Background: Heat shock protein 90 (Hsp90) is a pro-survival molecular chaperone that is nitrated only in pathological conditions.
Results: Hsp90 nitrated on tyrosine 33 down-regulates mitochondrial activity.
Conclusion: Differential nitration states of Hsp90 regulate distinct aspects of cellular metabolism.
Significance: This is the first demonstration of a site-specific-nitrated protein regulating mitochondrial metabolism.
Keywords: energy metabolism, heat shock protein 90 (Hsp90), mitochondria, reactive nitrogen species (RNS), redox signaling, peroxynitrite, tyrosine nitration
Abstract
Peroxynitrite production and tyrosine nitration are present in several pathological conditions, including neurodegeneration, stroke, aging, and cancer. Nitration of the pro-survival chaperone heat shock protein 90 (Hsp90) in position 33 and 56 induces motor neuron death through a toxic gain-of-function. Here we show that nitrated Hsp90 regulates mitochondrial metabolism independently of the induction of cell death. In PC12 cells, a small fraction of nitrated Hsp90 was located on the mitochondrial outer membrane and down-regulated mitochondrial membrane potential, oxygen consumption, and ATP production. Neither endogenous Hsp90 present in the homogenate nor unmodified and fully active recombinant Hsp90 was able to compete with the nitrated protein for the binding to mitochondria. Moreover, endogenous or recombinant Hsp90 did not prevent the decrease in mitochondrial activity but supported nitrated Hsp90 mitochondrial gain-of-function. Nitrotyrosine in position 33, but not in any of the other four tyrosine residues prone to nitration in Hsp90, was sufficient to down-regulate mitochondrial activity. Thus, in addition to induction of cell death, nitrated Hsp90 can also regulate mitochondrial metabolism, suggesting that depending on the cell type, distinct Hsp90 nitration states regulate different aspects of cellular metabolism. This regulation of mitochondrial homeostasis by nitrated Hsp90 could be of particular relevance in cancer cells.
Introduction
Tyrosine nitration is a biomarker of oxidative damage and a footprint left by peroxynitrite and other reactive nitrogen species. Peroxynitrite is the product of the diffusion-limited reaction of nitric oxide and superoxide (1). Nitrated tyrosine residues are detected in inflammation, neurodegeneration, and aging. We previously showed that tyrosine nitration plays a key role in the activation of apoptotic signaling pathways (2–4). Tyrosine-containing peptides protect PC12 cells from peroxynitrite-induced cell death, acting as scavengers of nitrating products of peroxynitrite decomposition (2, 4). Downstream of peroxynitrite, nitration of a single tyrosine residue on Hsp90 is sufficient to induce apoptosis in PC12 cells and motor neurons (3). Although Hsp90 is a ubiquitous pro-survival protein (5), nitrated Hsp90 is detected only in pathological conditions (3, 6). Interestingly, nitrotyrosine is also present in cancer where it is not associated with cell death but with growth and metastasis (7–9), suggesting that not all nitration is detrimental.
Hsp90 is a highly conserved member of the Hsp90 family of molecular chaperones (10). This chaperone constitutes 1–2% of the total cytosolic protein (11). Vertebrates express two cytosolic isoforms sharing 86% homology, Hsp90α (inducible form) and Hsp90β (constitutive form) (12, 13). Hsp90 plays a critical role in both the regulation of normal cellular homeostasis and the stress response (5, 14, 15). As a chaperone, it has >200 client proteins involved in an array of functions, including the regulation of cell survival and death (16, 17). Hsp90 interaction with client proteins leads to conformational changes that result in activation or inhibition of the client protein (5, 14, 18). In mammalian cells, Hsp90 plays a role in targeting and translocation of preproteins to mitochondria (19). In cancer cells, Hsp90 participates in a chaperone network that regulates mitochondrial integrity and homeostasis. Organelle-specific inhibition of this network causes the disruption of mitochondrial integrity and stimulates apoptosis (20).
Here we show that nitrated Hsp90 associates with mitochondria and regulates mitochondrial activity without inducing the release of cytochrome c. Site-specific nitration of tyrosine 33 on Hsp90 confers a gain-of-function responsible for the regulation of mitochondrial activity.
Experimental Procedures
Peroxynitrite Synthesis
Peroxynitrite was synthesized by rapidly mixing acidified hydrogen peroxide with sodium nitrite and quenching the reaction with sodium hydroxide as previously described (21).
PC12 Cell Culture and Treatment with Peroxynitrite
Undifferentiated PC12 cells were cultured as previously described (3). For subcellular fractionation, 1 × 107 cells were washed twice with warm phosphate buffer saline (PBS) and resuspended in 10 ml of 50 mm PBS. The cells were gently vortexed, and peroxynitrite was slowly added to a final concentration of 0.5 mm. The cells were then incubated for 3 min at 37 °C in 5% CO2/air and immediately processed for subcellular fractionation, or the buffer was replaced by complete RPMI medium, and the cells were incubated for an additional 2 h at 37 °C in 5% CO2/air before performing the subcellular fractionation. For high content imaging, 2 × 104 cells were plated per well in a 96-well glass-bottom plate and incubated overnight under normal culture conditions. After three washes with warm PBS, the cultures were incubated with peroxynitrite (0.5 mm) for 3 min at 37 °C in 5% CO2/air. The buffer was replaced by complete RPMI medium, and the cells were incubated for an additional 2 h before fixation and staining for high content imaging. For the reverse order addition treatment, peroxynitrite was incubated for 2 h at 37 °C before being added to the cells (decomposed peroxynitrite).
Subcellular Fractionation
PC12 cell homogenates were obtained by disruption of PC12 cells in ice-cold MT buffer (0.3 m mannitol, 10 mm HEPES) using a glass and Teflon Potter S Homogenizer (Sartorius, Bohemia, NY). The disrupted cells were centrifuged at 600 × g for 5 min at 4 °C, and the supernatant was centrifuged again at 600 × g for 5 min at 4 °C. The final supernatant corresponds to PC12 cell homogenates. For the experiments where cell homogenates were used, the amount of protein in the homogenates was assayed using the Qubit Fluorometer (Invitrogen), and the concentration was adjusted to 1 mg/ml with ice-cold MT buffer. To obtain the mitochondrial and cytosolic fractions, the cell homogenates were centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant (cytosolic fraction) was centrifuged for an additional 20 min at 12,000 × g, 4 °C. The pellet (mitochondrial fraction) was resuspended in ice-cold MT buffer and centrifuged for 10 min at 12,000 × g, 4 °C. The mitochondrial and cytosolic fractions were then prepared for Western blot analysis.
Release of Cytochrome c
PC12 cell homogenates (200 μg) were incubated with 2.5 μg of either recombinant unmodified or nitrated Hsp90 for 2 h at 37 °C with the addition of 0.5 mm potassium phosphate and in the presence of 4.2 mm succinate. The mitochondrial and cytosolic fractions were obtained by subcellular fractionation as detailed above and prepared for Western blot analysis.
Expression and Purification of Human Recombinant Hsp90β and Hsp90β with Nitrotyrosine in Selected Positions
Human recombinant unmodified Hsp90β was expressed and purified as previously described (3). Briefly, the proteins were expressed in bacterial cultures by induction with l-arabinose for 2 h at 37 °C and 220 rpm. The incorporation of a nitrotyrosine residue in a selected position of recombinant human Hsp90β was performed as previously described (3, 22, 23). Recombinant wild type and mutant Hsp90β were purified from the bacterial cultures using the nickel-nitrilotriacetic acid purification system (Invitrogen) according to the manufacturer's instructions.
Peroxynitrite Treatment of Recombinant Hsp90
Two microliters of a freshly prepared peroxynitrite dilution were slowly added to 30 μl of recombinant Hsp90 (1 mg/ml in PBS) under gentle vortexing to a final concentration of 0.5 mm peroxynitrite.
Intracellular Delivery of Proteins into PC12 Cells
The intracellular delivery of Hsp90 and nitrated Hsp90 was performed as described before (3). Briefly, a mixture containing 10 μg of the protein and 4 μl of the lipophilic Chariot (Active Motif, Carlsbad, CA) was prepared. A pellet corresponding to 1 × 106 PC12 cells was gently resuspended in the protein/Chariot mixture and RPMI 1640. After 1 h of incubation at 37 °C in 5% CO2/air, RPMI medium supplemented with 10% horse serum, 5% fetal bovine serum, and antibiotics was added, and the cells were incubated for an additional hour. After incubation, the cells were plated at the indicated concentrations.
Oxygen Consumption Rate
The oxygen consumption rate (OCR)2 was measured in adherent PC12 cells using a XF24 extracellular flux analyzer (Seahorse Bioscience, Billerica, MA). After intracellular delivery of nitrated Hsp90, the cells were seeded in a XF24- well cell culture microplate (Seahorse Bioscience) at a density of 7 × 104 cells per well and incubated for 5 h at 37 °C in 5% CO2. Then, the medium was replaced by 700 μl of seahorse running media (DMEM, 5.6 mm glucose, 1 mm pyruvate, 4 mm l-glutamine), and the cells were incubated for an additional hour. The OCR was determined under basal conditions and after the sequential addition of previously titrated oligomycin (3 μm), carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (FCCP, 10 μm), and antimycin A (10 μm) as previously described (24). OCR data from each well was normalized to protein levels in the same well, and the non-mitochondrial respiration after the addition of antimycin A was subtracted from all measurements. Results were expressed as % of cells incubated in the presence of Chariot alone.
