Abstract
Several independent studies have recently converged upon the conclusion that the human bacterial pathogen Mycobacterium tuberculosis encounters copper during infections. At least three independently regulated pathways respond to excess copper and are required for the full virulence of M. tuberculosis in animals. In this review, I will discuss the functions of the best-characterized copper-responsive proteins in M. tuberculosis, the potential sources of copper during an infection, and remaining questions about the interface between copper and tuberculosis.
Keywords: bacterial pathogenesis, copper, gene regulation, infectious disease, Mycobacterium tuberculosis, virulence
Introduction
Mycobacterium tuberculosis is the causative agent of the respiratory disease tuberculosis, infects nearly one-third of the world's population, and kills about 1.5 million people annually (WHO Global Tuberculosis Report 2014). M. tuberculosis is transmitted from person-to-person via aerosolized droplets created by coughing or sneezing, and only naturally infects humans. Although one might assume that M. tuberculosis does not have to adapt to widely variable external conditions like other microbes in the environment, the human body provides numerous challenges to which M. tuberculosis must adjust. These include toxic chemicals (reactive oxygen and nitrogen species), changes in temperature (e.g. from the inside of a human macrophage to an aerosol droplet in transit to the next host; fever), and accessibility to nutrients. Over the years the concept of “nutritional immunity” has become increasingly appreciated; several studies have determined that an infected host can sequester essential metals to prevent microbial pathogenesis (1). For example, Fe sequestration by mammalian host proteins during infections has been appreciated for decades. More recently, it has been determined that calprotectin, which is present in abundant amounts in neutrophils, is recruited to sites of infection to sequester Zn and Mn during Staphylococcus and Salmonella infections (2, 3). Strikingly, several studies almost simultaneously determined that the toxicity of excess Cu plays a critical role in suppressing M. tuberculosis infections and that different pathways contribute to Cu resistance in M. tuberculosis (4–8). Thus, in contrast to Fe, Mn, and Zn, Cu appears to co-localize with bacteria to limit their growth.
Early Clues of the Importance of Metal Transporters during M. tuberculosis Infections
The first evidence that Cu may play an important function during tuberculosis came from transcriptional analysis of M. tuberculosis grown in mice versus in culture. Several genes encoding putative cation transporters, which are integral cytoplasmic membrane proteins, are more highly expressed in bacteria isolated from mice than from broth culture and are found in an in vivo expressed genomic island (9). One of the genes in this locus was eventually determined to encode a Cu-sensing transcription factor known as CsoR for copper-sensitive operon repressor (8). CsoR is the founding member of a family of Cu-sensing transcriptional repressors in numerous Gram-positive and acid-fast bacterial species, and has a novel distinctive helical structure. In M. tuberculosis, CsoR binds to operator sequences as a dimer, with each monomer coordinating a +1 Cu ion (Cu+). Later studies determined that CsoR regulates a single promoter, csoRp, which controls the expression of a four-gene operon. Metallation of CsoR leads to a derepression of this operon, resulting in its expression in the presence of Cu. Included in this operon is ctpV (cation transport protein V), one of the cation transporter genes identified in the in vivo expressed genomic island (Fig. 1, dark green pathway) (6, 8). Several years earlier, ctpV was identified as a gene that is up-regulated in cultured human macrophages, further suggesting that it may be important during infection (10). Ultimately, ctpV was deleted and disrupted from M. tuberculosis and shown to have a Cu-sensitive phenotype in vitro and an attenuated growth phenotype in a guinea pig infection model. Interestingly, a ctpV mutant does not have a phenotype in a mouse model of infection (5); however, tuberculosis infections in mice do not always represent all aspects the human disease.
