Background: Bacillus subtilis is a model organism for analyzing bacterial biofilms, but the carbohydrate components are undescribed.
Results: Genes in the epsHIJK locus needed for biofilm formation encode proteins synthesizing the conserved bacterial polysaccharide poly-N-acetylglucosamine (PNAG).
Conclusion: PNAG is a major carbohydrate component of B. subtilis biofilms.
Significance: PNAG production is essential for formation of B. subtilis biofilms.
Keywords: bacteria, bacterial adhesion, bacterial genetics, carbohydrate biosynthesis, carbohydrate structure, glycobiology, glycosyltransferase, protein expression, protein structure
Abstract
Bacillus subtilis is intensively studied as a model organism for the development of bacterial biofilms or pellicles. A key component is currently undefined exopolysaccharides produced from proteins encoded by genes within the eps locus. Within this locus are four genes, epsHIJK, known to be essential for pellicle formation. We show they encode proteins synthesizing the broadly expressed microbial carbohydrate poly-N-acetylglucosamine (PNAG). PNAG was present in both pellicle and planktonic wild-type B. subtilis cells and in strains with deletions in the epsA–G and -L–O genes but not in strains deleted for epsH–K. Cloning of the B. subtilis epsH–K genes into Escherichia coli with in-frame deletions in the PNAG biosynthetic genes pgaA–D, respectively, restored PNAG production in E. coli. Cloning the entire B. subtilis epsHIJK locus into pga-deleted E. coli, Klebsiella pneumoniae, or alginate-negative Pseudomonas aeruginosa restored or conferred PNAG production. Bioinformatic and structural predictions of the EpsHIJK proteins suggest EpsH and EpsJ are glycosyltransferases (GT) with a GT-A fold; EpsI is a GT with a GT-B fold, and EpsK is an α-helical membrane transporter. B. subtilis, E. coli, and pga-deleted E. coli carrying the epsHIJK genes on a plasmid were all susceptible to opsonic killing by antibodies to PNAG. The immunochemical and genetic data identify the genes and proteins used by B. subtilis to produce PNAG as a significant carbohydrate factor essential for pellicle formation.
Introduction
Many microbial organisms produce biofilms, structurally complex multicellular communities inside an extracellular matrix of variable factors that can include exopolysaccharides (EPS),7 proteins, and nucleic acids (1, 2). Biofilms are probably the most common structures for microbial communities, as this state protects against many environmental stresses such as antimicrobial factors (3). Biofilms are also involved in the pathogenesis of many infectious diseases (1, 4, 5). Polysaccharides are often prominent components of biofilms but, like many factors in this structure, make a variable contribution depending on the microbial species, strain, growth conditions, and overall environment. Interestingly, numerous species have developed diverse metabolic pathways for production of EPS, alluding to the possibility that these systems have evolved independently. A common EPS associated with microbial biofilm formation is a polymeric β-1,6-linked N-acetylglucosamine (PNAG) structure that is highly conserved and expressed by a range of bacterial, fungal, and protozoan microorganisms (6, 7). PNAG was first isolated and characterized from Staphylococcus epidermidis (8), where it was referred to as the polysaccharide intercellular adhesin, and then later shown to be produced by Staphylococcus aureus (9, 10). In these two species, PNAG is synthesized by proteins encoded by four genes in the ica operon (9–11), and ica-deleted PNAG-deficient strains were unable to produce biofilms (9, 11). However, PNAG-independent biofilm formation in some staphylococcal strains has also been described (12). In general, depending on the strain and culture conditions, PNAG is often necessary, but not sufficient or essential, for biofilm formation.
PNAG synthesis also occurs in various Gram-negative organisms, including Escherichia coli (13), Acinetobacter baumannii (14) and Burkholderia spp. (15). In these organisms PNAG is synthesized by four proteins encoded by genes in the pga operon. More recently, Cywes-Bentley et al. (7) showed that a much wider spectrum of microbes can synthesize PNAG, including many human bacterial pathogens such as Streptococcus pneumoniae, Streptococcus pyogenes, Streptococcus agalactiae, Neisseria gonorrhoeae, and Neisseria meningitidis, the nontypable Haemophilus influenzae and Mycobacterium tuberculosis, and eukaryotic organisms such as fungal pathogens, and protozoan parasites such as Trichomonas vaginalis and murine and human Plasmodia spp. that cause malaria. Among many of these pathogens there are no readily identifiable genes homologous to those in the ica or pga loci. Nonetheless, the broad conservation of PNAG synthesis implies an important role in microbial biology, perhaps related to growth phases involving aggregation of microbes living in different environments or protection from anti-microbial factors.