Mitochondrial Complex Activities by Extracellular Flux Analysis
Mitochondrial complex activities were measured as previously described (25, 26). Briefly, after intracellular delivery of the recombinant proteins, PC12 cells were seeded at 3 × 104 cells per well in a XF-96-well plate (Seahorse Biosciences). The optimal cell density and concentration of permeabilizing agent (saponin, 25 μg/ml) was determined before assessing complex activities. Before measuring, the medium was removed and replaced with MAS buffer (70 mm sucrose, 220 mm mannitol, 10 mm KH2PO4, 5 mm MgCl2, 2 mm HEPES, 1 mm EGTA, pH 7.2). In addition to saponin and ADP (1 mm), specific substrates were added to measure complex activities. Complex I was 5 mm pyruvate, 2.5 mm malate. The addition of 1 μm rotenone was used to determine complex I rotenone-sensitive activity. Complex II was 10 mm succinate. Inhibition of complex III by the addition of 10 μm antimycin A was used to subtract the OCR not related to complex II activity. Complex III was 0.5 mm duroquinol. The addition of 10 μm antimycin A was used to determine complex III specific activity. Complex IV was 0.5 mm N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) and 2 mm ascorbate. The OCR measured in the presence of 20 mm azide was subtracted to complex IV activity. Cell number per well was comparable across the plate. The complex activities were calculated as: maximal OCR minus OCR in the presence of the specific inhibitors.
Immunofluorescence and High Content Imaging
To determine the presence of nitrated Hsp90 in mitochondria of PC12 cells, 20,000 cells per well were plated in a 96-well plate and incubated for 24 h at 37 °C in 5% CO2/air. The cells were then treated with 0.5 mm peroxynitrite or the products of peroxynitrite decomposition. The cultures were incubated for 2 min at room temperature and then fixed with 4% paraformaldehyde plus 0.1% glutaraldehyde (Sigma) on ice for 20 min before the staining was performed. The cells were incubated with an anti-nitrated Hsp90 monoclonal antibody (1:2000), as previously described (3). The localization of nitrated Hsp90 was determined by high content imaging and compartmental analysis using the Cellomics ArrayScan VTI (ThermoFisher Scientific, Pittsburgh, PA). Nitrated Hsp90 levels in cytosol or mitochondria were measured as the average mean intensity in a ring around DAPI-stained nuclear areas (ring) or spots in a ring around the nuclear areas (ring spots), respectively.
Quantitative Western Blotting Analysis
Western blots were performed as previously described (2, 3, 27). All Western blots were visualized, and the bands were quantified using the Odyssey System (Li-Cor Biosciences, Lincoln, NE). Quantification of endogenous Hsp90 was performed by loading 10 μg of total protein from PC12 cell homogenates together with a standard curve of recombinant human Hsp90 in a gel followed by SDS-PAGE, protein transfer to a PVDF membrane, and infrared detection using an anti-Hsp90 antibody (1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA). Detection of recombinant human Hsp90 was performed using an anti-myc-tag antibody (1:2,000, Cell Signaling Technology, Danvers, MA) or anti-V5-tag antibody (1:5,000, Invitrogen) and normalized using an anti-mitochondrial Complex I NDUFA9 subunit antibody (1:5,000, Molecular Probes/ThermoFisher Scientific, Eugene, OR) or anti-tubulin antibody (1:10,000, Sigma).
PC12 Cells Mitochondrial Content
PC12 cell mitochondrial content after intracellular delivery of the recombinant proteins was assayed by infrared Western blot for mitochondrial Complex I NDUFA9 subunit and MnSOD (anti-MnSOD antibody, 1:1000, EMD Millipore, Billerica, MA) and by measuring MitoTracker Green fluorescence according with the manufacturer's instructions (100 nm, Molecular Probes/ThermoFisher Scientific) using a Plate Runner (Trophos, Marseilles, France) and a Synergy H1 multi-mode reader (BioTek, Winooski, VT). When indicated, 10 μm FCCP was added to the cells together with the MitoTracker Green.
Mitochondrial Membrane Potential of PC12 Cells and Cell Homogenates
After intracellular delivery of the recombinant proteins, PC12 cells were plated (50,000 cells per well) and incubated for 2.5 h at 37 °C in 5% CO2/air before adding the probe JC-1 (1 μm, Molecular Probes/ThermoFisher Scientific). The cells were then incubated for an extra 30 min followed by a wash with Dulbecco's PBS. Fluorescence was measured at 560ex/590em (red) and 485ex/530em (green), and the data are expressed as the ratio of red/green signals. When indicated, 10 μm FCCP was added to the cells together with the JC-1. PC12 cell homogenates (1 mg/ml) were incubated with 5 and 50% recombinant unmodified Hsp90 (0.26 and 2.6 μg of recombinant protein per 200 μg of cell homogenate, respectively), peroxynitrite-treated Hsp90, or site-specific nitrated Hsp90 (3NT33, 3NT56, 3NT276, 3NT484, and 3NT596) for 30 min at 37 °C with the addition of 0.5 mm potassium phosphate, 4.2 mm succinate, and in the presence or absence of 2 μm FCCP. The probe JC-1 (75 nm) was then added, and the homogenate was incubated at 37 °C for additional 30 min. Fluorescence (560ex/590em) was read on a 96-well plate with 50 μg of total protein per well. For the measurement of isolated mitochondria, the cell homogenates (1 mg/ml) were centrifuged at 12,000 × g for 10 min at 4 °C before incubating with the recombinant proteins. The supernatant was replaced with the same volume of MT buffer supplemented with 0.5 mm potassium phosphate and in the presence or absence of 4.2 mm succinate. The pellet (isolated mitochondria) was gently resuspended and incubated with the recombinant proteins as described above. When indicated, mitochondria pellets were resuspended in their corresponding cytosolic fractions or supplemented with the same amount of recombinant Hsp90 as endogenous Hsp90 present in the original cell homogenate (5.2 μg of recombinant protein added to isolated mitochondria from 200 μg of cell homogenate).
ATP Production
PC12 cell homogenates (1 mg/ml) were incubated with 5 and 50% recombinant unmodified or nitrated Hsp90 for 45 min at 37 °C with the addition of 0.5 mm potassium phosphate, 4.2 mm succinate, and in the presence or absence of 2 μm FCCP. After incubation, the ATP production was stopped by the addition of 2 μm FCCP, and the ATP levels were measured in 10 μg of total protein from cell homogenates using the ATP Bioluminescence Assay kit HS II (Roche Diagnostics) according to the manufacturer's instructions. To assay the ATP produced during incubation with the recombinant proteins, the ATP level from cell homogenates incubated in the presence of 2 μm FCCP was used as basal level.
Mitochondrial Translocation and Competition Assays
PC12 cell homogenates (200 μg) or isolated mitochondria (50 μg) were incubated with 2.6 μg of either recombinant unmodified Hsp90, peroxynitrite-treated Hsp90, or site-specific nitrated Hsp90(3NT33) for 1 h at 37 °C with the addition of 0.5 mm potassium phosphate and in the presence or absence of 4.2 mm succinate or 2 μm FCCP. The mitochondrial fraction was then recovered by centrifuging at 12,000 × g for 10 min at 4 °C followed by two washes with ice-cold MT buffer. The resulting pellet was resuspended in 10 μl MT buffer with protease inhibitors for Western blot analysis. To determine the submitochondrial location of nitrated Hsp90, the mitochondrial fraction was resuspended in 50 μl of MT buffer and incubated with proteinase K (5 μg/ml) for an additional 25 min on ice. Proteinase K activity was then inhibited by the addition of 30 μm PMSF. For the competition assay, cell homogenates (200 μg) were incubated with 2 μg of peroxynitrite-treated Hsp90 or site-specific nitrated Hsp90(3NT33) and increasing concentrations of unmodified Hsp90 (0.2–20 μg) for 1 h at 37 °C.
Mitochondrial Complex Activities in Disrupted Mitochondria
The measurement of complex I, II+III, and IV activities was performed in disrupted mitochondria as previously described (28). PC12 cell homogenates were incubated in the presence of 5% recombinant unmodified Hsp90 or peroxynitrite-treated Hsp90, and the mitochondrial fraction was isolated and subjected to 3 cycles of freeze/thawing. Complex I activity was measured at 340 nm by the rotenone (10 μm)-sensitive reduction of 50 μm ubiquinone-1 in the presence of 1 mm potassium cyanide, 3 mg/ml fatty acid-free bovine serum albumin, and 200 μm NADH as electron donor. Activity of complexes II+III was determined by the antimycin A (20 μm)-sensitive reduction of 50 μm cytochrome c at 550 nm in the presence of 1 mm potassium cyanide and 10 mm succinate. Complex IV activity was determined by monitoring cytochrome c oxidation at 550 nm. The reaction rate was measured as the pseudo-first order reaction constant (k′) and expressed as k′/min/mg of protein.
Chaperone Activity of Hsp90
The chaperone activity of unmodified and modified recombinant Hsp90 (10 μg/ml) was measured at 43 °C by following aggregation of citrate synthase (5 μg/ml) in 40 mm HEPES, pH 7.5, for 1 h at 336 nm as previously described (29).
Statistical Analysis
For statistical analyses one-way ANOVA followed by Bonferroni multiple comparison tests or Kruskal-Wallis non parametric test were used. All tests were performed using the program Prism (GraphPad Software Inc., San Diego, CA).