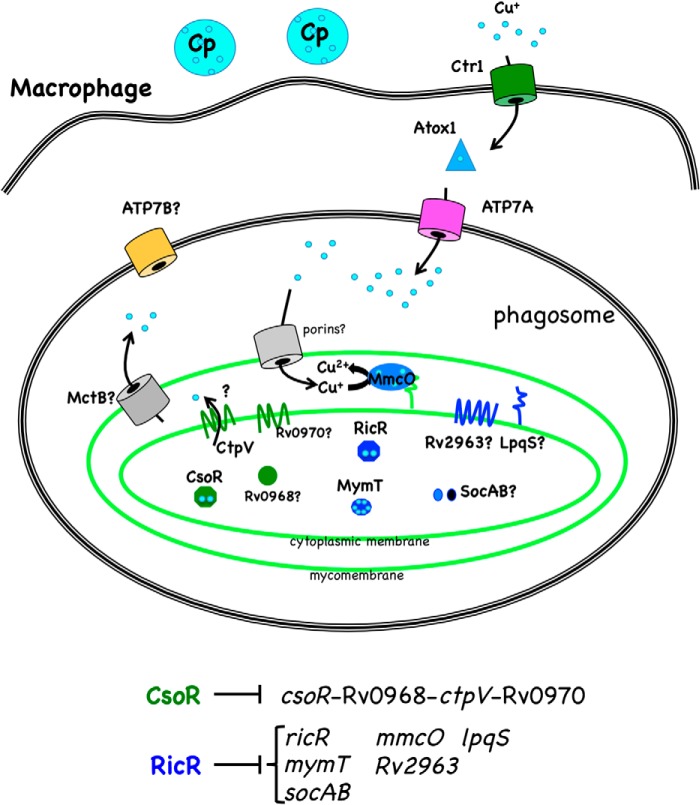
FIGURE 1.
M. tuberculosis and macrophage Cu homeostasis pathways. M. tuberculosis is found predominantly in phagocytic cells such as macrophages. Infection of macrophages by M. tuberculosis and other pathogens can lead to the production of IFNγ, which induces expression of Ctr1 and ATP7A on the host side of the interaction. Cu homeostasis on the host side proceeds as follows. Cp is a multicopper oxidase that converts ferrous iron (Fe2+) to ferric iron (Fe3+) throughout the body, Ctr1 is a high-affinity Cu+ transporter that transfers Cu to Atox1, a cytosolic Cu+ chaperone (reviewed in Ref. 30). Cu+ can then be transferred to the P-type ATPase ATP7A, which can pump Cu+ into the phagosome. ATP7B is another P-type ATPase that is essential for normal mammalian Cu homeostasis but has not yet been implicated in microbial pathogenesis (reviewed in Ref. 31). On the pathogen side, M. tuberculosis (light green) has at least three independent Cu-responsive pathways as described in the text. The CsoR regulon gene products are in dark green, RicR regulon gene products are in dark blue, and proteins of unknown regulation are in gray. Cu ions are depicted in light blue.
Based on the finding that a Cu-sensing regulator was induced in mice, Talaat and co-workers (11) hypothesized that Cu may induce the expression of other genes important for the pathogenesis of M. tuberculosis. Numerous genes are strongly induced when M. tuberculosis is treated with Cu sulfate, including several transcription factors and many genes of unknown function (11).
At about the same time Talaat's group (11) concluded that Cu played a role during tuberculosis infections, one of the first bacterial metallothioneins was discovered in M. tuberculosis (12). Metallothioneins are generally small, cysteine-rich proteins that bind metal ions and protect cells from metal overload. Nathan and colleagues (12) determined that expression of mymT (Mycobacterium metallothionein) in the non-pathogenic saprophyte Mycobacterium smegmatis confers resistance to a compound called ebselen (2-phenyl-1,2-bensioselenazol-3(2H)-one). Because Ebselen was known to interact with metallothioneins (13), this led to the hypothesis that MymT was a metallothionein. Indeed, the Nathan laboratory determined that MymT binds up to six Cu+ ions and protects M. tuberculosis from Cu toxicity in vitro. Although deletion of mymT results in increased Cu sensitivity in vitro, a mymT mutant is as virulent as wild type M. tuberculosis in mice (12).