Bacillus subtilis is a spore-forming Gram-positive bacillus wherein certain strains grown under specific conditions develop complex biofilms (16–18). A large amount of knowledge has been accumulated that explains how the production and composition of the B. subtilis biofilm are regulated (17), although many aspects of this process are not fully elucidated. Full biofilm formation by some strains of B. subtilis such as NCIB 3610 (3610) is dependent on 15 proteins encoded by genes in the eps locus (epsA–epsO) that are associated with the carbohydrate content and complexity of a surface pellicle (16) involved in the overall biofilm structure. The composition and structure of polysaccharides synthesized by the proteins within this complex are not well described, but mutations in most of the genes within the B. subtilis eps cluster lead to loss of biofilm formation (19). Some genes, such as epsE, have dual functions, affecting both polysaccharide synthesis and flagella-based motility (20, 21).
Given the ubiquity of PNAG synthesis among a range of microbial organisms, we evaluated B. subtilis biofilms for PNAG production and further examined the B. subtilis eps locus for genes potentially encoding PNAG biosynthetic proteins. Both biofilm and planktonic cells produced PNAG, and within the available annotated genome, we identified four genes, epsH–K, as potentially being responsible for PNAG synthesis. These genes are predicted to encode two glycosyltransferases (GT) (epsH and epsJ), separated by another GT with potential EPS modifying enzymatic activity (epsI), and a transporter/facilitator of synthesis (epsK). Cloning of the epsH–K genes into PNAG-deficient E. coli or Klebsiella pneumoniae (Δpga), or alginate-negative Pseudomonas aeruginosa, leads to synthesis of a polymer immunochemically equivalent to PNAG. Each of the four B. subtilis epsH–K genes could individually trans-complement E. coli strains deleted for the pgaA–D genes, respectively. Also the epsH and epsJ genes could partially complement E. coli strains deleted for the pgaC or pgaA genes, respectively. Extracts of both WT B. subtilis and E. coli (Δpga) complemented with B. subtilis epsHIJK contained PNAG-immunoreactive, hexosamine-containing material that was destroyed by treatment with both the PNAG-degrading enzyme dispersin B (22) and by sodium periodate, which can only hydrolyze polymeric hexosamines in a 1–6-linkage. Synthesis of PNAG in E. coli from the epsH–K genes resulted in susceptibility of cells to killing in an opsonophagocytic assay using antibodies specific to PNAG, indicative of functional conservation of PNAG properties when B. subtilis gene products direct synthesis of PNAG in E. coli. Overall, we identify the presence of PNAG in B. subtilis biofilms and the genes within the eps locus that encoded proteins that synthesize PNAG. B. subtilis lacking epsH–K genes are unable to form biofilms, indicating PNAG is essential for biofilm/pellicle formation by this organism.
Experimental Procedures
Bacterial Strains and Plasmids
Bacterial strains (B. subtilis, E. coli, K. pneumoniae, and P. aeruginosa) and plasmids used in this study are listed in Table 1. B. subtilis was grown on minimal medium glucose (MMG) agar (23) or lysogeny broth (LB) agar (24) for 3–5 days at room temperature, whereas the other bacterial strains were grown overnight at 37 °C in LB or on LB agar.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or ref. |
|---|---|---|
| Strain | ||
| E. coli sm10 | thi-1 relA1 thi-1 thr leu tonA lacy supE recA RP4–2-Tc::Mu Kmr | 57 |
| B. subtilis 3610 | Undomesticated wild strain | 58 |
| B. subtilis DS991 | sinR::kan tasA::Tn10 spec | 59 |
| B. subtilis DK1042 | comIQ12L | 60 |
| B. subtilis DS2569 | ΔpBS32 | 60 |
| B. subtilis DK1943 | ΔepsA comIQ12L | This study |
| B. subtilis DK1806 | ΔepsB comIQ12L | This study |