Results
Peroxynitrite-treated Hsp90 Associates with Mitochondria
Peroxynitrite can regulate both mitochondrial membrane potential and oxygen consumption (30–32). After treatment of undifferentiated PC12 cells with peroxynitrite, a fraction of nitrated Hsp90 was associated with mitochondria as determined using high content imaging and analysis as well as subcellular fractionation (Fig. 1, A and B). To investigate whether nitrated Hsp90 association with mitochondria was related to induction of cell death, we assessed the release of cytochrome c in vitro. We first determined the proportion of endogenous Hsp90 with respect to total cellular protein in PC12 cell homogenates by quantitative infrared Western blot (2.6 ± 0.7%, corresponding to 0.26 ± 0.07 μg in 10 μg of cell homogenate) (Fig. 1C). The release of cytochrome c from mitochondria was then assayed in PC12 cell homogenates incubated with 50% recombinant unmodified or peroxynitrite-treated Hsp90 compared with endogenous Hsp90 (2.6 μg of recombinant protein per 200 μg of cell homogenate) for 2 h. After incubation with peroxynitrite-treated Hsp90 there was no detectable release of cytochrome c from mitochondria (Fig. 1D) even when the amount of peroxynitrite-treated Hsp90 added was 10-fold higher than the amount needed to induce cell death (3). In contrast, incubation of the homogenates with 0.5 mm peroxynitrite induced the release of cytochrome c (Fig. 1D) as previously described (33). These results suggest that the translocation of nitrated Hsp90 to mitochondria is not related to induction of cell death but could have a role in the regulation of mitochondrial activity.
FIGURE 1.
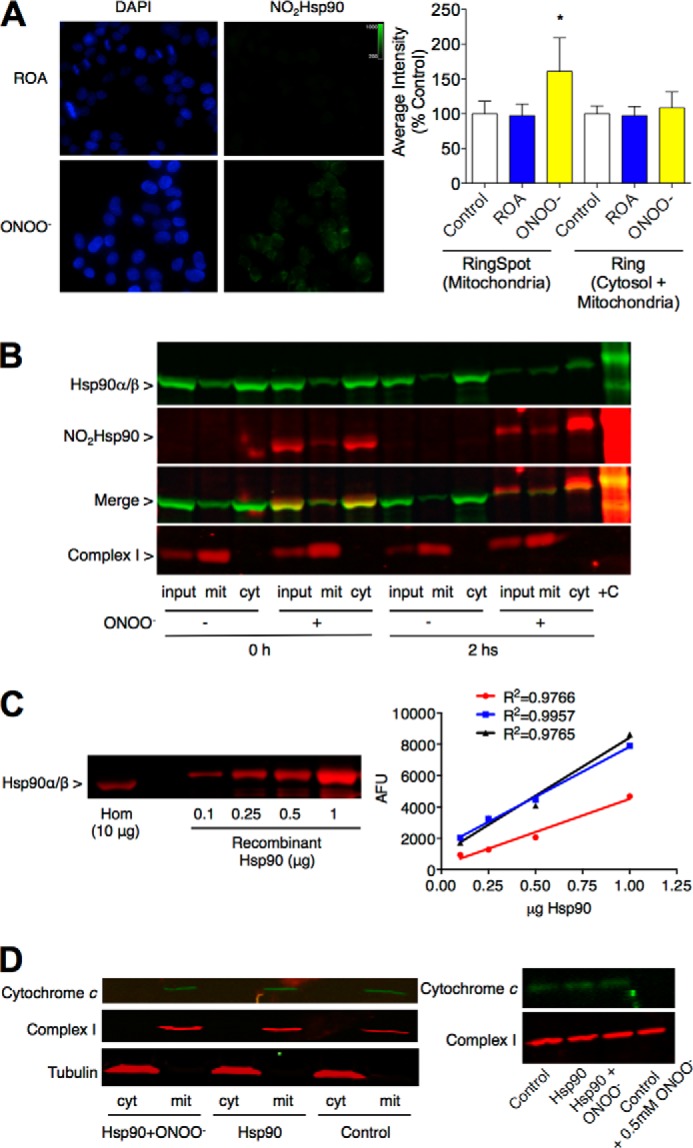
Nitrated Hsp90 localizes in mitochondria in PC12 cells. Endogenously nitrated Hsp90 (NO2Hsp90) localizes in mitochondria immediately after treatment of PC12 cells with 0.5 mm peroxynitrite (ONOO−) as shown by high content imaging and analysis (A) and subcellular fractionation (B). A, NO2Hsp90 levels (green) from cytosol or mitochondria were measured as the average mean intensity in a ring around blue DAPI-stained nuclear areas (ring) or spots in a ring around the nuclear areas (ring spots) as under “Experimental Procedures” (n = 3). On the left panel representative images of cells stained for nitrated Hsp90 (green) in the presence of a nuclear dye (DAPI, in blue) indicate the intracellular localization of the modified protein. *, p < 0.05 versus control (cells with no treatment) by ANOVA followed by Bonferroni multiple comparison test. Reverse order addition (ROA) corresponds to cells incubated in the presence of decomposed peroxynitrite. B, subcellular fractionation of peroxynitrite-treated PC12 cells. Input corresponds to the original cell homogenate from which the mitochondrial (mit) and cytosolic (cyt) fractions were obtained. The membrane was blotted for Hsp90 (green), nitrated Hsp90 (NO2Hsp90, red), and the mitochondrial complex I NDUFA9 subunit (red). Infrared detection allowed the merge of both signals (yellow, LiCor Biosciences). Peroxynitrite-treated recombinant Hsp90 was used as positive control (+C) (n = 3). C, the percentage of endogenous Hsp90 respect to total amount of protein in PC12 cell homogenates (Hom) was determined by Western blot (10 μg of total protein) blotted for Hsp90 (red). On the right, an increasing concentration of recombinant Hsp90 was used for the standard curves (linear regressions for three separate standard curves expressed as arbitrary fluorescence units (AFU) with their corresponding R2 values) (n = 3). D, on the left, the release of cytochrome c from mitochondria was assayed in vitro in PC12 cell homogenates incubated with unmodified (Hsp90) or peroxynitrite-treated Hsp90 (Hsp90+ONOO−). The mitochondrial and cytosolic fractions were loaded on a SDS-PAGE, and the membrane was blotted for cytochrome c (green), mitochondrial complex I NDUFA9 subunit (Complex I, red), and tubulin (red) (n = 3). On the right, PC12 cell homogenates were either incubated with Hsp90 and Hsp90+ONOO− or treated with 0.5 mm peroxynitrite before assaying the release of cytochrome c from the mitochondrial fraction.
Peroxynitrite-treated Hsp90 Decreases Mitochondrial Activity
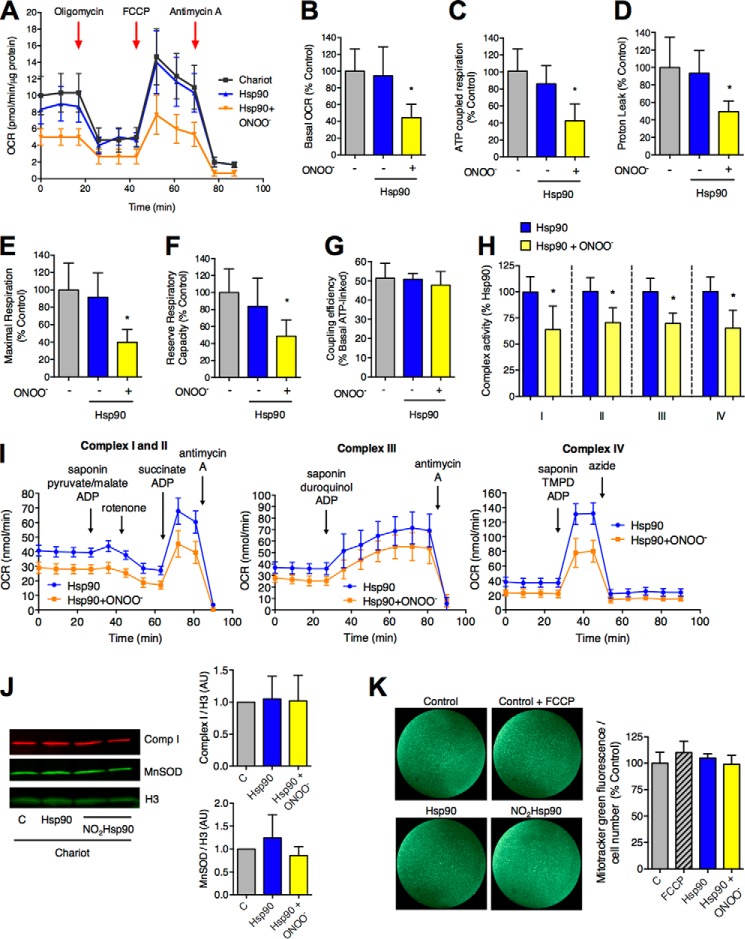
Nitration of only 5% Hsp90, the proportion of endogenous Hsp90 that is nitrated after treatment of the cells with peroxynitrite, is sufficient to induce PC12 cell death (3). To investigate if peroxynitrite-treated Hsp90 affected mitochondrial activity, 5% of recombinant human peroxynitrite-treated Hsp90β with respect to endogenous Hsp90 was intracellularly delivered into PC12 cells as previously described (3), and the mitochondrial OCR was measured 6 h post-delivery in the XF24 extracellular flux analyzer (Seahorse Bioscience) (24). Peroxynitrite-treated Hsp90 decreased the mitochondrial basal OCR as early as 6 h after the intracellular delivery of the peroxynitrite-treated chaperone when compared with the control (Fig. 2, A and B). Incubation with unmodified Hsp90 had no effect on basal OCR. The mitochondrial basal respiration combines two different parameters, the ATP-coupled OCR and proton leak. The addition of the ATP synthase inhibitor oligomycin allows the dissection of both parameters by inhibiting ATP-coupled respiration but not the proton leak. Upon intracellular delivery of peroxynitrite-treated Hsp90, there was a significant decrease of both ATP-coupled OCR and proton leak (Fig. 2, C and D). Similar results were observed for the maximal mitochondrial respiration and the reserve respiratory capacity (difference between maximal and basal mitochondrial respiration) after the addition of the uncoupler FCCP (Fig. 2, E and F). In addition, the mitochondrial coupling efficiency, calculated as the percentage of basal respiration linked to ATP production (34), was not affected by the delivery of the recombinant proteins (Fig. 2G), implying that peroxynitrite-treated Hsp90 does not induce mitochondria uncoupling. Together these results suggest that peroxynitrite-treated Hsp90 regulates mitochondrial activity by inhibiting the electron transport chain or by limiting substrate supply.
