Abstract
Numerous pathogenic microorganisms secrete small molecule chelators called siderophores defined by their ability to bind extracellular ferric iron, making it bioavailable to microbes. Recently, a siderophore produced by uropathogenic Escherichia coli, yersiniabactin, was found to also bind copper ions during human infections. The ability of yersiniabactin to protect E. coli from copper toxicity and redox-based phagocyte defenses distinguishes it from other E. coli siderophores. Here we compare yersiniabactin to other extracellular copper-binding molecules and review how copper-binding siderophores may confer virulence-associated gains of function during infection pathogenesis.
Keywords: copper, host-pathogen interaction, microbial pathogenesis, siderophore, superoxide dismutase (SOD)
Siderophores
Siderophores (Greek for iron carrier) are a diverse group of specialized ferric iron(III)-binding metabolites that function to supply iron to the microbe and are the subject of classic works in microbial metabolism (1–3). Iron is an essential metal for most bacteria, acting as an essential cofactor in diverse physiological processes including respiration, deoxynucleotide biosynthesis, and DNA replication (4). The evolution of siderophores is viewed as a response to the appearance of O2 in the early atmosphere, which resulted in oxidation of soluble ferrous iron (Fe(II)) to its relatively insoluble ferric form (Fe(III)) (5).
Siderophores are widely synthesized among microbes, with over 500 different siderophores described. These specialized metabolites use a chemically diverse array of metal-binding prosthetic groups, often deployed in varying combinations within a single siderophore. Bacterial siderophore biosynthesis and transporter proteins are translated when intracellular iron is low, classically following transcriptional derepression by the global iron regulator, Fur (ferric uptake repressor) (6, 7). Siderophore biosynthetic pathways can be classified as non-ribosomal peptide synthetase/polyketide synthase (NRPS/PKS)2-dependent or -independent systems. NRPS/PKS are large (>350 kDa for the yersiniabactin biosynthesis protein irp1 (8)) multi-enzyme complexes that use a wide range of cellular substrates to synthesize various metabolites including catecholate and phenolate type siderophores.
Once secreted into the extracellular space, siderophores can bind oxidized metal ions with remarkably high affinities (9). Ferric siderophore complexes are subsequently recognized and internalized through siderophore-specific transport machinery in bacterial inner and outer membranes. Transport through the Gram-negative outer membrane transporters occurs through specialized proteins associated with the TonB protein complex, which transduces energy from the cytoplasmic proton motive force (3, 10, 11). Inner membrane ATP-binding cassette transporters in both Gram-negative and Gram-positive organisms transport ferric siderophore complexes, or the iron released from them, to the cytoplasm (12–16). Once ferric siderophores are internalized, their intracellular fate varies between different siderophores, with reported examples of Fe(III) reduction to Fe(II) or Fe(III)-siderophore hydrolysis. All or part of the siderophore may be recycled and used again after releasing its metal ion cargo (2, 18).
Although pathogens often carry multiple iron acquisition systems that are redundant in laboratory culture conditions, siderophore-dependent systems appear to play specialized roles at the host-pathogen interface (2, 16, 19, 20). Specifically, siderophores have been ascribed roles in liberating ferric ions bound to host iron storage proteins or sequestered away from pathogens within distinctive compartments. As such, siderophores have long been considered a microbial solution to low iron availability imposed by microenvironments in the infected host (21).
Uropathogenic Escherichia coli Siderophores
E. coli recovered from infected patients or animals often synthesize different siderophore types spanning three chemical families in addition to the prototypical, genetically conserved enterobactin siderophore system (Fig. 1) (22). Although unnecessary for growth in standard culture conditions, multiple siderophore genes are up-regulated during E. coli urinary tract infections (23, 24), and siderophore biosynthesis in this disease has been demonstrated by direct mass spectrometric detection (25). The multiplicity of siderophores synthesized by these uropathogenic E. coli strains raises the question of why bacteria would commit scarce cellular resources to synthesize chemically distinctive siderophores with a common recognized function (26).
FIGURE 1.
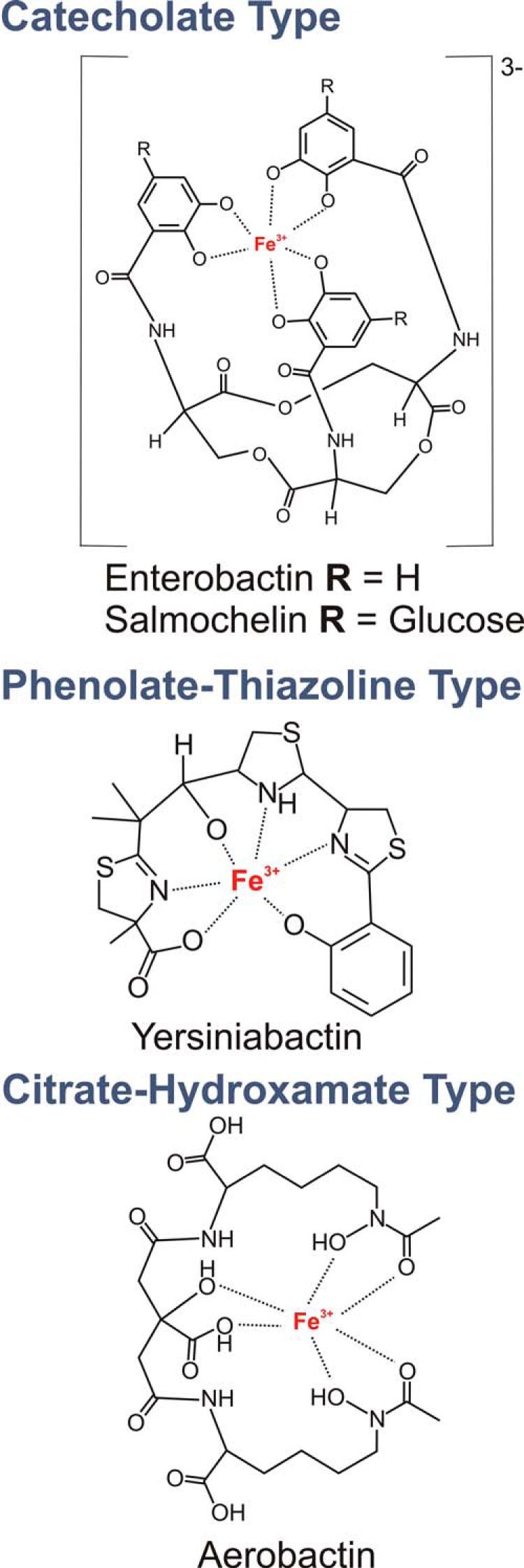
Uropathogenic E. coli siderophores. Pathogenic E. coli isolates synthesize multiple siderophore types known to coordinate Fe(III) with a variety of functional groups (22).