| B. subtilis DK1807 | ΔepsC comIQ12L | This study |
| B. subtilis DS4248 | ΔepsD | This study |
| B. subtilis DS2152 | ΔepsE | 20 |
| B. subtilis DS4164 | ΔepsF | This study |
| B. subtilis DS7499 | ΔepsG | This study |
| B. subtilis DS6776 | ΔepsH | This study |
| B. subtilis DK1758 | ΔepsI comIQ12L | This study |
| B. subtilis DS4166 | ΔepsJ | This study |
| B. subtilis DK2055 | ΔepsK comIQ12L | This study |
| B. subtilis DS7432 | ΔepsL | This study |
| B. subtilis DS4901 | ΔepsM | This study |
| B. subtilis DS4900 | ΔepsN | This study |
| B. subtilis DK1759 | ΔepsO comIQ12L | This study |
| E. coli E11 | K1 capsule type; clinical isolate | K. Sik-Kim |
| E. coli E11 Δpga | E. coli E11 with in-frame deletion of entire pga locus | Baltimore, MD |
| E. coli E11 ΔpgaA | E. coli E11 in-frame deletion of pgaA | This study |
| E. coli E11 ΔpgaB | E. coli E11 in-frame deletion of pgaB | This study |
| E. coli E11 ΔpgaC | E. coli E11 in-frame deletion of pgaC | This study |
| E. coli E11 ΔpgaD | E. coli E11 in-frame deletion of pgaD | This study |
| K. pneumoniae | K2 capsule type; clinical isolate | |
| K. pneumoniae Δpga | K. pneumoniae K2 with in-frame deletion of entire pga locus | This study |
| P. aeruginosa | WT strain FRD1 | 31 |
| P. aeruginosa Tn::algF | FRD1 Tn501::algF, alginate-deficient | 31 |
| Plasmid | ||
| pRED/ET | Red/ET expression plasmid | 61 |
| pCP20 | Helper plasmid; FLP+, temperature-sensitive, AmpR, CmR | 62 |
| pUCP18Tc | Broad host range vector pUCP18 derivative; TetR | 63 |
| pUCP18Tc-pga | TetR, pgaABCD (pga) locus from E. coli E11 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-pgaA | TetR, pgaA from E. coli E11 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-pgaB | TetR, pgaB from E. coli E11 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-pgaC | TetR, pgaC from E. coli E11 cloned into pUCP18Tc at SacI-XbaI sites | This study |
| pUCP18Tc-pgaD | TetR, pgaD from E. coli E11 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-epsHIJK | TetR, epsHIJK from B. subtilis DS991 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-epsH | TetR, epsH locus from B. subtilis DS991 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-epsI | TetR, epsI locus from B. subtilis DS991 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-epsJ | TetR, epsJ locus from B. subtilis DS991 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pUCP18Tc-epsK | TetR, epsK locus from B. subtilis DS991 cloned into pUCP18Tc at XbaI-SbfI sites | This study |
| pMiniMAD | oriBsTs amp mls | 64 |
| pMP201 | ΔepsA mls amp | This study |
| pMP202 | ΔepsB mls amp | This study |
| pMP203 | ΔepsC mls amp | This study |
| pKB90 | ΔepsD mls amp | This study |
| pKB91 | ΔepsF mls amp | This study |
| pSG52 | ΔepsG mls amp | This study |
| pSG37 | ΔepsH mls amp | This study |
| pMP204 | ΔepsI mls amp | This study |
| pKB92 | ΔepsJ mls amp | This study |
| pMP215 | ΔepsK mls amp | This study |
| pSG53 | ΔepsL mls amp | This study |
| pMP6 | ΔepsM mls amp | This study |
| pMP5 | ΔepsN mls amp | This study |
| pMP206 | ΔepsO mls amp | This study |
| Cassette | ||
| FRT-PGK-gb2-neo-FRT | PGK-gb2-neo cassette flanked by FRT sites | Gene Bridges GmbH, Heidelberg Germany |
BLAST (Basic Local Alignment Search Tool) and Structural Analysis
Sequences of eps genes in B. subtilis strain 168 were obtained from the NCBI website (reference sequence, NZ_CM000487.1). Amino acid sequences from four eps genes (epsH, epsI, epsJ, and epsK) were mapped on E. coli and S. aureus sequences using BLAST from the NCBI website. Amino acid sequences from the four pga genes of E. coli IHE3034 were similarly mapped on S. aureus sequences. The amino acid sequences of B. subtilis epsH–K were analyzed by the Protein Homology/analogY Recognition Engine Version 2 (PHYRE2) (25) for structural predictions as to their function.