FIGURE 2.
Peroxynitrite-treated Hsp90 down-regulates mitochondrial activity. A, the mitochondrial OCR was studied in PC12 cells in culture using extracellular flux analysis 6 h after intracellular delivery of either 5% Hsp90 or peroxynitrite-treated Hsp90 (Hsp90+ONOO−) with respect to total endogenous Hsp90. Three measurements were obtained for basal OCR followed by inhibition of ATP synthase with oligomycin (3 μm). Mitochondrial maximal respiratory capacity was studied by uncoupling the mitochondrial inner membrane with FCCP (10 μm). The difference between maximal and basal respiration indicates the mitochondrial spare respiratory capacity. Finally, electron flux was inhibited with antimycin A (10 μm) at complex III. The remaining oxygen consumption is from sources other than mitochondria (non-mitochondrial OCR). OCR data from each well was normalized to protein levels in the same well (mean ± S.E., n = 3). For comparison purposes, the mitochondrial basal OCR (B), ATP-coupled respiration (C), proton leak (D), maximal respiration (E), and reserve respiratory capacity (F) and were expressed as % control (Chariot alone) after subtracting non-mitochondrial respiration (mean ± S.D.). *, p < 0.05 versus control by ANOVA followed by Bonferroni multiple comparison test or by Kruskal-Wallis non-parametric test. Columns represent the mean ± S.D. G, the coupling efficiency was calculated as the percentage of basal respiration linked to ATP production. H and I, the activities of complex I to IV were measured in saponin-permeabilized PC12 cells 6 h after intracellular delivery of either unmodified Hsp90 or peroxynitrite-treated Hsp90 (Hsp90+ONOO−) by extracellular flux analysis. Specific substrates and inhibitors were added in the presence of 1 mm ADP as described under “Experimental Procedures.” TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine. In H, complex I to IV activities were expressed as the percentage of activity after delivery of unmodified Hsp90 for comparison purposes. *, p < 0.05 versus Hsp90 by t test. J and K, the mitochondrial content of PC12 cells 6 h after intracellular delivery of either 5% Hsp90 or peroxynitrite-treated Hsp90 with respect to total endogenous Hsp90 was determined by Western blot (J) and after incubation with MitoTracker Green (K). J, the membranes were blotted for mitochondrial complex I NDUFA9 subunit (comp I, red), manganese SOD (MnSOD, green), and histone 3 (H3, green). The quantitation of the complex I and MnSOD infrared signals was normalized to H3 (mean ± S.D., n = 3). AU, arbitrary units. K, PC12 cells were incubated with MitoTracker Green (100 nm) for 30 min before measuring fluorescence at 485ex/528em. When indicated, FCCP (10 μm) was added together with the MitoTracker Green. Fluorescence intensity of entire wells (96-well plates) was measured using a Plate Runner. On the left, pictures of representative wells stained with MitoTracker Green (n = 3) are shown.
Thus, the effect of peroxynitrite-treated Hsp90 on the mitochondrial electron transport chain activity was assayed in permeabilized PC12 cells using extracellular flux analysis (26). This methodology allows the study of mitochondrial complex activities by measuring the OCR after supplying permeabilized cells with specific substrates for each complex. In agreement with the down-regulation of mitochondrial respiration, intracellular delivery of peroxynitrite-treated Hsp90 decreased the activity of mitochondrial complex I to IV to the same extent (Fig. 2, H and I). Because specific substrates were provided for the different complexes, these results suggest that the inhibitory effect lies in the end of the respiratory transport chain by inhibition of cytochrome c oxidase (complex IV) activity. There were no detectable changes in mitochondrial content upon the addition of peroxynitrite-treated Hsp90 as determined by mitochondrial complex I NDUFA9 subunit and manganese SOD content (Fig. 2J) and MitoTracker Green fluorescence (Fig. 2K). The addition of FCCP (2 μm) was used as a control for MitoTracker Green mitochondrial membrane potential independent uptake (Fig. 2K). Together, these findings suggest that peroxynitrite-treated Hsp90 regulates mitochondrial respiration by partially inhibiting complex IV activity.
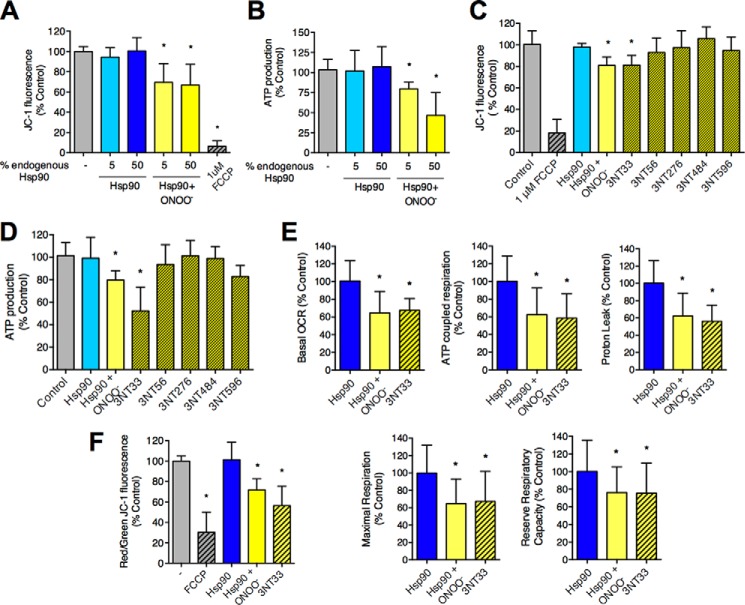
To further study the role of peroxynitrite-treated Hsp90 in the regulation of mitochondrial activity, PC12 cell homogenates were incubated in the presence or absence of recombinant-unmodified and peroxynitrite-treated Hsp90. Incubation of homogenates with 5 and 50% peroxynitrite-treated Hsp90 with respect to total endogenous Hsp90 present in the homogenate (0.26 and 2.6 μg of recombinant protein per 200 μg cell homogenate, respectively) significantly reduced the mitochondrial membrane potential compared with that of mitochondria in homogenates incubated without recombinant protein (control). No effect was observed upon incubation with unmodified chaperone (Fig. 3A). Similar results were observed for mitochondrial ATP production (Fig. 3B), indicating that a small proportion of peroxynitrite-treated Hsp90 with respect to total fully active Hsp90 (ratio 1:20) is enough to regulate mitochondrial activity. These observations suggest that peroxynitrite-treated Hsp90 regulation of mitochondrial activity is the result of a gain-of-function.
FIGURE 3.
Hsp90 with nitrotyrosine in position 33 down-regulates mitochondrial activity. Mitochondrial membrane potential (A) and ATP production (B) were assayed in vitro in PC12 cell homogenates incubated with 5 and 50% of Hsp90 or peroxynitrite-treated Hsp90 (Hsp90+ONOO−) compared with total endogenous Hsp90 (0.26 and 2.6 μg of recombinant protein per 200 μg homogenate, respectively). Mitochondrial membrane potential was measured after 30 min of incubation at 37 °C followed by an additional 30 min upon the addition of 75 nm JC-1 and expressed as JC-1 fluorescence. FCCP (2 μm) was added together with JC-1 in the indicated samples. ATP production was assayed using the ATP Bioluminescence Assay kit HS II (Roche Diagnostics). JC-1 fluorescence (C) and ATP production (D) were assayed as described above in the presence of 5% Hsp90, Hsp90+ONOO−, or five modified Hsp90 proteins in which one of the tyrosine residues prone to nitration was replaced by nitrotyrosine (3NT33, 3NT56, 3NT276, 3NT484, and 3NT596) (0.26 μg of modified recombinant protein per 200 μg of homogenate). Results are expressed as % control (homogenate alone) (mean ± S.D., n = 5–10). Mitochondrial OCR (E) and JC-1 fluorescence (F) were studied in PC12 cells 6 h (OCR) and 3 h (JC-1 fluorescence) after the intracellular delivery of either unmodified Hsp90, Hsp90+ONOO−, or Hsp90(3NT33). E, OCR was measured as described in Fig. 2. Results are expressed as % control (unmodified Hsp90) (mean ± S.D., n = 3). F, PC12 cells were incubated with JC-1 (1 μm) for 30 min, and fluorescence was measured at 560ex/590em (red) and 485ex/530em (green). When indicated, FCCP (10 μm) was added together with the JC-1. Results are expressed as % control of the red/green signals ratio (mean ± S.D., n = 3). *, p < 0.05 versus control by ANOVA followed by Bonferroni multiple comparison test. Columns represent the mean ± S.D.