One reason pathogenic bacteria may synthesize multiple siderophore types emerged from the finding that the inflammation-associated protein siderocalin (SCN, also known as lipocalin 2 (Lcn2), NGAL, or 24p3) can tightly bind the ferric complex of the prototypical E. coli siderophore enterobactin, rendering its iron cargo inaccessible to bacteria (27). In contrast, functional data with siderophores synthesized from the three genetically non-conserved E. coli siderophore systems (salmochelin, yersiniabactin, and aerobactin) suggest that these do not bind to SCN, leading to their categorization as “stealth siderophores” along with other non-E. coli siderophores with this property. SCN-deficient mice exhibit increased susceptibility to bloodstream infections with E. coli that produce enterobactin as its sole siderophore, consistent with a physiologic role for this SCN-enterobactin binding interaction (28, 29). In the human urinary environment, this situation can be reversed, with enterobactin becoming necessary to liberate iron from SCN-bound ferric complexes that form with urinary catechol metabolites (30).
An additional rationale for synthesizing salmochelin (a more polar, glucose-conjugated version of enterobactin) is suggested by measurements showing that this siderophore partitions into cellular membranes less than enterobactin, which may allow it to more effectively scavenge ferric ions in membrane-rich host environments (31). Finally, using siderophores with different pH optima for ferric ion binding may allow uropathogenic E. coli to adapt to a wider range of host environments. Aerobactin, for example, may be a better iron scavenger in low pH environments where protonation diminishes the ferric ion affinity of catecholate siderophores (32, 33). Together, these findings suggest that siderophores' remarkable chemical diversity, even within a single bacterial strain, may represent chemical co-evolutionary responses to numerous selective pressures within vertebrate hosts.
Cupric Yersiniabactin in Human Infections
The phenolate/thiazoline-based yersiniabactin (Ybt) siderophore system was first described in pathogenic Yersinia species and subsequently found to be the most frequently expressed, genetically non-conserved siderophore system in uropathogenic E. coli (22, 34–36). Ybt is synthesized by a large NRPS/PKS complex that has been successfully reconstituted in vitro and assembles Ybt through a succession of phosphopantetheine-bound intermediates (37, 38). These biosynthetic genes are encoded alongside metabolic, transport, and transcription factor genes in the genetically mobile, multi-operon Yersinia high pathogenicity island (11, 39).
Although siderophores are known for ferric metal binding, in vitro binding of non-ferric metal ions has been widely noted (40–44). To identify the metals that bind Ybt in a biologically relevant environment, Chaturvedi et al. (25) devised an LC-constant neutral loss mass spectrometric screen using a yersiniabactin-specific MS/MS fragmentation pathway. When Ybt was mixed with pooled urine samples from healthy human donors, LC-constant neutral loss screen identified a chromatographic peak corresponding to a new metal-yersiniabactin complex. Accurate mass values and isotope pattern matched that of a cupric yersiniabactin (Cu(II)-Ybt) complex. This complex was replicated using defined components in vitro when Ybt was titrated with cupric sulfate solution, producing a visibly blue-colored complex. LC-MS/MS analysis detected Cu(II)-Ybt in urine from urinary tract infection patients infected with yersiniabactin-producing E. coli, demonstrating the physiologic relevance of copper binding by yersiniabactin. Cu(II)-Ybt was also detected in urine and bladder tissue from mice experimentally infected with UTI89, a model yersiniabactin-producing uropathogenic E. coli strain. Interestingly, the Cu(II)-Ybt/Fe(III)-Ybt molar ratio in both human and mouse specimens was consistently greater than 1, suggesting that Ybt binds host-derived copper at least as extensively as Fe(III) during infections (25).
These findings provide direct mass spectrometric evidence that yersiniabactin is produced during human urinary tract infections and that metal availability and yersiniabactin metal affinities are such that copper complexes exceed iron complexes. The observed excess copper complexes may reflect host responses that limit siderophore-accessible iron (nutritional immunity) as well as inflammation-associated processes that increase copper availability at the host-pathogen interface (19, 45, 46). The presence of a virulence-associated, copper-binding siderophore in E. coli may reflect an adaptive response to the distinctive physiologic metal composition that arises in infection microenvironments (21). Yersiniabactin may play a similar role in Yersinia pestis, where it is linked to virulence in experimental animal models (12, 47). Common structural features of yersiniabactin and other biological copper-binding molecules may facilitate identification of analogous biological copper carriers in other microbial pathogens.
Structural Features of Biological Copper-binding Molecules
Although yersiniabactin is currently the best-described example of a siderophore shown to bind copper in the host, other examples may await discovery. Structural features of well known extracellular copper carriers may assist identification of currently unappreciated copper binders among pathogen-associated siderophores. In the mammalian host, the prototypical extracellular copper carrier is ceruloplasmin, which carries up to 95% of circulating copper and rises and falls with copper excess and deficiency, respectively (48). An ∼2-kDa urinary copper-binding molecule has also been observed in copper-overloaded mice but has not been structurally characterized (49). Copper ions bound to ceruloplasmin are critical components of its multi-copper oxidase function, which is essential for normal mammalian iron homeostasis. Ceruloplasmin harbors six copper atoms in three distinct copper sites, as is typical of the conserved multi-copper oxidase family (50). Ceruloplasmin contains three type I “blue copper” sites, two of which coordinate a single copper ion using a cysteine residue, a methionine residue, and two histidine residues (Fig. 2a). The other three copper ions form a histidine-rich trinuclear copper cluster in which four histidine imidazole nitrogens coordinate one type II copper and three histidine residues coordinate two type III copper ions (51). These six copper atoms are required to achieve the final conformation-driven state and are incorporated during ceruloplasmin biosynthesis late in the secretory pathway (52). Although an array of negatively charged groups might effectively coordinate copper, it is notable that cuproproteins often use histidine imidazole nitrogens for this purpose. Indeed this is a useful basis for identifying new cuproproteins from genomic databases (53, 54). Incorporation of analogous nitrogen-containing heterocycles into protein and non-protein microbial products may confer a similar copper binding preference.