Strain Construction in Gram-negative Bacteria
Deletions of individual pga genes in a clinical isolate of an E. coli K1 strain from a child with meningitis, designated E11 and kindly provided by Kwan Sik-Kim of Johns Hopkins University School of Medicine, and deletion of the entire 4-gene pga locus in both E. coli E11 and K. pneumoniae NTUH-K2044 (26) were constructed as described previously (27). Briefly, a kanamycin resistance cassette flanked by FLP recombinase recognition target sites and homology arms to replace the DNA segments of interest in-frame were generated by PCR with deletion primers (supplemental Table 1). Recombination within the targeted chromosomal sequences was mediated by the red recombinase encoded on pRdET (28), resulting in the replacement of the targeted sequence with a kanamycin-resistant cassette; all allelic replacements were confirmed by PCR. Subsequently, the kanamycin marker was removed, using the FLP expression plasmid pCP20 (29).
Strain Construction in B. subtilis
All constructs were either directly integrated and resolved in the competent strain DK1042 (comIQ12L) or integrated in the competent strain DS2569 (ΔpBS32), transferred to the 3610 background using SPP1-mediated generalized phage transduction, and resolved (30). All strains and plasmids used in this study are listed in Table 1. Primer sequences are delineated in supplemental Table 1.
In-frame Deletions
To generate the in frame marker-less deletion constructs, each plasmid was introduced by single crossover integration at the restrictive temperature for plasmid replication (37 °C) using mls resistance as a selection. To evict the plasmid, the strain was incubated in 3 ml of LB broth at a permissive temperature for plasmid replication (22 °C) for 14 h. Cells were then serially diluted and plated on LB agar at 37 °C. Individual colonies were patched onto LB agar plates and LB agar plates containing mls to identify mls-sensitive colonies that had evicted the plasmid. Chromosomal DNA from colonies that had excised the plasmid was purified and screened by PCR to determine which isolate had retained the deletion allele.
To generate the ΔepsA in-frame marker-less deletion construct, the region upstream of epsA was amplified by PCR using the primer pair 3971/3972 and digested with EcoRI and XhoI, and the region downstream of epsA was amplified by PCR using the primer pair 3973/3974 and digested with XhoI and BamHI. The two fragments were then simultaneously ligated into the EcoRI and BamHI sites of pMiniMAD that carries a temperature-sensitive origin of replication and an erythromycin resistance cassette to generate pMP201.
The same method was used to generate the other eps gene in-frame marker-less deletion constructs. Plasmids are detailed in Table 1 and primers in supplemental Table 1.
Genetic Complementation
Wild-type alginate overproducing mucoid P. aeruginosa FRD1 and an alginate-deficient strain (P. aeruginosa (Tn501::algF)) due to a polar effect of the Tn501 insert in the algF gene on alginate synthesis (31) were used to express the B. subtilis epsHIJK locus (29).
For complementation of the various mutations, pga or eps genes from E. coli K1 strain E11 or B. subtilis DS991, respectively, were amplified by PCR with complementation primers (see Table 2) using chromosomal DNA as a template. PCR products were cloned into the broad host range vector pUCP18Tc as XbaI/SbfI or SacI/XbaI fragments and transformed into E. coli Sm10 with selection on LB agar containing tetracycline (10 mg/liter). All constructs were confirmed by sequencing. E. coli and K. pneumoniae strains were complemented with individual plasmids by electroporation and selection on LB agar supplemented with tetracycline (10 mg/liter). For P. aeruginosa, complementation plasmids were conjugated from E. coli Sm10 as described previously (32). The complemented P. aeruginosa strains were selected on LB agar containing irgasan (25 mg/liter) and tetracycline (75 mg/liter).
TABLE 2.