Tyrosine Nitration Is Sufficient for the Peroxynitrite-treated Hsp90 Role in Mitochondria
Five tyrosine residues are target for nitration in Hsp90. In human Hsp90β these residues correspond to tyrosine 33, 56, 276, 484, and 596 (3). The presence of a nitrotyrosine at either position 33 or 56 induces a gain-of-function responsible for the toxic activity of nitrated Hsp90 in motor neurons (3). However, in addition to tyrosine residues, peroxynitrite reacts and oxidizes other amino acids such as cysteine and tryptophan. To investigate if tyrosine nitration was sufficient for the mitochondrial function of peroxynitrite-treated Hsp90, mitochondrial activity was assessed in the presence of recombinant Hsp90 in which either tyrosine 33, 56, 276, 484, or 596 was replaced by nitrotyrosine using a non-natural amino acid incorporation bacterial system (3, 22, 23). To this end PC12 cell homogenates were incubated with 5% unmodified or modified Hsp90 with respect to total endogenous Hsp90. Out of the five tyrosine residues prone to nitration, only nitrotyrosine in position 33 (3NT33) as the sole modification on the protein was sufficient to significantly decrease mitochondrial membrane potential (Fig. 3C) and ATP production (Fig. 3D) at the same levels observed for peroxynitrite-treated Hsp90. Incorporation of a nitrotyrosine at any of the other four positions had no effect on mitochondrial activity (Fig. 3, C and D). Similarly and as observed for peroxynitrite-treated Hsp90, intracellular delivery of Hsp90(3NT33) into PC12 cells significantly decreased basal and ATP-coupled respiration, proton leak, maximal respiration, and the reserve respiratory capacity (Fig. 3E). In agreement, the mitochondrial membrane potential was also decreased in the presence of peroxynitrite-treated Hsp90 and Hsp90(3NT33) as soon as 3 h after intracellular delivery of the proteins (Fig. 3F). Thus, nitration of a single tyrosine residue in position 33 is sufficient to support the mitochondrial activity of nitrated Hsp90.
Nitrated Hsp90 Has Increased Affinity for a Mitochondrial Client
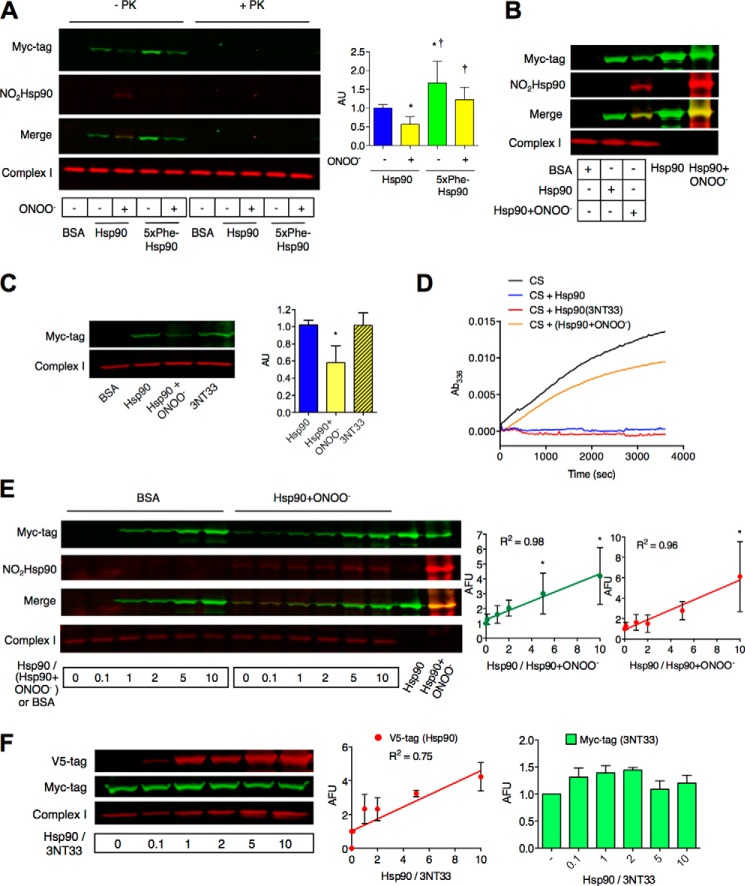
To determine if nitrated Hsp90 had a direct interaction with mitochondria, we studied the kinetics of the peroxynitrite-treated chaperone mitochondrial translocation. Homogenates of PC12 cells were incubated with recombinant unmodified or peroxynitrite-treated Hsp90 carrying a myc-6xhistidine tag in the C-terminal end for 1 h at 37 °C, and the mitochondrial and cytosolic fractions were obtained by differential centrifugation. Peroxynitrite-treated Hsp90 retained ∼50% capability to translocate to mitochondria when compared with the unmodified protein (Fig. 4A). Similar results were observed upon incubation of the nitrated chaperone with isolated mitochondria (Fig. 4B), suggesting that both unmodified and peroxynitrite-treated Hsp90 binds to mitochondria independently of its interaction with cytosolic proteins. Both peroxynitrite-treated and unmodified Hsp90 were associated with the mitochondrial outer membrane, as both chaperones were susceptible to degradation by proteinase k (Fig. 4A). On the other hand, the levels of Hsp90(3NT33) detected in mitochondria upon incubation were similar to those of unmodified Hsp90 (Fig. 4C), suggesting that nitration on tyrosine 33 alone does not affect the translocation process of the modified protein to mitochondria. In agreement, although treatment with peroxynitrite decreases 50% of Hsp90 chaperone activity (3), measured as the protection from unfolding and aggregation afforded to citrate synthase in conditions of heat shock, Hsp90(3NT33) was fully protective (Fig. 4D), suggesting that the presence of nitrotyrosine in position 33 does not affect the chaperone function of Hsp90.
FIGURE 4.
Nitrated Hsp90 has increased affinity for mitochondria. A, the translocation to mitochondria and submitochondrial location of recombinant Hsp90, peroxynitrite-treated Hsp90, Hsp90 in which the five tyrosine residues prone to nitration were replaced by phenylalanine (5xPhe-Hsp90), and peroxynitrite-treated 5xPhe-Hsp90 were studied in vitro in mitochondrial fractions from homogenates of PC12 cells by infrared Western blot. The homogenates were incubated with the recombinant proteins for 1 h at 37 °C, and the mitochondrial fraction was isolated by differential centrifugation. The recombinant proteins were detected using an anti-myc-tag antibody (green) and nitrated Hsp90 (red). To determine the submitochondrial location of the recombinant proteins, mitochondrial fractions were incubated in the presence or absence of proteinase k (PK). The mitochondrial complex I NDUFA9 subunit (red) was used to normalize and to confirm mitochondrial integrity after treatment with proteinase K. AU, arbitrary units. B, the translocation of recombinant Hsp90 and peroxynitrite-treated Hsp90 to mitochondria was studied in vitro in isolated mitochondria from PC12 cells incubated with the recombinant proteins as described in A. C, translocation of Hsp90(3NT33) to mitochondria was studied as described in A. D, the chaperone activity of unmodified Hsp90, peroxynitrite-treated Hsp90 (Hsp90+ONOO−), and Hsp90(3NT33) was assayed at 336 nm by following protection of citrate synthase (CS) from aggregation at 43 °C for 1 h (n = 4). E and F, representative Western blots of competition assays performed in PC12 cell homogenates incubated with increasing amounts of unmodified Hsp90 and a fixed amount of either bovine serum albumin (BSA) or peroxynitrite-treated Hsp90 (Hsp90+ONOO−) (E) or Hsp90(3NT33) (F) in a ratio 0 to 10 unmodified Hsp90 compared with either Hsp90+ONOO− or Hsp90(3NT33). In E both Hsp90 and Hsp90+ONOO− carrying a C-terminal myc tag were detected using an ant-myc-tag antibody (green). In red, Hsp90+ONOO− was detected using a monoclonal anti-nitrated Hsp90 antibody that recognizes nitrotyrosine at position 56 in Hsp90 (NO2Hsp90). F, to detect unmodified Hsp90 and Hsp90(3NT33) independently, recombinant proteins carrying different C-terminal tags were used. Unmodified Hsp90 carrying a V5 tag was detected using an anti-V5-tag antibody (red) and Hsp90(3NT33) carrying an myc tag was detected using an anti-myc-tag antibody (green). Right, quantitation of the infrared signals corresponding to total Hsp90 (Hsp90 and Hsp90+ONOO−, green) and NO2Hsp90 (red) (E) or unmodified Hsp90 (red) and Hsp90(3NT33) (green) (F) expressed as arbitrary fluorescence units (AFU). Linear regressions are shown for the signals corresponding to total Hsp90 and NO2Hsp90 (E) and unmodified Hsp90 (F) with their corresponding R2 values. The mitochondrial complex I NDUFA9 subunit (red) was used to normalize the infrared signals. *, p < 0.05 versus control; †, versus Hsp90+ONOO− by ANOVA followed by Bonferroni multiple comparison test. Columns represent the mean ± S.D. of at least three independent experiments.