FIGURE 2.
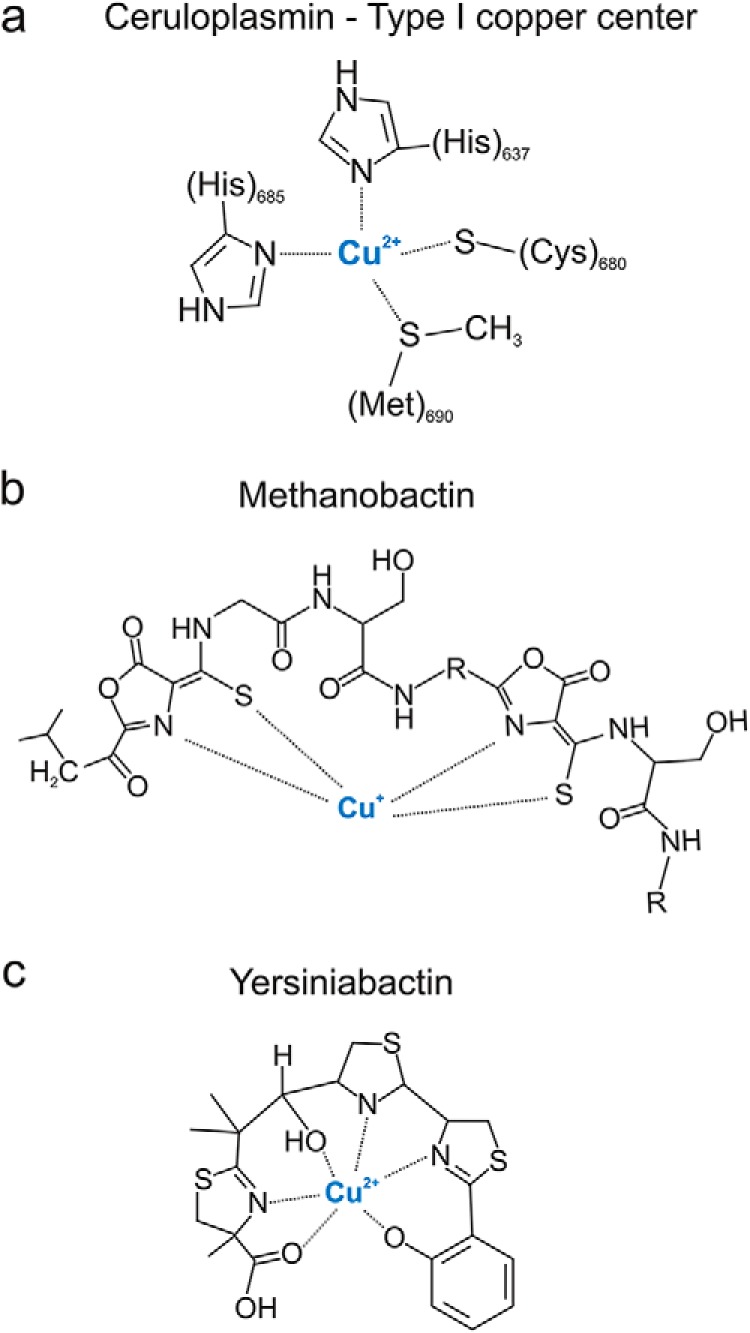
Extracellular copper-binding molecules. a, ceruloplasmin has six copper-binding sites of which three are type I copper centers coordinated with two histidine residues, a cysteine residue and a methionine residue (48). b, methanobactin structures contain a recognizable peptide backbone with distinctive oxazolone heterocycles, which are implicated in copper ion coordination (43). c, yersiniabactin has three heterocyclic nitrogens (two thiazoline and one thiazolidine) that contribute to a square planar configuration around Cu(II) alongside the phenolate oxygen (61).
Among prokaryotes, a new family of copper binding, siderophore-like, natural products has recently been described among methanotrophic Gram-negative bacteria from environmental sources. Methanobactins (Mb) are a family of small molecules secreted into the extracellular space to scavenge copper for nutritional purposes (for detailed review, see Ref. 42). These chalkophores (Greek for copper carriers) help satisfy the high copper demand resulting from biosynthesis of particulate methane monooxygenase, which permits methanotrophs to use methane as their sole carbon source (55, 56). There are numerous similarities between Mb and Gram-negative bacterial siderophore systems. Gram-negative siderophores and Mb are actively transported through the outer membrane by TonB-dependent transporters and possess distinctive heterocyclic structural elements involved in metal binding (43, 57). As in most cuproproteins, biophysical studies of Mb implicate heterocyclic nitrogens (oxazolones for Mb) in copper ion coordination (Fig. 2b). Use of these chemical groups to bind copper in Mb raises the possibility that previously described mechanisms for nitrogen heterocycle incorporation into other peptides and natural products (reviewed by Walsh et al. (58)) may facilitate copper binding in some of these molecules.