Results of blast of protein sequences
| Organism strain | Gene | Location | No of amino acids | Known or putative annotated protein function | Similar to | Coverage | Amino acid identities | Positive amino acids |
|---|---|---|---|---|---|---|---|---|
| % | % | |||||||
| E. coli E11 IHE3034 | pgaA | 1,119,576–1,121,999 | 808 | Putative outer membrane-β-PNAG translocation/docking protein | Unclear | None | ||
| E. coli E11 IHE3034 | pgaB | 1,117,549–1,119,567 | 673 | β-PNAG N-deacetylase, belonging to CAZy family CE4 | S. aureus IcaB | 37 | 22 | 42 |
| E. coli E11 IHE3034 | pgaC | 1,116,231–1,117,556 | 442 | PNAG N-glycosyltransferase belonging to CAZy family GT2, and PNAG co-transporter | S. aureus IcaA | 92 | 40 | 63 |
| E. coli E11 IHE3034 | pgaD | 1,115,816–1,116,229 | 138 | Inner membrane c-di-GMP receptor and PNAG co-transporter | Unclear | None | ||
| B. subtilis DS991 | epsH | 3,521,111–3,522,145 | 345 | Putative glycosyltransferase involved in biofilm formation | S. aureus IcaA | 28 | 29 | 54 |
| E. coli PgaC | 28 | 31 | 53 | |||||
| B. subtilis DS991 | epsI | 3,520,030–3,521,106 | 359 | Putative polysaccharide pyruvyl transferase | E. coli WffR | 73 | 25 | 41 |
| B. subtilis DS991 | epsJ | 3,518,999–3,520,033 | 345 | Putative glycosyltransferase | S. aureus IcaA | 26 | 31 | 52 |
| E. coli PgaC | 26 | 42 | 60 | |||||
| B. subtilis DS991 | epsK | 3,517,485–3,519,002 | 506 | Putative extracellular matrix component exporter | E. coli Wzx | 60–97 | 28 | 48 |
Immunochemical Detection of PNAG on Microbial Cells
To detect PNAG in B. subtilis biofilms, strain DS991 was inoculated into 1 ml of MMG broth in 12-well tissue culture plates and left for 1 week at room temperature in a humidified environment. Bacterial biofilms were removed with as little remaining media as possible and placed onto glass slides in a demarcated well. Samples were allowed to air dry before fixing for 5 min at room temperature with ice-cold methanol, which was removed by gently tipping the slide onto its side and absorbing residual methanol with a tissue.
Planktonic B. subtilis strains were grown in MMG media at 25 °C for 3–10 days at room temperature, whereas the other bacterial species were grown on LB agar overnight at 37 °C and then held at room temperature for 48 h to promote PNAG expression. Strains harboring one of the pUC vectors were grown on LB agar plates with tetracycline (10 mg/liter) under the same conditions with protection from light. Microbial samples were suspended in PBS, then spotted onto microscope slides, air-dried, and fixed for 1 min at room temperature with ice-cold methanol. After washing, slides were reacted with control or PNAG-specific mAbs directly conjugated to Alexa Fluor 488 at 5.2 μg/ml along with nucleic acid stain, Syto 83 (Molecular Probes) (7). After 2 h at room temperature or overnight at 4 °C, slides were washed and evaluated by confocal microscopy. For enzymatic and periodate treatments, samples fixed to slides were incubated in Tris-buffered saline (pH 6.4) containing either 50 μg of dispersin B/ml (digests PNAG) or 50 μg of chitinase/ml (no effect on PNAG) overnight at 37 °C or in 0.4 m periodate (destroys PNAG) for 2 h at 37 °C in a humidified environment. After washing, cells were treated with the Alexa Fluor 488 directly conjugated mAbs.
Extraction and Detection of PNAG
B. subtilis DS991 and E. coli Δpga (pUCP18Tc-epsHIJK) were grown as described above on MMG agar or LB agar plates, respectively, and cells were used for extraction and immunologic detection of PNAG by a slot blot, as described previously (14), and chemical detection of hexosamines, as described previously (33).
B. subtilis DS991, B. subtilis DK2055 ΔepsK, E. coli WT, and E. coli Δpga as a negative control were tested for intracellular PNAG by a direct-binding ELISA using lysates of these cells prepared by sonication. Briefly, cells from blood agar plates grown overnight at 37 °C were suspended in normal saline, washed, and then suspended in Tris-buffered saline containing 12.5 μg/ml dispersin B to remove surface PNAG during a 24-h 37 °C incubation step. After washing, bacterial cells were suspended in 0.04 m phosphate buffer (pH 7.2) at an absorbance at 650 nm (A650 nm) of 0.8, sonicated with 10-s bursts, 10 times, and insoluble debris was removed by centrifugation, and dilutions of the lysate were used to directly sensitize Immulon 4 ELISA plates. A standard curve was generated using purified PNAG (0.015–0.1 μg/ml) to sensitize ELISA wells in duplicate, and samples were probed with 10 μg/ml mAb F598 followed by an alkaline phosphatase-conjugated goat antibody to human IgG. After assay development with the p-nitrophenyl phosphate substrate, A405 nm readings were obtained; a standard curve of the purified PNAG concentration versus A405 nm was calculated by linear regression, and the amount of PNAG in bacterial cell lysates was calculated after subtracting out the background from the negative control. Samples of cells taken before and after dispersin B treatment were probed as described above for immunochemical detection of PNAG n microbial cells.