Both peroxynitrite-treated Hsp90 and Hsp90(3NT33) affected mitochondrial activity even when competing with the fully active endogenous unmodified protein in a ratio 1:20 (5% nitrated recombinant protein versus 100% endogenous Hsp90). These results may be explained by an increased affinity of peroxynitrite-treated Hsp90 for a mitochondrial client or a new interaction of peroxynitrite-treated Hsp90 in the mitochondria. These alternative hypotheses were investigated using a competition assay. Homogenates from PC12 cells were incubated with a fixed amount of peroxynitrite-treated Hsp90 or Hsp90(3NT33) and increased amounts of unmodified Hsp90. Even in 10-fold excess, the unmodified chaperone was not able to compete with nitrated Hsp90 (Fig. 4E) or Hsp90(3NT33) (Fig. 4F) for the binding to mitochondria. Moreover, the translocation of nitrated Hsp90 to mitochondria was linearly enhanced by increased concentrations of unmodified Hsp90 (Fig. 4E). These results suggest that nitration on tyrosine 33 in Hsp90 may confer the modified protein binding to a new mitochondrial client and that unmodified Hsp90 facilitates peroxynitrite-treated Hsp90 association with mitochondria.
Nitrated Hsp90 Activity in Mitochondria Depends on the Presence of Unmodified Hsp90
Due to the role of Hsp90 in translocation of proteins to mitochondria, there is a fraction of the chaperone normally associated with the mitochondrial outer membrane (19). Additionally, nitrated Hsp90 was detected in mitochondria of PC12 cells immediately after peroxynitrite treatment (Fig. 1B), implying that the mitochondrial fraction of Hsp90 may be susceptible to nitration and/or the translocation kinetics of nitrated Hsp90 from cytosol to mitochondria is very fast. We investigated next whether the Hsp90 fraction normally associated with mitochondria was susceptible to nitration by peroxynitrite. Isolated mitochondria from PC12 cells were treated with increasing concentrations of peroxynitrite, and the nitration of Hsp90 was assayed by Western blot. Nitrated Hsp90 was detected in isolated mitochondria after treatment with 0.5 mm peroxynitrite (Fig. 5A), suggesting that the mitochondrial pool of Hsp90 is also targeted by the oxidant. Furthermore, in whole cell homogenates the translocation of nitrated Hsp90 to mitochondria occurred within seconds, reaching equilibrium in <10 min (Fig. 5B). Therefore, both the cytosolic and the mitochondrial fractions of Hsp90 are targets for nitration and may play a role in the regulation of mitochondrial activity. However, neither peroxynitrite-treated Hsp90 nor Hsp90(3NT33) had an effect on mitochondrial membrane potential of isolated mitochondria (Fig. 5C), indicating that a cytosolic client or co-chaperone may translocate to mitochondria in a complex with the nitrated proteins and that this complex is responsible for the regulatory effect on mitochondrial activity. Indeed, peroxynitrite-treated Hsp90 decreased mitochondrial membrane potential when isolated mitochondria were supplemented with the cytosolic fraction (Fig. 5C).
FIGURE 5.
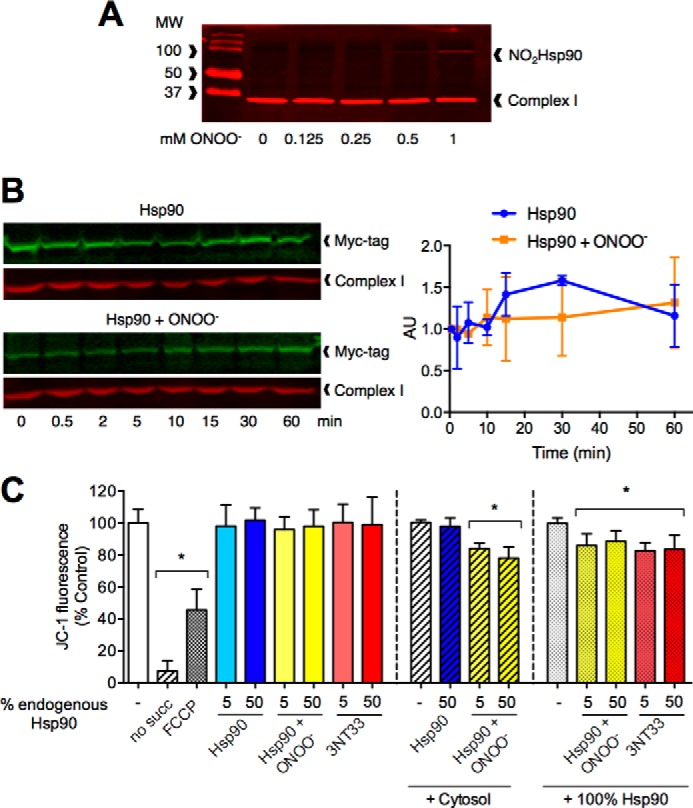
Nitrated Hsp90 mitochondrial activity regulation is supported by unmodified Hsp90. A, representative infrared Western blot of the isolated mitochondrial fraction from PC12 cells treated with increasing concentrations of peroxynitrite. The Western blot membranes were blotted for nitrated Hsp90 (NO2Hsp90) and mitochondrial complex I NDUFA9 subunit (n = 3). B, representative infrared Western blots of unmodified Hsp90 and peroxynitrite-treated Hsp90 (Hsp90+ONOO−) mitochondrial translocation time courses. The translocation time-course was assayed by infrared Western blot of PC12 cell homogenates incubated with the recombinant proteins for the indicated times followed by isolation of the mitochondrial fraction. The membranes were blotted for an myc tag (green), and the infrared signal was normalized to mitochondrial complex I NDUFA9 subunit (red). Right, quantitation of the normalized infrared signals for unmodified Hsp90 and Hsp90+ONOO− time courses expressed as arbitrary units (AU). Each curve was normalized to its corresponding time 0 (n = 3). C, JC-1 fluorescence of isolated mitochondria from PC12 cells incubated with 5 and 50% of either Hsp90, Hsp90+ONOO−, or Hsp90(3NT33) compared with total endogenous Hsp90 was assayed as described in Fig. 2. Mitochondria incubated with the recombinant proteins in the absence of succinate (no succ) were used as the positive control for decreased mitochondrial membrane potential. Isolated mitochondria were supplemented with either the cytosolic fraction (+ Cytosol) or the same amount of recombinant unmodified Hsp90 as endogenous Hsp90 present in the original homogenate (+ 100% Hsp90). Results are expressed as % control (isolated mitochondria alone, isolated mitochondria + cytosol, or isolated mitochondria + 100% Hsp90) (n = 3–7). *, p < 0.05 versus control by ANOVA followed by Bonferroni multiple comparison test. Columns represent the mean ± S.D. of at least three independent experiments.
Because unmodified Hsp90 did not compete with peroxynitrite-treated Hsp90 or Hsp90(3NT33) for the binding to mitochondria but facilitated translocation of the nitrated chaperone to the organelle (Fig. 4C), unmodified Hsp90 may be the cytosolic partner required for peroxynitrite-treated Hsp90 and Hsp90(3NT33) to mimic the effect on mitochondrial activity observed in cell homogenates. Supplementing isolated mitochondria with the same concentration of unmodified Hsp90 present in cell homogenates, corresponding to 2.6% of total protein (5.2 μg of recombinant Hsp90 added to mitochondria isolated from 200 μg of homogenate), was enough to support the decrease of mitochondrial membrane potential induced by both peroxynitrite-treated Hsp90 and Hsp90(3NT33) (Fig. 5C), implying that unmodified Hsp90 is required for the nitrated chaperone mitochondrial activity.
Discussion
Tyrosine nitration is found in neurodegenerative diseases, inflammation processes, and aging. Recently, we showed that tyrosine nitration is not collateral damage but plays an actual role in the induction of cell death in PC12 cells and motor neurons. Nitration of a single tyrosine residue on Hsp90 is sufficient to turn the pro-survival chaperone into a toxic protein (3). Here we show that nitrated Hsp90 regulated mitochondrial metabolism by partially inhibiting cytochrome c oxidase activity independently of induction of cell death.
In PC12 cells, peroxynitrite induces cell death by simultaneously activating pro-apoptotic pathways and inactivating pro-survival pathways (27). The activation of the pro-apoptotic pathways ultimately leads to release of cytochrome c from mitochondria and activation of the intrinsic apoptotic pathway (27). Downstream of peroxynitrite, nitrated Hsp90 induces PC12 cells death through a toxic gain-of-function (3). However, depending on the concentration, peroxynitrite also regulates mitochondrial function (30–32). Interestingly, we found a fraction of nitrated Hsp90 associated with mitochondria, down-regulating mitochondrial activity (Figs. 1 and 2). The regulation of mitochondrial function by nitrated Hsp90 was not related to the toxic activity of the oxidized chaperone because nitrated Hsp90 did not stimulate detectable release of cytochrome c from mitochondria (Fig. 1D).
Peroxynitrite oxidizes tyrosine, cysteine, and tryptophan residues (35–37). In addition to tyrosine nitration, the presence of nitrotryptophan was reported on Hsp90α after treatment of PC12 cells with peroxynitrite (38). However, nitration of one single tyrosine residue in position 33 on Hsp90 is sufficient for the mitochondrial regulatory role of the nitrated chaperone. A recombinant protein produced by non-natural amino acid incorporation carrying a nitrotyrosine in position 33 as the sole oxidative modification on the protein had the same effect on mitochondrial oxygen consumption rate, JC-1 fluorescence, and ATP production than the peroxynitrite-treated chaperone (Fig. 3). Conversely, the presence of nitrotyrosine in position 56, 276, 484, and 596 had no effect on mitochondrial activity (Fig. 3). Interestingly, tyrosine 56 is involved in the induction of motor neuron death, but it played no role in the regulation of mitochondrial metabolism (3), whereas the presence of nitrotyrosine in position 33 is involved in both processes. Collectively these data suggest that differential nitration states of Hsp90 regulate different aspects of cell metabolism.