Yersiniabactin is an NRPS/PKS product that lacks the peptide backbone of Mb and possesses three nitrogen-sulfur heterocycles whose nitrogens participate in co-planar ferric ion binding identified in crystallographic structures (59, 60). Quantum mechanical modeling using density functional theory predicts a similar coordination environment for Cu(II)-Ybt, in which the three heterocyclic nitrogens (two thiazoline and one thiazolidine) and the phenolate oxygen contribute to a typical square planar configuration around Cu(II) (Fig. 2c). However, in contrast to other metal-Ybt complexes, Cu(II)-Ybt is predicted to exist as two closely related linkage isomers with one having an open penta-coordinate site arising from differential interaction with the terminal carboxylic acid group (61, 62). A similar phenolate-thiazoline role in copper binding may occur with pyochelin, a related siderophore from the opportunistic pathogen Pseudomonas aeruginosa, in which UV-visible analysis indicates formation of an in vitro Cu(II) complex (41). The consistent appearance of nitrogen-containing heterocycles in ceruloplasmin, methanobactin, and yersiniabactin metal-binding sites suggests a particularly important role for this structural element in binding copper. More examples of copper-binding molecules are necessary to properly evaluate this association and to predict biologically meaningful copper binding activity among microbial siderophores.
Copper-binding Siderophores as a Defense Against Host-derived Copper Toxicity
Secreted copper-binding molecules may have evolved in pathogens to neutralize the antibacterial activity of copper (for recent reviews, see Refs. 46, 63, and 64). Although inflammatory responses classically limit transition metal bioavailability, copper appears to be a notable exception. Several works report unchanged or increased systemic and local copper availability during infections (65–68). More recently, White et al. (45) have shown ATP7A-mediated copper trafficking from the Golgi apparatus to E. coli-containing phagosomes, consistent with a bactericidal system that targets phagocytosed microorganisms. Within this restricted phagosomal space (∼1.2 × 10−15 liters (69)), toxic levels of copper sufficient to kill microbes may be rapidly achieved.
Consistent with an important role for copper-based host defenses, uropathogenic E. coli isolates have been observed to exhibit greater copper resistance than intestinal isolates in urinary tract infection patients. Yersiniabactin synthesis was associated with copper resistance in these isolates, and a causative association was supported by a significant decrease in resistance by yersiniabactin-null mutants and a significant increase in resistance following supplementation with purified yersiniabactin (25). A similar relationship was observed in Pseudomonas aeruginosa, where synthesis of the virulence-associated siderophores pyochelin and pyoverdine (70, 71) increased copper resistance (72). Furthermore P. aeruginosa with deficiency in copper detoxification system showed reduced virulence, although connections with pyochelin and pyoverdine were unexplored (73).
Further studies will be necessary to examine how Cu(I) in the phagosome is oxidized to Cu(II) for yersiniabactin binding. The bacterial periplasmic multi-copper oxidase, CueO, has been suggested to oxidize Cu(I) to Cu(II) (74, 75). These copper defense systems may thus work in tandem with yersiniabactin and related siderophores to sequester Cu(II) ions.
Not all siderophores protect against copper toxicity. Catecholate siderophores in E. coli sensitized them to copper toxicity, an effect linked to Cu(II) reduction by catechols to the more cytotoxic Cu(I) ion (25, 74, 76). When bound to yersiniabactin, catechols were unable to reduce Cu(II) (25). In the presence of copper ions, yersiniabactin may thus protect E. coli by preventing enterobactin-mediated Cu(I) formation. (Fig. 3). It is however unclear whether aerobactin is similarly involved in strains that produce this siderophore.
FIGURE 3.
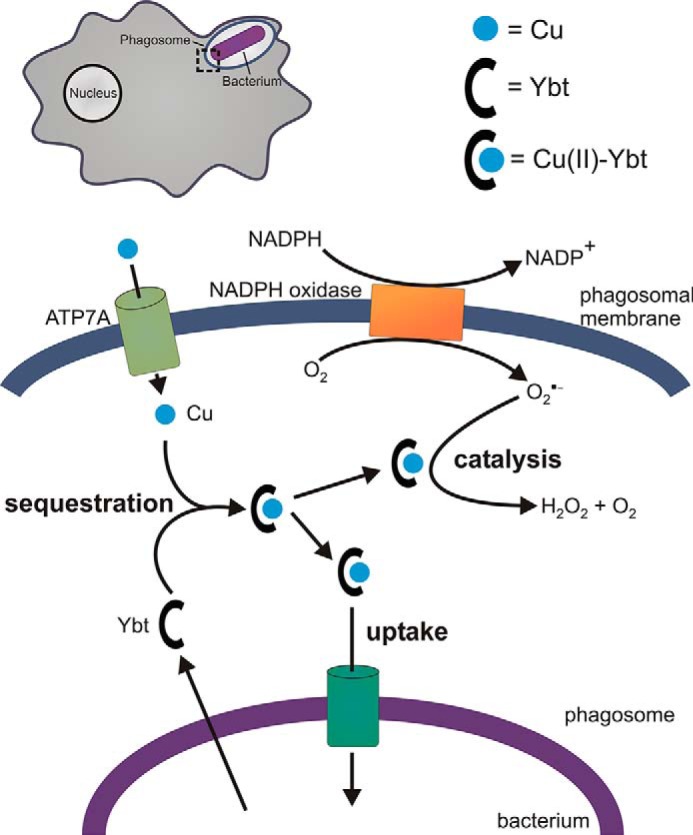
Copper-binding siderophore functions at the host-pathogen interface. Within the phagosomal compartments of host phagocytic cells (upper left), yersiniabactin may protect bacterial pathogens by sequestering toxic copper ions translocated by ATP7A into the phagosome (25, 45). The resulting Cu(II)-Ybt complex may further protect the host by catalyzing superoxide dismutation to counter superoxide-based host defenses (61). In phagosomal or non-phagosomal environments characterized instead by copper scarcity, E. coli may import Cu(II)-Ybt as a nutritional copper source.
The mechanism of copper toxicity in bacteria is incompletely understood. Cu(I) toxicity through Fenton reaction-mediated hydroxyl radical (OH•) generation has long been proposed to be an important mechanism, although observations that copper-loaded E. coli are more, not less, resistant to hydrogen peroxide suggest that Fenton chemistry alone is an inadequate explanation (77). Recent works instead point to a major role for cytoplasmic Cu(I) in dissociating iron-sulfur clusters and thereby interfering with multiple metabolic processes including heme and branched chain amino biosynthesis (78–80). This mechanism finds additional support from the finding that metal ions that do not act as Fenton reagents but also disrupt iron-sulfur clusters exert similar toxicity (81, 82). Although further studies are needed to dissect the toxic activities of copper, these results point to the ability of yersiniabactin to prevent free copper from reaching the cytoplasm as an important virulence function.