Opsonophagocytic Assay
Opsonophagocytic killing (OPK) of B. subtilis DS991, E. coli WT, E. coli (Δpga), and E. coli (Δpga) (pUCP18Tc-epsHIJK) was carried out, as described previously (34), except differentiated human promyelocytic HL60 cells were used as the phagocyte source (35). The percent killing mediated by antibodies in immune sera raised to a conjugate of nine residues of β-1–6-linked glucosamine and tetanus toxoid (TT; 9GlcNH2-TT vaccine) (36) was calculated by dividing the colony-forming units (cfu) in the test sera by those in the corresponding dilution of the nonimmune control serum.
Results
Detection of PNAG in B. subtilis Biofilms/Pellicles
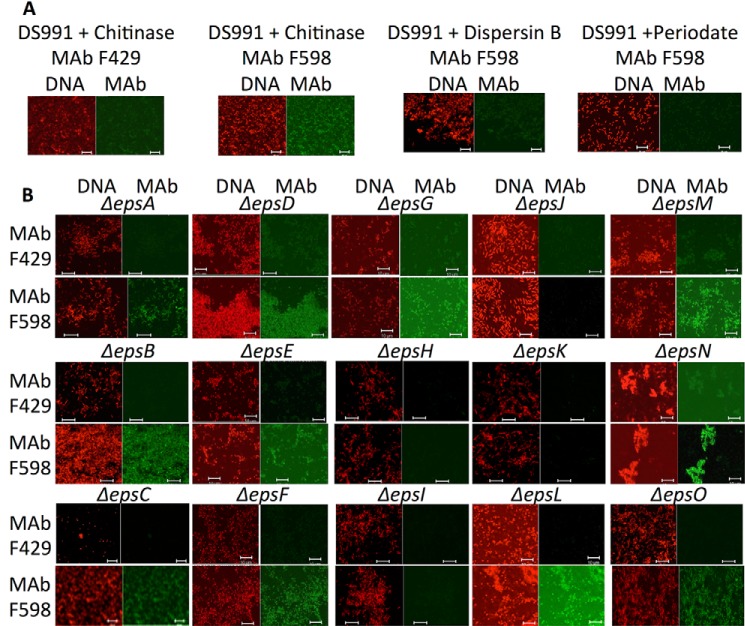
B. subtilis pellicles formed over 10 days of growth at the air-liquid interface in MMG medium were reacted with control mAb F429 or mAb F598 to PNAG, both directly conjugated to Alexa Fluor 488 and visualized for immunofluorescence by confocal microscopy. Bacilli embedded in a strongly immunoreactive matrix of PNAG were readily observed, and binding to mAb F598 was lost after treating the B. subtilis biofilms with the PNAG-degrading enzyme dispersin B or with PNAG-hydrolyzing sodium periodate (Fig. 1).
FIGURE 1.
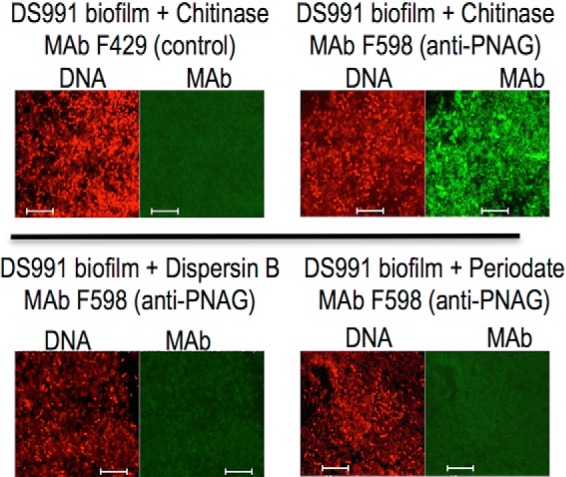
PNAG production in 10-day-old pellicles of B. subtilis. Pellicles/biofilms formed at the air-liquid interface of MMG medium over 10 days at room temperature were analyzed via immunofluorescence for the presence of PNAG within these structures. Cells were identified using Syto 83 DNA stain and PNAG by reactivity with mAb F598 directly conjugated to Alexa Fluor 488. Loss or immunologic reactivity following digestion with dispersin B or exposure to 0.04 m periodate are both treatments that degrade PNAG due to their enzymatic and chemical specificity to attack the β-1–6-linked N-acetylglucosamine polymer. Bars, 10 μm.