Gatekeepers of Cu Entry
Mycobacteria have an unusual and non-canonical outer membrane referred to as the mycobacterial outer membrane (“MOM”).2 Although this feature might categorize Mycobacterium as a Gram-negative genus, mycobacteria do not colorize properly using Gram-staining methods and are thus referred to as “acid-fast” organisms. Despite lacking a traditional Gram-negative outer membrane, the Niederweis group (14) determined that some mycobacteria possess outer membrane channel-forming proteins including porins. Among the putative MOM proteins, the Niederweis group (4) identified MctB (mycobacterial copper transport protein B), which is required for Cu resistance (Fig. 1). Although it was initially believed to be a MOM protein, a more recent study suggests that it is a cytoplasmic membrane protein (15). The link between MctB function and Cu resistance was a fortuitous discovery; deletion of mctB in M. smegmatis results in a strong growth defect when the bacteria are inoculated onto a specialized mycobacterial medium called Middlebrook 7H10 (“7H10”) agar, but not when they are inoculated onto Luria-Bertani (LB) agar. Niederweis and colleagues (4) ultimately determined that it was the presence of Cu in 7H10 agar that limited the growth of the mctB mutant. A deletion mutation in mctB of M. tuberculosis, a species that does not grow on LB agar, also results in a growth defect on 7H10 agar. More importantly, the mctB mutant is less fit for growth in mice and guinea pigs. Similar to the ctpV data, the attenuated phenotype of the mctB mutant is more dramatic in guinea pigs than in mice. Supplementation of the animals' drinking water with Cu, however, decreases the bacterial burden of the mctB mutant, whereas the parental M. tuberculosis strain is not dramatically affected for growth under these conditions.
Wolschendorf et al. (4) determined that MctB is important for maintaining normal Cu levels in the cytosol of mycobacteria. Interestingly, the authors of this study also determined that M. tuberculosis has about 100 times less Cu in the cytosol than M. smegmatis (4). It is possible that this reflects proteomic differences between the bacteria; M. smegmatis has a genome that is about twice the size of the M. tuberculosis genome, and may thus encode more Cu-binding proteins or other Cu-chelating molecules.
More recently, the Wolschendorf group (15) determined that porins are important for Cu homeostasis in M. smegmatis. Although porin genes have not yet been identified in M. tuberculosis, there is some evidence to suggest this pathogen uses one or more porins for Cu uptake. Speer et al. (15) showed that incubation of M. tuberculosis with spermine, which can block porin-mediated transport, protects bacteria in media with otherwise toxic Cu levels, suggesting that one or more porins exist in M. tuberculosis that allow Cu into this bacterium.
Cu Regulation, Resistance, and Alien Genes
In yet another independent but coincidental study, my laboratory identified a third Cu homeostasis system called the regulated in copper repressor (RicR) regulon (6). In a microarray analysis of three M. tuberculosis H37Rv strains defective for proteasomal protein degradation, my colleagues and I identified several genes that are repressed for expression when compared with expression in a wild type strain. Proteasome-dependent protein degradation is essential for the pathogenesis of M. tuberculosis, and it is likely that proteolysis is linked to several pathways responsible for virulence. Ultimately, my colleagues and I determined that several of the genes repressed in proteasome-defective M. tuberculosis are also highly expressed in the presence of Cu (11). One gene, RicR (Rv0190), is a homologue of M. tuberculosis CsoR. RicR regulates the expression of genes from six different promoters, including its own, distributed throughout the M. tuberculosis genome. Under low Cu conditions, RicR represses the expression of the genes encoding a multicopper oxidase (mmcO), two putative membrane proteins (lpqS and Rv2963), Mycobacterium metallothionein (mymT), and two putative open reading frames called socAB (Fig. 1). All of these genes include a palindromic sequence near the −10 region of their promoters that is required for RicR binding and repression in the absence of Cu (6).