A pathogenic role for yersiniabactin copper binding was assessed in a version of the macrophage survival assay first described by White et al. (45). In this study, a yersiniabactin-deficient uropathogenic E. coli was profoundly sensitized to intracellular killing by macrophage-like RAW264.7 cells relative to its yersiniabactin-proficient isogenic wild type control (61). The yersiniabactin-null phenotype was abolished when copper-deficient RAW264.7 cells were used. This sensitization phenotype was similar to that observed for a K12 E. coli strain lacking the copper efflux pump CopA (83). Overall, these results are consistent with yersiniabactin sequestration of phagosomal copper ions as a contributor to intracellular survival. Together, these findings support a role for non-reducing, copper-binding siderophores in protecting pathogens from copper-based host defenses.
Copper-binding Siderophores as a Countermeasure Against Superoxide-based Host Defenses
Phagocytes are a classic host defense against invading microbial pathogens, and the copper-dependent ATP7A bactericidal system described by White et al. (45) is one of several chemical defenses deployed by these cells. Also notable among these defenses is the “respiratory burst,” in which a membrane-bound NADPH oxidase generates superoxide anion (O2⨪) within the phagosome (84).
Yersiniabactin synthesis was implicated in resisting superoxide-dependent host defenses through the observation that the yersiniabactin-dependent intracellular survival phenotype requires both copper and NADPH oxidase activity (61). Subsequent studies identified a soluble superoxide dismutase-like activity in uropathogenic Escherichia coli (UPEC)-conditioned medium that was similarly dependent upon yersiniabactin biosynthesis and copper supplementation. Chromatographic fractionation and mass spectrometric analysis associated this soluble SOD-like activity to Cu(II)-Ybt. Purified Cu(II)-Ybt exhibited SOD-like activity that was also observed upon substitution of Cu(II) with Fe(III) but not with non-redox active gallium (III), consistent with a catalytic requirement for redox active metal. Unlike previously described cupric salicylate complexes with SOD-like activity, Cu(II)-Ybt retained its activity in the presence of protein, which acts as a competitive copper chelator (61). These data are consistent with the ability of non-protein Cu(II)-Ybt complexes to catalyze superoxide dismutation as follows.
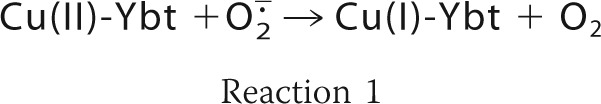 |
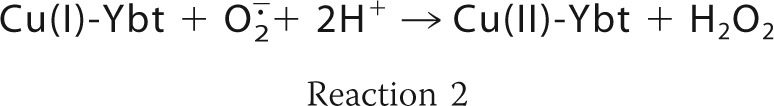 |
Density functional theory-based quantum simulations of this catalytic cycle supported a key role for copper redox cycling during coordination by the phenolate, thiazoline, and thiazolidine groups in yersiniabactin (61). There is evidence that the thiolate and heteroaromatic ring coordination sites of methanobactin may support a similar catalytic cycle (85). Although the catalytic activity of Cu(II)-Ybt does not exceed that of periplasmic Cu,Zn-SOD, the extracellular localization of yersiniabactin would be expected to place it in closer proximity to the superoxide source (the host NADPH oxidase complex) where it may be more effective. Cu(II)-Ybt catalysis may further benefit the pathogen through its lower metabolic cost of production and its likely ability to avoid proteolysis, which may permit substantial accumulation within the phagosome. Siderophore-based catalysts that take advantage of host-supplied copper may therefore be well suited to this distinctive host environment (86) (Fig. 3).
An analogous protective role for an extracellular copper-binding protein was recently suggested by Gleason et al. (87), in the human fungal pathogen Candida albicans. Candidal SOD5 was found to possess an open active site that readily captures extracellular copper for its activity. Although it lacks the zinc-binding site, it is otherwise structurally homologous to Cu,Zn-SODs and rapidly reacts with superoxide with rates approaching the dismutase limit (87). Yersiniabactin and SOD5 may thus use similar strategies to catalyze superoxide dismutation through spontaneous capture and immobilization of host-supplied, extracellular copper. A more detailed investigation of copper-based dismutation catalysts in this context will require an improved biochemical understanding of the role of superoxide in microbial killing. Nevertheless, it appears likely that additional examples of this survival strategy await discovery among invasive, disease-associated microorganisms.
The surprising ability of yersiniabactin production to modulate copper-dependent intracellular survival underscores that there is still much to learn about transition metal interactions at the host-pathogen interface. As the phagosome matures, metal content, pH, and redox potential may change dramatically (for recent reviews, see Refs. 88 and 89). Future measurements of the binding affinity of yersiniabactin for Cu(II) and Cu(I), both of which may be present within the phagosome, would aid these investigations.
Copper-binding Siderophores as Copper Scavengers
Methanobactin is a notable precedent for a siderophore-like small molecule that scavenges and delivers copper (a chalkophore) to bacteria. Although methanotrophic Gram-negative bacteria use methanobactin to support exceptional copper demands associated with particulate methane monooxygenase biosynthesis (56, 57), it remains unclear whether pathogenic bacteria benefit from a similar strategy. Copper is an important cofactor in multiple bacterial enzymes including NADH dehydrogenase, cytochrome oxidase, and copper/zinc superoxide dismutases (53). Gram-negative cuproenzymes such as Cu,Zn-SOD have been described to receive copper in the periplasmic compartment (17), and it therefore appears plausible that virulence-associated chalkophore systems could serve a nutritional function in low copper host environments.