Expression of PNAG by WT and eps-mutant B. subtilis
Planktonic cells of B. subtilis that overproduced EPS (due to mutation of the master repressor SinR) and also defective for the biofilm-organizing protein TasA (DS991) were also positive for PNAG expression by immunofluorescence microscopy (Fig. 2A), and immunoreactivity was lost following treatment with dispersin B or periodate (Fig. 2A). When PNAG production was evaluated in B. subtilis strains with in-frame deletions in all 15 identified eps genes (Table 1), most of which are unable to form biofilms, only disruption of the epsH–K genes led to loss of PNAG production (Fig. 2B). The epsH and epsJ genes are annotated as putative GTs and show similarity to proteins encoded by S. aureus icaA and E. coli pgaC (Table 2). The epsI gene is currently annotated as a putative polysaccharide pyruvyltransferase (Table 2), but as shown below, its structural predictions suggest other functions. The epsK gene is currently annotated as a putative extracellular matrix component exporter similar to the wzx proteins in E. coli (Table 2). Two other eps genes (epsE and epsF) are annotated as putative GTs, but deletions in these genes did not lead to loss of PNAG production (Fig. 2B). Similarly, an epsG deletion mutant, known to be deficient in biofilm production like the epsH mutant (17), was still able to produce PNAG (Fig. 2B). As with other microbial species, PNAG production appears to be necessary, but not sufficient, for full biofilm formation in B. subtilis.
FIGURE 2.
Demonstration of PNAG production by B. subtilis WT and deletion strains by immunofluorescence microscopy. A, parental strain B. subtilis DS991 failed to react with mAb F429 to P. aeruginosa alginate after chitinase treatment but did bind mAb F598 to PNAG. Binding was lost after treatment of cells with dispersin B or sodium periodate. B, binding of mAb F429 (control) or F598 to PNAG to indicated B. subtilis strain with a deletion of in the indicated gene in the eps locus (Δeps). DNA was stained with Syto 83 to visualize cells, and mAbs were conjugated directly to Alexa Fluor 488 (green). White bars, 10 μm.
Bioinformatic Analysis of B. subtilis PNAG Biosynthetic eps Genes and Proteins
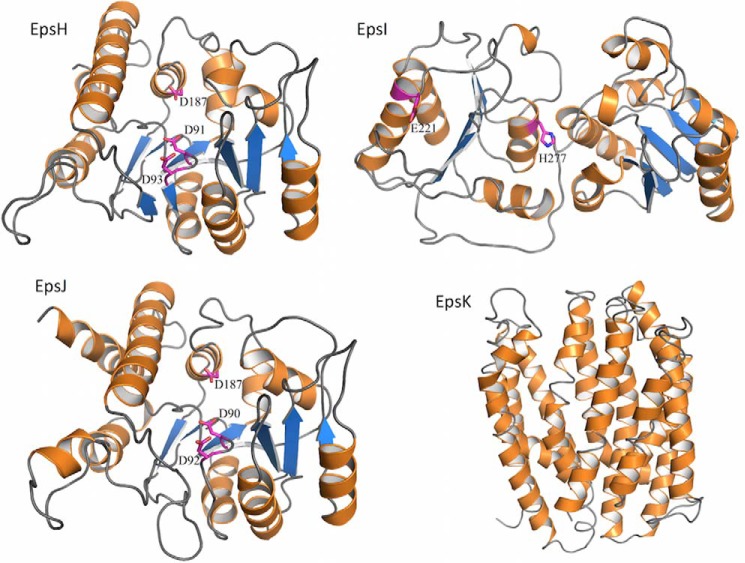
The above results, along with bioinformatic analysis of the B. subtilis eps locus, indicated the genes required for PNAG production, epsH–K, could comprise a four-gene locus somewhat similar to the S. aureus icaADBC and E. coli pgaABCD loci encoding the PNAG biosynthetic proteins in these two organisms (Table 2) (11, 13). To obtain a more precise idea of the functions of the EpsH–K proteins, as well as other proteins within the eps locus, the predicted protein structures were analyzed using the PHYRE2 server (Fig. 3) (25). Overall, the entire eps locus contains genes encoding proteins known to be involved in polysaccharide synthesis using either the Wzx/Wzy or ABC transporter pathways for lipopolysaccharides, capsules, colonic acid, and S-layer components (37–39). Both EpsH and EpsJ have nearly identical predicted structures (Fig. 3) with homology to the GT domains of IcaA and PgaC proteins used in S. aureus and E. coli for PNAG synthesis, respectively (13, 40), as well as the BcsA protein used for cellulose (β-1–4-linked glucose) synthesis (41). With 60–70% sequence coverage and 15–22% sequence identity, EpsH and EpsJ are predicted with 100% confidence to be UDP-N-acetylglucosamine transferases with a GT-A fold. They are not predicted to have any transmembrane domains and therefore would be unable to translocate the synthesized polymer across the membrane.