RicR is conserved in many Gram-positive bacterial species and, as mentioned previously, highly similar to CsoR. Interestingly, RicR is more similar to CsoR orthologues in other bacteria than M. tuberculosis CsoR. The crystal structure of the RicR orthologue in Streptomyces lividans (CsoR) revealed a dimer of tetramers configuration (16). Based on this, my colleagues and I presume that RicR binds to DNA as a dimer of tetramers in M. tuberculosis, with each monomer capable of binding a Cu+ ion, leading to their release from DNA.
The RicR regulon genes, with the notable exception of ricR itself, are almost all exclusively found in pathogenic mycobacteria, suggesting that the RicR Cu response is important during infections. Almost nothing is known about lpqS, a putative lipoprotein gene, or Rv2963, a putative permease gene. Genetic disruption of lpqS or Rv2963 alone does not lead to Cu sensitivity, and their roles in pathogenesis are unclear; transposon mutations in either gene result in hypervirulence. However, this phenotype could not be rescued by restoring a wild type copy of the respective genes into the mutants. Thus, it remains to be determined what the functions of these putative membrane proteins are in M. tuberculosis physiology.
In contrast to LpqS and Rv2963, MmcO has high similarity to several well characterized multicopper oxidases (MCO)/ferroxidases in other domains of life (17, 18). In eukaryotes from yeast to humans, MCOs oxidize Fe2+ to Fe3+, making it less toxic to cells. MCOs such as Saccharomyces cerevisiae Fet3p facilitate the receptor-mediated transport of Fe into cells (19). Interestingly, MmcO is lipidated (18) and, like Fet3p in yeast, is membrane-anchored in M. tuberculosis, although most if not all known bacterial MCOs are soluble periplasmic proteins. Fet3p, along with the Fe permease Ftr1p, is involved in Fe transport and also provides Cu resistance in yeast (20); therefore, my colleagues and I speculated that MmcO may perform similar functions in M. tuberculosis. It was previously determined that a related periplasmic MCO in the Gram-negative bacterium Pseudomonas aeruginosa was important for Fe acquisition (21). However, experiments in my laboratory to test this hypothesis have so far proved negative for M. tuberculosis.3 Furthermore, deletion of mmcO in M. tuberculosis results in a Cu-sensitive phenotype that is observed using an agar plate-based assay, but not a liquid-based assay. This result differs from the more robust Cu-sensitive phenotype observed with a mymT mutant that has phenotypes both in liquid and on solid medium (12, 18, 22). Like a mymT mutant, an mmcO mutant is not attenuated for virulence in a mouse infection model (22).
Because mymT and mmcO mutants have Cu-sensitive phenotypes in vitro but no apparent virulence defect in vivo, Shi et al. tested a double mymT mmcO mutant for these phenotypes. Although a double mutant is more sensitive to Cu in vitro than either single mutant, it is still as virulent as wild type M. tuberculosis in mice (22). Taken together, it appears that the functions of MymT and MmcO are to combat excess extracellular Cu but that they are not critical for virulence in at least one animal infection model.
MymT was not annotated prior to the study by Gold et al. (12), most likely due to its small size (53 amino acids) and lack of homology to any known protein. Like mymT, socAB was also not annotated in the M. tuberculosis H37Rv genome, and only socB was annotated as an open reading frame in the M. tuberculosis CDC1551 genome (6). socAB encodes what are predicted to be two highly basic small proteins, each of about 50–60 residues. Of all of the RicR-regulated genes, socAB are only found in the “tuberculosis complex” of mycobacteria that are generally associated with human and other mammalian infections. A transposon insertion mutation in socA, which is likely to inactivate socB as well, does not affect Cu resistance or virulence of M. tuberculosis H37Rv. Thus, like lpqS and Rv2963, the functions of these genes remain to be determined.
