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duxbury M. S., Ashley S. W., Whang E. E. (2005) RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem. Biophys. Res. Commun. 331, 459–463 [DOI] [PubMed] [Google Scholar]
- 13. Elhassan M. O., Christie J., Duxbury M. S. (2012) Homo sapiens systemic RNA interference-defective-1 transmembrane family member 1 (SIDT1) protein mediates contact-dependent small RNA transfer and microRNA-21-driven chemoresistance. J. Biol. Chem. 287, 5267–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pratt A. J., Rambo R. P., Lau P. W., MacRae I. J. (2012) Preparation and characterization of the extracellular domain of human Sid-1. PLoS One 7, e33607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren R., Xu X., Lin T., Weng S., Liang H., Huang M., Dong C., Luo Y., He J. (2011) Cloning, characterization, and biological function analysis of the SidT2 gene from Siniperca chuatsi. Dev. Comp. Immunol. 35, 692–701 [DOI] [PubMed] [Google Scholar]
- 16. Leahy D. J., Dann C. E., 3rd, Longo P., Perman B., Ramyar K. X. (2000) A mammalian expression vector for expression and purification of secreted proteins for structural studies. Protein Expr. Purif. 20, 500–506 [DOI] [PubMed] [Google Scholar]
- 17. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 18. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 19. Blom N., Gammeltoft S., Brunak S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 20. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poirot O., O'Toole E., Notredame C. (2003) Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31, 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jordan G. E., Piel W. H. (2008) PhyloWidget: web-based visualizations for the tree of life. Bioinformatics 24, 1641–1642 [DOI] [PubMed] [Google Scholar]
- 23. Sarov-Blat L., Livneh Z. (1998) The mutagenesis protein MucB interacts with single strand DNA binding protein and induces a major conformational change in its complex with single-stranded DNA. J. Biol. Chem. 273, 5520–5527 [DOI] [PubMed] [Google Scholar]
- 24. Serna-Rico A., Salas M., Meijer W. J. (2002) The Bacillus subtilis phage φ29 protein p16.7, involved in φ29 DNA replication, is a membrane-localized single-stranded DNA-binding protein. J. Biol. Chem. 277, 6733–6742 [DOI] [PubMed] [Google Scholar]
- 25. Beattie T. R., Bell S. D. (2012) Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 31, 1556–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Schott D. H., Cureton D. K., Whelan S. P., Hunter C. P. (2005) An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 102, 18420–18424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Draper D. E., von Hippel P. H. (1978) Nucleic acid binding properties of Escherichia coli ribosomal protein S1: II. co-operativity and specificity of binding site II. J. Mol. Biol. 122, 339–359 [DOI] [PubMed] [Google Scholar]
- 29. Morrone S. R., Wang T., Constantoulakis L. M., Hooy R. M., Delannoy M. J., Sohn J. (2014) Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc. Natl. Acad. Sci. U.S.A. 111, E62–E71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T., Hur S. (2011) Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U.S.A. 108, 21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maillard P. V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S. W., Voinnet O. (2013) Antiviral RNA interference in mammalian cells. Science 342, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Lu J., Han Y., Fan X., Ding S. W. (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jialin G., Xuefan G., Huiwen Z. (2010) SID1 transmembrane family, member 2 (Sidt2): a novel lysosomal membrane protein. Biochem. Biophys. Res. Commun. 402, 588–594 [DOI] [PubMed] [Google Scholar]