Hsp90 conformational changes necessary for its activity are tightly regulated by post-translational modifications, implying that in different cellular environments Hsp90 activity is differentially regulated through specific post-translational modifications (39). Tyrosine residue 33 is particularly important for the intra and intermonomer interactions of Hsp90 dimer, controlling the ATPase activity of the chaperone (40). In addition, phosphorylation of tyrosine 33 by Wee1 kinase affects Hsp90 interaction with client proteins and co-chaperones (41). Interestingly, peroxynitrite-treated Hsp90 showed a significant decrease in its chaperone activity when compared with the unmodified protein, which may explain its decreased translocation to mitochondria (Fig. 4) (3). After replacing the 5 tyrosine residues prone to nitration by phenylalanine, there was still a 50% decrease in the amount of translocated chaperone upon treatment with peroxynitrite. This observation suggests that in addition to tyrosine nitration, the translocation process may be regulated by other oxidative modifications as well as other post-translational modification affecting tyrosine residues, such as phosphorylation. Independently of the regulation of nitrated Hsp90 translocation to mitochondria, specific tyrosine nitration of the chaperone is sufficient to decrease mitochondrial activity.
In cell culture as well as in in vitro assays, the addition of a small proportion of either peroxynitrite-treated Hsp90 or Hsp90(3NT33) compared with total endogenous unmodified Hsp90 (1:20 recombinant protein versus endogenous Hsp90) was sufficient to induce a decrease in mitochondrial activity, indicating that peroxynitrite-treated Hsp90 and Hsp90(3NT33) had a new role in the mitochondria that could not be compensated by the fully active endogenous unmodified Hsp90 (Figs. 2 and 3). Moreover, nitration of tyrosine 33 was key for the increased mitochondrial affinity of the nitrated protein (Fig. 4). There was also an increase in the translocation of peroxynitrite-treated Hsp90 to mitochondria as the amount of unmodified Hsp90 increased, suggesting that when endogenously oxidized by peroxynitrite, Hsp90 may rely on cytosolic Hsp90 to translocate to mitochondria. However, translocation of nitrated Hsp90 to mitochondria was not enough to regulate mitochondrial metabolism but required the presence of unmodified Hsp90. Indeed, the addition of recombinant unmodified Hsp90 to isolated mitochondria supported the decrease in mitochondrial activity in the presence of either peroxynitrite-treated Hsp90 or Hsp90(3NT33), implying that unmodified Hsp90 is necessary for nitrated Hsp90 mitochondrial activity (Fig. 5). This effect could be due to unmodified Hsp90 acting as a co-chaperone for nitrated Hsp90, stabilizing the modified protein, thus supporting nitrated Hsp90 gain-of-function. Another possibility is the formation of asymmetric dimers containing one nitrated and one unmodified Hsp90 monomer through subunit exchange. The exchange process can occur within minutes of incubation in vitro (42, 43). Recent reports show that asymmetric post-translational modifications and interactions with co-chaperones regulate Hsp90 chaperone cycle as well as its interaction with client proteins (44, 45).
Using extracellular flux analysis in non-permeabilized and permeabilized PC12 cells, we identified the last step of the electron transport chain at the level of cytochrome c/cytochrome c oxidase as a target of nitrated Hsp90 mitochondrial activity inhibition (Fig. 2). Complex IV is one of the main points of regulation of mitochondrial oxidative metabolism (46). Its activity can be inhibited or activated at different levels, including transcriptional regulation, the assembly of subunits into functional complexes, phosphorylation, substrate availability, and binding of molecules such as nitric oxide, carbon monoxide, hydrogen sulfide, and hydrogen cyanide (47, 48). The fact that nitrated Hsp90 down-regulates mitochondrial activity shortly after intracellular delivery or after a short incubation period in vitro suggests the level of regulation to be downstream of complex IV assembly. Alternatively, nitrated Hsp90 could regulate the cytochrome c redox state. In addition, nitrated Hsp90 localizes in the mitochondrial outer membrane (Fig. 4), whereas complex IV subunits are embedded in the mitochondrial inner membrane, and cytochrome c locates in the mitochondrial intermembrane space predominantly as an inner membrane-bound pool (49, 50). Together these observations suggest that nitrated Hsp90 gain-of-function may require the formation of a complex with one or more mitochondrial proteins in order to regulate the electron transport to complex IV. Indeed, peroxynitrite-treated Hsp90 had no effect on the activities of complex I, II+III, and IV in disrupted mitochondria isolated from PC12 cell homogenates incubated with 5% peroxynitrite-treated Hsp90 or unmodified Hsp90 (not shown). Together these observations suggest that mitochondrial integrity is required for nitrated Hsp90 regulatory activity. Further studies are needed to understand the nature of this regulation by nitrated Hsp90.
PC12 cells are derived from a malignant transplantable rat pheochromocytoma, a tumor of the adrenal glands (51). The results presented here may be of particular relevance in cancer, a pathology in which the mitochondrial homeostasis is tightly regulated to favor cell growth and survival (52, 53). Tyrosine nitration is present in vivo in malignant gliomas (7) and in invasive breast carcinomas (9) where anti-angiogenic therapies have had very limited success (54, 55). The regulation of mitochondrial homeostasis by nitrated Hsp90 may be associated with pro-survival processes in cancer. Our observations suggest that nitration of Hsp90 may play a key role in decreasing oxygen consumption by cancer cells, thereby increasing their resistance to hypoxia.
Author Contributions
M. C. F. conceived the project, and designed and performed most of the experiments, and analyzed the data. K. C. R. performed the extracellular flux analysis and analyzed data. A. S. G. participated in experiments in isolated mitochondria and analyzed data. C. N. D. and P. A. N. participated in the determination of endogenous Hsp90 content and the intracellular delivery of recombinant proteins. M. S. J. and M. C. F. performed the high content analysis and analyzed data. R. A. M. led the production of modified Hsp90β. A. L. led the extracellular flux analysis. A. G. E. supervised the project, designed experiments, and analyzed data. M. C. F. wrote the manuscript with comments from co-authors. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Sarah B. Gitto and Jonathan Virgilio for technical assistance and Victor Darley-Usmar and Paul S. Brookes for constructive criticism on the manuscript. The Oregon State University Cell Imaging and Analysis Facilities of the Environmental Health Sciences Center is supported by Grant P30 ES00210 (to R. A. M.).
This work was supported, in whole or in part, by National Institutes of Health Grants NS36761 (to A. G. E.). This work was also supported by an OHSU-MRF grant (to R. A. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- OCR
- oxygen consumption rate
- FCCP
- p-(trifluoromethoxy)phenylhydrazone
- SOD
- superoxide dismutase
- ANOVA
- analysis of variance.