It is noteworthy that the socAB locus is so “alien” that we cannot identify related sequences in any organism or bacteriophage to date. That RicR controls its expression is intriguing. Did the tuberculosis complex acquire socAB to allow it to be a better pathogen in humans and other mammals? Perhaps these genes, as well as others in the RicR regulon, are required during specific phases of infection in higher mammals.
Because inactivation of one or two RicR-regulated genes has no effect on virulence in mice, Shi et al. tested the hypothesis that the entire regulon needs to be inactivated to observe a phenotype. Based on the CsoR structure, Shi et al. could predict which residues in RicR might be important for coordinating Cu; mutagenesis of one or more of these amino acids could render RicR “Cu-blind” (and thus DNA-bound even in the presence of Cu), resulting in the constitutive repression of the entire regulon. Because RicR represses the ricR promoter, Shi et al. made a construct where the RicR-binding site was mutated to allow constitutive expression (“ricRpc”), and where the ricR coding sequence was mutated to prevent Cu binding (“RicRC38A”); thus, RicRC38A would be constitutively expressed to repress the other RicR-regulated promoters. This construct, “ricRpc-ricRC38A”results in M. tuberculosis that is highly sensitive to Cu in vitro and, more importantly, attenuated in mice (22). Interestingly, a control strain that constitutively expresses wild type ricR (“ricRpc-ricR+) is more resistant to Cu than wild type M. tuberculosis; this may be due to elevated levels of RicR, which could act as a sink for Cu+.
It is important to consider that not all Cu-regulated genes may be necessary for Cu homeostasis. For example, Cu may act as a signal to indicate that M. tuberculosis has entered an environment that requires the expression of specific genes needed to deal with Cu-independent stresses within a macrophage. Because LpqS, Rv2963, and SocAB do not appear to be required for Cu resistance, perhaps they counteract other yet-to-be-identified antimicrobial factors in the host.
Copper, Copper Everywhere
Where exactly does M. tuberculosis (or any pathogen) encounter Cu? Several studies in the last few years have strongly indicated that microbes sense Cu and other metals during infections. Petris and colleagues (23) showed early on that the eukaryotic Cu transporter ATP7A contributes to restricting bacterial growth in cultured macrophages. The Petris group showed that cultured macrophages in which ATP7A gene expression is silenced do not control the growth of a non-pathogenic Escherichia coli strain as well as control macrophages can. Because M. tuberculosis is mainly found in phagocytic cells, it would not be surprising if ATP7A were important for controlling mycobacterial growth as well, a hypothesis my colleagues and I are currently testing.
Another potential source of Cu is ceruloplasmin (Cp). Cp is a multicopper oxidase that oxidizes Fe2+ to Fe3+, and is found in blood plasma and thus in most if not all parts of the body (24). Although the Cu in Cp has not been previously considered as a source of antimicrobial activity, it could nonetheless have the potential to affect infection. For example, phagocytes may take up Cp, after which phagolysosomal proteases could break down Cp and release its Cu. If bacteria happen to be in the same compartment, the released Cu could potentially kill the microbe in parallel to the ATP7A-dependent mechanism proposed by Petris and colleagues (23). Because microbes typically induce an immune response that leads to the acidification of the phagolysosomal compartment, one could also imagine that the acidic pH would maintain Cu in its most toxic form (Cu+). Thus, it will be interesting to see if animals that are deficient in Cp are more susceptible to infection by M. tuberculosis or other intracellular pathogens such as Salmonella.