The outer membrane ferric yersiniabactin importer gene (fyuA (11, 12, 60)) was found to be unnecessary for protection from copper toxicity, consistent with the prominent role for extracellular copper binding described above. Nevertheless, FyuA was recently found to act as a promiscuous transporter of metal-yersiniabactin complexes, including Cu(II)-Ybt. Relative to non-cupric complexes, Cu(II)-Ybt import reached a maximal level at low concentrations. Under competitive import conditions, Fe(III)-Ybt import was favored and notably uninhibited by high Cu(II)-Ybt concentrations. These observations raise the possibility that yersiniabactin exhibits a copper-scavenging functionality that can adapt to a high copper, low iron environment such as the phagosome by limiting copper import and prioritizing iron import (62). Further studies are necessary to determine how FyuA distinguishes between different metal-yersiniabactin complexes and whether bacteria use imported Cu(II)-Ybt to provide copper to cuproproteins.
Summary
Bacterial siderophores and siderophore-like molecules with characteristic structural features have been found to form stable complexes with extracellular copper ions in vitro and in vivo during human infections. In the case of yersiniabactin, this property may protect uropathogenic E. coli from innate antibacterial immune responses based on copper and superoxide generation. Because the chemical environment during infections is highly variable and is manipulated by the host, multiple roles for copper-binding siderophores are possible. Improved understanding of fundamental chemical properties of pathogenic siderophores as well as the biochemical environments in which they are deployed is necessary to identify new virulence-associated copper binders and to better understand their role in bacterial pathogenesis.
This work was supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund and National Institutes of Health Grant R01DK099534 (to J. P. H.) through the NIDDK. This is the fourth article in the Thematic Minireview series “Metals at the Host-Pathogen Interface.” The authors declare that they have no conflicts of interest with the contents of this article.
- NRPS/PKS
- non-ribosomal peptide synthase/polyketide synthase
- Ybt
- yersiniabactin
- SCN
- siderocalin
- Mb
- methanobactins
- SOD
- superoxide dismutase.
References
- 1. Neilands J. B. (1995) Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726 [DOI] [PubMed] [Google Scholar]
- 2. Miethke M., Marahiel M. A. (2007) Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., Slavinskaya Z., Vogel H. J. (2010) Siderophore uptake in bacteria and the battle for iron with the host: a bird's eye view. Biometals 23, 601–6111 [DOI] [PubMed] [Google Scholar]
- 4. Andreini C., Bertini I., Cavallaro G., Holliday G. L., Thornton J. M. (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 13, 1205–1218 [DOI] [PubMed] [Google Scholar]
- 5. Neilands J. B. (1981) Microbial iron compounds. Annu. Rev. Biochem. 50, 715–731 [DOI] [PubMed] [Google Scholar]
- 6. Bagg A., Neilands J. B. (1987) Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477 [DOI] [PubMed] [Google Scholar]
- 7. Fetherston J. D., Bearden S. W., Perry R. D. (1996) YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22, 315–325 [DOI] [PubMed] [Google Scholar]
- 8. Gehring A. M., DeMoll E., Fetherston J. D., Mori I., Mayhew G. F., Blattner F. R., Walsh C. T., Perry R. D. (1998) Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5, 573–586 [DOI] [PubMed] [Google Scholar]
- 9. Harris W. R., Carrano C. J., Cooper S. R., Sofen S. R., Avdeef A. E., McArdle J. V., Raymond K. N. (1979) Coordination chemistry of microbial iron transport compounds. 19. Stability constants and electrochemical behavior of ferric enterobactin and model complexes. J. Am. Chem. Soc. 101, 6097–6104, 10.1021/ja00514a037 [DOI] [Google Scholar]
- 10. Noinaj N., Guillier M., Barnard T. J., Buchanan S. K. (2010) TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perry R. D., Fetherston J. D. (2011) Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 13, 808–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fetherston J. D., Bertolino V. J., Perry R. D. (1999) YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32, 289–299 [DOI] [PubMed] [Google Scholar]
- 13. Raymond K. N., Dertz E. A., Kim S. S. (2003) Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schalk I. J. (2008) Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 102, 1159–1169 [DOI] [PubMed] [Google Scholar]
- 15. Brem D., Pelludat C., Rakin A., Jacobi C. A., Heesemann J. (2001) Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147, 1115–1127 [DOI] [PubMed] [Google Scholar]
- 16. Rodriguez G. M., Smith I. (2006) Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188, 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Osman D., Patterson C. J., Bailey K., Fisher K., Robinson N. J., Rigby S. E., Cavet J. S. (2013) The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P1B-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87, 466–477 [DOI] [PubMed] [Google Scholar]
- 18. Brickman T. J., McIntosh M. A. (1992) Overexpression and purification of ferric enterobactin esterase from Escherichia coli: demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267, 12350–12355 [PubMed] [Google Scholar]
- 19. Hood M. I., Skaar E. P. (2012) Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 10, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fetherston J. D., Kirillina O., Bobrov A. G., Paulley J. T., Perry R. D. (2010) The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 78, 2045–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prentice A. M., Ghattas H., Cox S. E. (2007) Host-pathogen interactions: can micronutrients tip the balance? J. Nutr. 137, 1334–1337 [DOI] [PubMed] [Google Scholar]
- 22. Henderson J. P., Crowley J. R., Pinkner J. S., Walker J. N., Tsukayama P., Stamm W. E., Hooton T. M., Hultgren S. J. (2009) Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 5, e1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagan E. C., Lloyd A. L., Rasko D. A., Faerber G. J., Mobley H. L. T. (2010) Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6, e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reigstad C. S., Hultgren S. J., Gordon J. I. (2007) Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282, 21259–21267 [DOI] [PubMed] [Google Scholar]