References
- 1. Beckman J. S., Beckman T. W., Chen J., Marshall P. A., Freeman B. A. (1990) Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. U.S.A. 87, 1620–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ye Y., Quijano C., Robinson K. M., Ricart K. C., Strayer A. L., Sahawneh M. A., Shacka J. J., Kirk M., Barnes S., Accavitti-Loper M. A., Radi R., Beckman J. S., Estévez A. G. (2007) Prevention of peroxynitrite-induced apoptosis of motor neurons and PC12 cells by tyrosine-containing peptides. J. Biol. Chem. 282, 6324–6337 [DOI] [PubMed] [Google Scholar]
- 3. Franco M. C., Ye Y., Refakis C. A., Feldman J. L., Stokes A. L., Basso M., Melero Fernández de Mera R. M., Sparrow N. A., Calingasan N. Y., Kiaei M., Rhoads T. W., Ma T. C., Grumet M., Barnes S., Beal M. F., Beckman J. S., Mehl R., Estévez A. G. (2013) Nitration of Hsp90 induces cell death. Proc. Natl. Acad. Sci. U.S.A. 110, E1102–E1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sahawneh M. A., Ricart K. C., Roberts B. R., Bomben V. C., Basso M., Ye Y., Sahawneh J., Franco M. C., Beckman J. S., Estévez A. G. (2010) Cu,Zn superoxide dismutase (SOD) increases toxicity of mutant and Zn2+-deficient superoxide dismutase by enhancing protein stability. J. Biol. Chem. 285, 33885–33897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao R., Davey M., Hsu Y.-C., Kaplanek P., Tong A., Parsons A. B., Krogan N., Cagney G., Mai D., Greenblatt J., Boone C., Emili A., Houry W. A. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell 120, 715–727 [DOI] [PubMed] [Google Scholar]
- 6. Viera L., Radmilovich M., Vargas M. R., Dennys C. N., Wilson L., Barnes S., Franco M. C., Beckman J. S., Estévez A. G. (2013) Temporal patterns of tyrosine nitration in embryo heart development. Free Radic. Biol. Med. 55, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cobbs C. S., Samanta M., Harkins L. E., Gillespie G. Y., Merrick B. A., MacMillan-Crow L. A. (2001) Evidence for peroxynitrite-mediated modifications to p53 in human gliomas: possible functional consequences. Arch Biochem. Biophys. 394, 167–172 [DOI] [PubMed] [Google Scholar]
- 8. MacMillan-Crow L. A., Greendorfer J. S., Vickers S. M., Thompson J. A. (2000) Tyrosine nitration of c-SRC tyrosine kinase in human pancreatic ductal adenocarcinoma. Arch. Biochem. Biophys. 377, 350–356 [DOI] [PubMed] [Google Scholar]
- 9. Nakamura Y., Yasuoka H., Tsujimoto M., Yoshidome K., Nakahara M., Nakao K., Nakamura M., Kakudo K. (2006) Nitric oxide in breast cancer: induction of vascular endothelial growth factor-C and correlation with metastasis and poor prognosis. Clin. Cancer Res. 12, 1201–1207 [DOI] [PubMed] [Google Scholar]
- 10. Borkovich K. A., Farrelly F. W., Finkelstein D. B., Taulien J., Lindquist S. (1989) hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9, 3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Didelot C., Schmitt E., Brunet M., Maingret L., Parcellier A., Garrido C. (2006) Heat shock proteins: endogenous modulators of apoptotic cell death. In Molecular Chaperones in Health and Disease (Gaestel M., ed.) pp. 171–198, Springer, Berlin: [DOI] [PubMed] [Google Scholar]
- 12. Sreedhar A. S., Kalmár E., Csermely P., Shen Y. F. (2004) Hsp90 isoforms: functions, expression, and clinical importance. FEBS Lett. 562, 11–15 [DOI] [PubMed] [Google Scholar]
- 13. Gupta R. S. (1995) Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol. Biol. Evol. 12, 1063–1073 [DOI] [PubMed] [Google Scholar]
- 14. Wandinger S. K., Richter K., Buchner J. (2008) The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477 [DOI] [PubMed] [Google Scholar]
- 15. Dezwaan D. C., Freeman B. C. (2008) HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle 7, 1006–1012 [DOI] [PubMed] [Google Scholar]
- 16. Pearl L. H., Prodromou C. (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Ann. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 17. Pratt W. B., Morishima Y., Osawa Y. (2008) The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 283, 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiech H., Buchner J., Zimmermann R., Jakob U. (1992) Hsp90 chaperones protein folding in vitro. Nature 358, 169–170 [DOI] [PubMed] [Google Scholar]
- 19. Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 20. Kang B. H., Plescia J., Dohi T., Rosa J., Doxsey S. J., Altieri D. C. (2007) Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell 131, 257–270 [DOI] [PubMed] [Google Scholar]
- 21. Robinson K. M., Beckman J. S. (2005) Synthesis of peroxynitrite from nitrite and hydrogen peroxide. Methods Enzymol. 396, 207–214 [DOI] [PubMed] [Google Scholar]
- 22. Neumann H., Hazen J. L., Weinstein J., Mehl R. A., Chin J. W. (2008) Genetically encoding protein oxidative damage. J. Am. Chem. Soc. 130, 4028–4033 [DOI] [PubMed] [Google Scholar]
- 23. Cooley R. B., Feldman J. L., Driggers C. M., Bundy T. A., Stokes A. L., Karplus P. A., Mehl R. A. (2014) Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 53, 1916–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dranka B. P., Benavides G. A., Diers A. R., Giordano S., Zelickson B. R., Reily C., Zou L., Chatham J. C., Hill B. G., Zhang J., Landar A., Darley-Usmar V. M. (2011) Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med. 51, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clerc P., Polster B. M. (2012) Investigation of mitochondrial dysfunction by sequential microplate-based respiration measurements from intact and permeabilized neurons. PLoS ONE 7, e34465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salabei J. K., Gibb A. A., Hill B. G. (2014) Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 9, 421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shacka J. J., Sahawneh M. A., Gonzalez J. D., Ye Y. Z., D'Alessandro T. L., Estévez A. G. (2006) Two distinct signaling pathways regulate peroxynitrite-induced apoptosis in PC12 cells. Cell Death Differ. 13, 1506–1514 [DOI] [PubMed] [Google Scholar]
- 28. Franco M. C., Antico Arciuch V. G., Peralta J. G., Galli S., Levisman D., López L. M., Romorini L., Poderoso J. J., Carreras M. C. (2006) Hypothyroid phenotype is contributed by mitochondrial complex I inactivation due to translocated neuronal nitric-oxide synthase. J. Biol. Chem. 281, 4779–4786 [DOI] [PubMed] [Google Scholar]
- 29. Jakob U., Lilie H., Meyer I., Buchner J. (1995) Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J. Biol. Chem. 270, 7288–7294 [DOI] [PubMed] [Google Scholar]
- 30. Radi R., Rodriguez M., Castro L., Telleri R. (1994) Inhibition of mitochondrial electron transport by peroxynitrite. Arch. Biochem. Biophys. 308, 89–95 [DOI] [PubMed] [Google Scholar]
- 31. Brookes P. S., Land J. M., Clark J. B., Heales S. J. (1998) Peroxynitrite and brain mitochondria: evidence for increased proton leak. J. Neurochem. 70, 2195–2202 [DOI] [PubMed] [Google Scholar]
- 32. Gadelha F. R., Thomson L., Fagian M. M., Costa A. D., Radi R., Vercesi A. E. (1997) Ca2+-independent permeabilization of the inner mitochondrial membrane by peroxynitrite is mediated by membrane protein thiol cross-linking and lipid peroxidation. Arch. Biochem. Biophys. 345, 243–250 [DOI] [PubMed] [Google Scholar]
- 33. Borutaite V., Morkuniene R., Brown G. C. (1999) Release of cytochrome c from heart mitochondria is induced by high Ca2+ and peroxynitrite and is responsible for Ca2+-induced inhibition of substrate oxidation. Biochim. Biophys. Acta 1453, 41–48 [DOI] [PubMed] [Google Scholar]
- 34. Brand M. D., Nicholls D. G. (2011) Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ischiropoulos H., Zhu L., Chen J., Tsai M., Martin J. C., Smith C. D., Beckman J. S. (1992) Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch. Biochem. Biophys. 298, 431–437 [DOI] [PubMed] [Google Scholar]
- 36. Alvarez B., Ferrer-Sueta G., Freeman B. A., Radi R. (1999) Kinetics of peroxynitrite reaction with amino acids and human serum albumin. J. Biol. Chem. 274, 842–848 [DOI] [PubMed] [Google Scholar]
- 37. Alvarez B., Rubbo H., Kirk M., Barnes S., Freeman B. A., Radi R. (1996) Peroxynitrite-dependent tryptophan nitration. Chem. Res. Toxicol. 9, 390–396 [DOI] [PubMed] [Google Scholar]
- 38. Kawasaki H., Ikeda K., Shigenaga A., Baba T., Takamori K., Ogawa H., Yamakura F. (2011) Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells. Free Radic. Biol. Med. 50, 419–427 [DOI] [PubMed] [Google Scholar]
- 39. Xu W., Mollapour M., Prodromou C., Wang S., Scroggins B. T., Palchick Z., Beebe K., Siderius M., Lee M. J., Couvillon A., Trepel J. B., Miyata Y., Matts R., Neckers L. (2012) Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol. Cell 47, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cunningham C. N., Krukenberg K. A., Agard D. A. (2008) Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J. Biol. Chem. 283, 21170–21178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mollapour M., Tsutsumi S., Donnelly A. C., Beebe K., Tokita M. J., Lee M. J., Lee S., Morra G., Bourboulia D., Scroggins B. T., Colombo G., Blagg B. S., Panaretou B., Stetler-Stevenson W. G., Trepel J. B., Piper P. W., Prodromou C., Pearl L. H., Neckers L. (2010) Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol. Cell 37, 333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richter K., Muschler P., Hainzl O., Buchner J. (2001) Coordinated ATP hydrolysis by the Hsp90 dimer. J. Biol. Chem. 276, 33689–33696 [DOI] [PubMed] [Google Scholar]
- 43. Soroka J., Wandinger S. K., Mäusbacher N., Schreiber T., Richter K., Daub H., Buchner J. (2012) Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol. Cell 45, 517–528 [DOI] [PubMed] [Google Scholar]
- 44. Flynn J. M., Mishra P., Bolon D. N. (2015) Mechanistic Asymmetry in Hsp90 Dimers. J. Mol. Biol. 10.1016/j.jmb.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayer M. P., Le Breton L. (2015) Hsp90: Breaking the Symmetry. Mol. Cell 58, 8–20 [DOI] [PubMed] [Google Scholar]
- 46. Arnold S. (2012) The power of life: cytochrome c oxidase takes center stage in metabolic control, cell signalling, and survival. Mitochondrion 12, 46–56 [DOI] [PubMed] [Google Scholar]
- 47. Srinivasan S., Avadhani N. G. (2012) Cytochrome c oxidase dysfunction in oxidative stress. Free Radic. Biol. Med. 53, 1252–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cooper C. E., Brown G. C. (2008) The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40, 533–539 [DOI] [PubMed] [Google Scholar]
- 49. Ott M., Robertson J. D., Gogvadze V., Zhivotovsky B., Orrenius S. (2002) Cytochrome c release from mitochondria proceeds by a two-step process. Proc. Natl. Acad. Sci. U.S.A. 99, 1259–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ow Y. P., Green D. R., Hao Z., Mak T. W. (2008) Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9, 532–542 [DOI] [PubMed] [Google Scholar]
- 51. Greene L. A., Tischler A. S. (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U.S.A. 73, 2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cairns R. A., Harris I. S., Mak T. W. (2011) Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 [DOI] [PubMed] [Google Scholar]
- 54. Reardon D. A., Herndon J. E., 2nd, Peters K., Desjardins A., Coan A., Lou E., Sumrall A., Turner S., Sathornsumetee S., Rich J. N., Boulton S., Lipp E. S., Friedman H. S., Vredenburgh J. J. (2012) Outcome after bevacizumab clinical trial therapy among recurrent grade III malignant glioma patients. J. Neurooncol. 107, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bergers G., Hanahan D. (2008) Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 8, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]