Lots of Remaining Questions
Although much research has revealed that M. tuberculosis uses several independent pathways to deal with Cu toxicity, several key questions remain. For one, mctB is not CsoR- or RicR-regulated and therefore represents a third independent Cu resistance pathway. The mechanism of regulation of mctB is unknown. Furthermore, the mechanism of action of many Cu-responsive proteins has yet to be determined. CtpV appears to be required for Cu export, but this has not been shown definitively. All Ctp proteins are P-type ATPases, and there are 11 Ctp proteins in M. tuberculosis; although not all of them are regulated in a Cu-dependent manner, we cannot rule out that one or more of them function in Cu efflux. Interestingly, CtpA and CtpB are predicted to be Cu-binding ATPases, (25, 26). If they do indeed bind Cu, it is possible these are required for metallating extra-cytoplasmic Cu-binding proteins. Also, it is curious as to why an organism such as M. tuberculosis requires two distinct Cu-responsive regulons, which begs to ask if there is a biphasic, fine-tuned, temporal response to Cu in the host. Additionally, a Cu chaperone, with features of well characterized Cu chaperones (27), has not been identified in M. tuberculosis. It is possible that there is no protein chaperone, or that there is a non-classical type of chaperone, or that mycobacteria use one or more small molecules such as mycothiol to mobilize Cu. However, this last option seems unlikely as it would not be clear how this could be carefully regulated. Along these lines, although it is generally presumed that most Cu-binding proteins work outside of the cytoplasm, Cu nonetheless enters the bacterial cytoplasm and interacts with proteins such as the transcriptional regulators CsoR and RicR and the metallothionein MymT; how then is the metal displaced from the regulator and disposed of to restore transcriptional repression? It seems unlikely that mycothiol or similar molecules would be sufficient to deal with the necessary rapid changes in Cu-regulated gene expression.
With the advent of improved technologies for quantifying metals in biological systems (28) as well as a growing interest in the role of nutritional immunity in host-pathogen interactions (29), there is little doubt that the understanding of how microbes and their hosts regulate metal homeostasis will lead to the improved treatment of many devastating diseases.
Acknowledgments
I thank Ricky Festa, Mick Petris, and Dennis Thiele for lively discussions about copper and bacterial pathogenesis, and Ricky Festa for reading a draft version of this review.
This work was supported by National Institutes of Health Grant HL92774 (to K. H. D.). This is the third article in the Thematic Minireview series “Metals at the Host-Pathogen Interface.” The author declares that she has no conflicts of interest with the contents of this article.
X. Shi and K. H. Darwin, unpublished observations.
- MOM
- mycobacterial outer membrane
- Cp
- ceruloplasmin
- MCO
- multicopper oxidase.
References
- 1. Weinberg E. D. (1975) Nutritional immunity: host's attempt to withold iron from microbial invaders. JAMA 231, 39–41 [DOI] [PubMed] [Google Scholar]
- 2. Corbin B. D., Seeley E. H., Raab A., Feldmann J., Miller M. R., Torres V. J., Anderson K. L., Dattilo B. M., Dunman P. M., Gerads R., Caprioli R. M., Nacken W., Chazin W. J., Skaar E. P. (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965 [DOI] [PubMed] [Google Scholar]
- 3. Liu J. Z., Jellbauer S., Poe A. J., Ton V., Pesciaroli M., Kehl-Fie T. E., Restrepo N. A., Hosking M. P., Edwards R. A., Battistoni A., Pasquali P., Lane T. E., Chazin W. J., Vogl T., Roth J., Skaar E. P., Raffatellu M. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolschendorf F., Ackart D., Shrestha T. B., Hascall-Dove L., Nolan S., Lamichhane G., Wang Y., Bossmann S. H., Basaraba R. J., Niederweis M. (2011) Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 108, 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward S. K., Abomoelak B., Hoye E. A., Steinberg H., Talaat A. M. (2010) CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77, 1096–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Festa R. A., Jones M. B., Butler-Wu S., Sinsimer D., Gerads R., Bishai W. R., Peterson S. N., Darwin K. H. (2011) A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wagner D., Maser J., Lai B., Cai Z., Barry C. E., 3rd, Höner Zu Bentrup K., Russell D. G., Bermudez L. E. (2005) Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174, 1491–1500 [DOI] [PubMed] [Google Scholar]