- 25. Chaturvedi K. S., Hung C. S., Crowley J. R., Stapleton A. E., Henderson J. P. (2012) The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lv H., Hung C. S., Henderson J. P. (2014) Metabolomic analysis of siderophore cheater mutants reveals metabolic costs of expression in uropathogenic Escherichia coli. J. Proteome Res. 13, 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 28. Berger T., Togawa A., Duncan G. S., Elia A. J., You-Ten A., Wakeham A., Fong H. E., Cheung C. C., Mak T. W. (2006) Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 103, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 30. Shields-Cutler R. R., Crowley J. R., Hung C. S., Stapleton A. E., Aldrich C. C., Marschall J., Henderson J. P. (2015) Human urinary composition controls siderocalin's antibacterial activity. J. Biol. Chem. 290, 15949–15960, 10.1074/jbc.M115.645812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luo M., Lin H., Fischbach M. A., Liu D. R., Walsh C. T., Groves J. T. (2006) Enzymatic tailoring of enterobactin alters membrane partitioning and iron acquisition. ACS Chem. Biol. 1, 29–32 [DOI] [PubMed] [Google Scholar]
- 32. Valdebenito M., Crumbliss A. L., Winkelmann G., Hantke K. (2006) Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 296, 513–520 [DOI] [PubMed] [Google Scholar]
- 33. Abergel R. J., Warner J. A., Shuh D. K., Raymond K. N. (2006) Enterobactin protonation and iron release: structural characterization of the salicylate coordination shift in ferric enterobactin. J. Am. Chem. Soc. 128, 8920–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haag H., Hantke K., Drechsel H., Stojiljkovic I., Jung G., Zähner H. (1993) Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol. 139, 2159–2165 [DOI] [PubMed] [Google Scholar]
- 35. Heesemann J., Hantke K., Vocke T., Saken E., Rakin A., Stojiljkovic I., Berner R. (1993) Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol. 8, 397–408 [DOI] [PubMed] [Google Scholar]
- 36. Mabbett A. N., Ulett G. C., Watts R. E., Tree J. J., Totsika M., Ong C.-l. Y., Wood J. M., Monaghan W., Looke D. F., Nimmo G. R., Svanborg C., Schembri M. A. (2009) Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299, 53–63 [DOI] [PubMed] [Google Scholar]
- 37. Mazur M. T., Walsh C. T., Kelleher N. L. (2003) Site-specific observation of acyl intermediate processing in thiotemplate biosynthesis by Fourier transform mass spectrometry: the polyketide module of yersiniabactin synthetase. Biochemistry 42, 13393–13400 [DOI] [PubMed] [Google Scholar]
- 38. Miller D. A., Luo L., Hillson N., Keating T. A., Walsh C. T. (2002) Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 9, 333–344 [DOI] [PubMed] [Google Scholar]
- 39. Benedek O., Schubert S. (2007) Mobility of the Yersinia High-Pathogenicity Island (HPI): transfer mechanisms of pathogenicity islands (PAIS) revisited (a review). Acta Microbiol. Immunol. Hung. 54, 89–105 [DOI] [PubMed] [Google Scholar]
- 40. Hannauer M., Braud A., Hoegy F., Ronot P., Boos A., Schalk I. J. (2012) The PvdRT-OpmQ efflux pump controls the metal selectivity of the iron uptake pathway mediated by the siderophore pyoverdine in Pseudomonas aeruginosa. Environ. Microbiol. 14, 1696–1708 [DOI] [PubMed] [Google Scholar]
- 41. Brandel J., Humbert N., Elhabiri M., Schalk I. J., Mislin G. L., Albrecht-Gary A. M. (2012) Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton Trans. 41, 2820–2834 [DOI] [PubMed] [Google Scholar]
- 42. Kenney G. E., Rosenzweig A. C. (2012) Chemistry and biology of the copper chelator methanobactin. ACS Chem. Biol. 7, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H. J., Graham D. W., DiSpirito A. A., Alterman M. A., Galeva N., Larive C. K., Asunskis D., Sherwood P. M. (2004) Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 44. Ecker D. J., Emery T. (1983) Iron uptake from ferrichrome A and iron citrate in Ustilago sphaerogena. J. Bacteriol. 155, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. White C., Lee J., Kambe T., Fritsche K., Petris M. J. (2009) A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu Y., Chang F. M., Giedroc D. P. (2014) Copper transport and trafficking at the host-bacterial pathogen interface. Acc. Chem. Res. 47, 3605–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bearden S. W., Fetherston J. D., Perry R. D. (1997) Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65, 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hellman N. E., Gitlin J. D. (2002) Ceruloplasmin metabolism and function. Annu. Rev. Nutr. 22, 439–458 [DOI] [PubMed] [Google Scholar]
- 49. Gray L. W., Peng F., Molloy S. A., Pendyala V. S., Muchenditsi A., Muzik O., Lee J., Kaplan J. H., Lutsenko S. (2012) Urinary copper elevation in a mouse model of Wilson's disease is a regulated process to specifically decrease the hepatic copper load. PLoS One 7, e38327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Messerschmidt A., Huber R. (1990) The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur. J. Biochem. 187, 341–352, 10.1111/j.1432-1033.1990.tb15311.x [DOI] [PubMed] [Google Scholar]
- 51. Zaitseva I., Zaitsev V., Card G., Moshkov K., Bax B., Ralph A., Lindley P. (1996) The x-ray structure of human serum ceruloplasmin at 3.1 Å: nature of the copper centres. J. Biol. Inorg. Chem. 1, 15–23 [Google Scholar]
- 52. Hellman N. E., Kono S., Mancini G. M., Hoogeboom A. J., De Jong G. J., Gitlin J. D. (2002) Mechanisms of copper incorporation into human ceruloplasmin. J. Biol. Chem. 277, 46632–46638 [DOI] [PubMed] [Google Scholar]
- 53. Ridge P. G., Zhang Y., Gladyshev V. N. (2008) Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One 3, e1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y., Gladyshev V. N. (2011) Comparative genomics of trace element dependence in biology. J. Biol. Chem. 286, 23623–23629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Balasubramanian R., Smith S. M., Rawat S., Yatsunyk L. A., Stemmler T. L., Rosenzweig A. C. (2010) Oxidation of methane by a biological dicopper centre. Nature 465, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Knapp C. W., Fowle D. A., Kulczycki E., Roberts J. A., Graham D. W. (2007) Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc. Natl. Acad. Sci. U.S.A. 104, 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Balasubramanian R., Kenney G. E., Rosenzweig A. C. (2011) Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem. 286, 37313–37319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walsh C. T., Haynes S. W., Ames B. D. (2012) Aminobenzoates as building blocks for natural product assembly lines. Nat. Prod. Rep. 29, 37–59 [DOI] [PubMed] [Google Scholar]