- 8. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 9. Talaat A. M., Lyons R., Howard S. T., Johnston S. A. (2004) The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U.S.A. 101, 4602–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Graham J. E., Clark-Curtiss J. E. (1999) Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U.S.A. 96, 11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ward S. K., Hoye E. A., Talaat A. M. (2008) The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gold B., Deng H., Bryk R., Vargas D., Eliezer D., Roberts J., Jiang X., Nathan C. (2008) Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 4, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jacob C., Maret W., Vallee B. L. (1998) Ebselen, a selenium-containing redox drug, releases zinc from metallothionein. Biochem. Biophys. Res. Comm. 248, 569–573 [DOI] [PubMed] [Google Scholar]
- 14. Niederweis M., Ehrt S., Heinz C., Klöcker U., Karosi S., Swiderek K. M., Riley L. W., Benz R. (1999) Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 33, 933–945 [DOI] [PubMed] [Google Scholar]
- 15. Speer A., Shrestha T. B., Bossmann S. H., Basaraba R. J., Harber G. J., Michalek S. M., Niederweis M., Kutsch O., Wolschendorf F. (2013) Copper-boosting compounds: a novel concept for antimycobacterial drug discovery. Antimicrob. Agents Chemother. 57, 1089–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan B. G., Vijgenboom E., Worrall J. A. (2014) Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucleic Acids Res. 42, 1326–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kosman D. J. (2010) Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J. Biol. Inorg. Chem. 15, 15–28 [DOI] [PubMed] [Google Scholar]
- 18. Rowland J. L., Niederweis M. (2013) A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J. Bacteriol. 195, 3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taylor A. B., Stoj C. S., Ziegler L., Kosman D. J., Hart P. J. (2005) The copper-iron connection in biology: structure of the metallo-oxidase Fet3p. Proc. Natl. Acad. Sci. U.S.A. 102, 15459–15464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi X., Stoj C., Romeo A., Kosman D. J., Zhu Z. (2003) Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 278, 50309–50315 [DOI] [PubMed] [Google Scholar]
- 21. Huston W. M., Jennings M. P., McEwan A. G. (2002) The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 22. Shi X., Festa R. A., Ioerger T. R., Butler-Wu S., Sacchettini J. C., Darwin K. H., Samanovic M. I. (2014) The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis cirulence. mBio. 5, e00876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White C., Lee J., Kambe T., Fritsche K., Petris M. J. (2009) A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kono S. (2013) Aceruloplasminemia: an update. Int Rev. Neurobiol. 110, 125–151 [DOI] [PubMed] [Google Scholar]
- 25. Novoa-Aponte L., León-Torres A., Patiño-Ruiz M., Cuesta-Bernal J., Salazar L. M., Landsman D., Mariño-Ramírez L., Soto C. Y. (2012) In silico identification and characterization of the ion transport specificity for P-type ATPases in the Mycobacterium tuberculosis complex. BMC structural biology 12, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agranoff D. D., Krishna S. (1998) Metal ion homeostasis and intracellular parasitism. Mol. Microbiol. 28, 403–412 [DOI] [PubMed] [Google Scholar]
- 27. Robinson N. J., Winge D. R. (2010) Copper metallochaperones. Ann. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ward S. K., Heintz J. A., Albrecht R. M., Talaat A. M. (2012) Single-cell elemental analysis of bacteria: quantitative analysis of polyphosphates in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hood M. I., Skaar E. P. (2012) Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hatori Y., Lutsenko S. (2013) An expanding range of functions for the copper chaperone/antioxidant protein Atox1. Antioxid. Redox Signal. 19, 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lutsenko S. (2008) Atp7b−/− mice as a model for studies of Wilson's disease. Biochem. Soc. Trans. 36, 1233–1238 [DOI] [PubMed] [Google Scholar]