- 59. Miller M. C., Parkin S., Fetherston J. D., Perry R. D., Demoll E. (2006) Crystal structure of ferric-yersiniabactin, a virulence factor of Yersinia pestis. J. Inorg. Biochem. 100, 1495–1500 [DOI] [PubMed] [Google Scholar]
- 60. Lukacik P., Barnard T. J., Keller P. W., Chaturvedi K. S., Seddiki N., Fairman J. W., Noinaj N., Kirby T. L., Henderson J. P., Steven A. C., Hinnebusch B. J., Buchanan S. K. (2012) Structural engineering of a phage lysin that targets Gram-negative pathogens. Proc. Natl. Acad. Sci. U.S.A. 109, 9857–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaturvedi K. S., Hung C. S., Giblin D. E., Urushidani S., Austin A. M., Dinauer M. C., Henderson J. P. (2014) Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol. 9, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koh E. I., Hung C. S., Parker K. S., Crowley J. R., Giblin D. E., Henderson J. P. (2015) Metal selectivity by the virulence-associated yersiniabactin metallophore system. Metallomics 7, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chaturvedi K. S., Henderson J. P. (2014) Pathogenic adaptations to host-derived antibacterial copper. Front. Cell. Infect. Microbiol. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hodgkinson V., Petris M. J. (2012) Copper homeostasis at the host-pathogen interface. J. Biol. Chem. 287, 13549–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crocker A., Lee C., Aboko-Cole G., Durham C. (1992) Interaction of nutrition and infection: effect of copper deficiency on resistance to Trypanosoma lewisi. J. Natl. Med. Assoc. 84, 697–706 [PMC free article] [PubMed] [Google Scholar]
- 66. Matousek de Abel de la Cruz A. J., Burguera J. L., Burguera M., Añez N. (1993) Changes in the total content of iron, copper, and zinc in serum, heart, liver, spleen, and skeletal muscle tissues of rats infected with Trypanosoma cruzi. Biol. Trace Elem. Res. 37, 51–70 [DOI] [PubMed] [Google Scholar]
- 67. Ilbäck N. G., Benyamin G., Lindh U., Friman G. (2003) Sequential changes in Fe, Cu, and Zn in target organs during early Coxsackievirus B3 infection in mice. Biol. Trace Elem. Res. 91, 111–124 [DOI] [PubMed] [Google Scholar]
- 68. Tufft L. S., Nockels C. F., Fettman M. J. (1988) Effects of Escherichia coli on iron, copper, and zinc metabolism in chicks. Avian Dis. 32, 779–786 [PubMed] [Google Scholar]
- 69. Winterbourn C. C., Hampton M. B., Livesey J. H., Kettle A. J. (2006) Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J. Biol. Chem. 281, 39860–39869 [DOI] [PubMed] [Google Scholar]
- 70. Wang J., Mushegian A., Lory S., Jin S. (1996) Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. U.S.A. 93, 10434–10439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lamont I. L., Beare P. A., Ochsner U., Vasil A. I., Vasil M. L. (2002) Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99, 7072–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Braud A., Geoffroy V., Hoegy F., Mislin G. L., Schalk I. J. (2010) Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ. Microbiol. Rep. 2, 419–425 [DOI] [PubMed] [Google Scholar]
- 73. Schwan W. R., Warrener P., Keunz E., Stover C. K., Folger K. R. (2005) Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. Int. J. Med. Microbiol. 295, 237–242 [DOI] [PubMed] [Google Scholar]
- 74. Grass G., Thakali K., Klebba P. E., Thieme D., Müller A., Wildner G. F., Rensing C. (2004) Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186, 5826–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Outten F. W., Huffman D. L., Hale J. A., O'Halloran T. V. (2001) The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677 [DOI] [PubMed] [Google Scholar]
- 76. Beswick P. H., Hall G. H., Hook A. J., Little K., McBrien D. C., Lott K. A. (1976) Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem. Biol. Interact. 14, 347–356 [DOI] [PubMed] [Google Scholar]
- 77. Macomber L., Rensing C., Imlay J. A. (2007) Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189, 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Macomber L., Imlay J. A. (2009) The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chillappagari S., Seubert A., Trip H., Kuipers O. P., Marahiel M. A., Miethke M. (2010) Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192, 2512–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Djoko K. Y., McEwan A. G. (2013) Antimicrobial action of copper is amplified via inhibition of heme biosynthesis. ACS Chem. Biol. 8, 2217–2223 [DOI] [PubMed] [Google Scholar]
- 81. Jozefczak M., Remans T., Vangronsveld J., Cuypers A. (2012) Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 13, 3145–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu F. F., Imlay J. A. (2012) Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 78, 3614–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rensing C., Fan B., Sharma R., Mitra B., Rosen B. P. (2000) CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U.S.A. 97, 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Beaman L., Beaman B. L. (1984) The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu. Rev. Microbiol. 38, 27–48 [DOI] [PubMed] [Google Scholar]
- 85. Choi D. W., Semrau J. D., Antholine W. E., Hartsel S. C., Anderson R. C., Carey J. N., Dreis A. M., Kenseth E. M., Renstrom J. M., Scardino L. L., Van Gorden G. S., Volkert A. A., Wingad A. D., Yanzer P. J., McEllistrem M. T., de la Mora A. M., DiSpirito A. A. (2008) Oxidase, superoxide dismutase, and hydrogen peroxide reductase activities of methanobactin from types I and II methanotrophs. J. Inorg. Biochem. 102, 1571–1580 [DOI] [PubMed] [Google Scholar]
- 86. Kim B., Richards S. M., Gunn J. S., Slauch J. M. (2010) Protecting against antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192, 2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gleason J. E., Galaleldeen A., Peterson R. L., Taylor A. B., Holloway S. P., Waninger-Saroni J., Cormack B. P., Cabelli D. E., Hart P. J., Culotta V. C. (2014) Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U.S.A. 111, 5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Soldati T., Neyrolles O. (2012) Mycobacteria and the intraphagosomal environment: take it with a pinch of salt(s)! Traffic 13, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 89. Nunes P., Demaurex N., Dinauer M. C. (2013) Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic 14, 1118–1131 [DOI] [PubMed] [Google Scholar]