FIGURE 3.
Structural models of the EpsH–K proteins. Predicted model of EpsH based on the cellulose synthase BcsA (PDB code 4HG6) shows a GT-A fold. Predicted model of EpsI based on the family 9 GT (PDB code 3TOV) shows a GT-B fold. Predicted model of EpsJ based on a chondroitin polymerase (PDB code 2Z86) shows a GT-A fold. Predicted model of EpsK based on DinF (PDB code 4LZ9) shows a MATE transporter fold. The structure models were generated using PHYRE2 (25) and are shown in schematic representations with α-helices, β-strands, and loops colored orange, blue, and gray, respectively. Predicted active site residues for the EpsH–J proteins are shown as magenta sticks.
BLAST analysis showed that EpsK belongs to the polysaccharide biosynthesis protein family, pfam01943. Members of this family are integral membrane proteins that encode for multiple antimicrobial extrusion protein (MATE)-like transporters, such as the O-antigen flippase Wzx. EpsK was predicted to be an α-helical membrane transporter with 14 transmembrane helices using PHYRE2 and various transmembrane domain prediction servers, respectively (Fig. 3) (42). PHYRE2 predicts EpsK to be structurally similar to DinF, a member of the MATE family, covering 84% of the sequence with 10–12% amino acid identity and 100% confidence of the fold prediction.
The analysis of the EpsI protein (Fig. 3) indicated it was not related structurally to the PNAG deacetylases IcaB or PgaB (43, 44) or any members of the family 4 carbohydrate esterases (deacetylases), but rather it is predicted to be a cytoplasmic GT with homology to genes in lipopolysaccharide biosynthesis. There were no predicted signal sequences or transmembrane helices but almost complete coverage (80–90%) of the EpsI sequence with 95% confidence in fold prediction to known lipopolysaccharide transferases.
Functional Equivalence of B. subtilis EpsH–K Proteins to E. coli PNAG Biosynthetic Proteins
To determine whether each of these four B. subtilis eps genes encoded proteins that could replace those encoded by the pga operon, we constructed individual in-frame mutations in each of the four E. coli pga genes, pgaA–D, and complemented each of these different mutants with an eps-related gene judged to most likely encode a protein of potential similar function. As controls, we also complemented some of the E. coli mutants with a mismatched gene from B. subtilis. The WT E. coli strain produced PNAG, and deletion of any of the four individual pga genes abolished PNAG production (Table 3). Complementation with an empty vector (pUCP18Tc) did not restore the phenotype in any strain with an in-frame pga mutation. Complementation of the E. coli-pga mutant strains with individual clones of B. subtilis epsH, epsI, epsJ, or epsK resulted in restoration of PNAG production in each E. coli mutant strain with the gene from the B. subtilis locus judged most likely to be a functional equivalent (Table 3). Interestingly, we found that PNAG production in the E. coli ΔpgaC strain could be restored by complementation with either the B. subtilis epsJ gene and, to a lesser extent, the epsH gene (Table 3). Similarly, we could achieve phenotypic complementation of PNAG production in the E. coli ΔpgaA mutant strain most strongly with the B. subtilis epsH gene and, to a lesser extent, the epsJ gene. The E. coli pgaB mutant was efficiently complemented with the B. subtilis epsI gene, and the E. coli pgaD mutant was complemented with the B. subtilis epsK gene (Table 3). Cloning of the B. subtilis epsI and epsK genes into the E. coli ΔpgaA mutant did not result in PNAG production. Finally, when the entire pga locus was deleted from either E. coli or K. pneumoniae, the loss of PNAG production could be restored in both strains by complementation with the entire B subtilis epsHIJK locus in pUCP18Tc-epsHIJK (Table 3).
TABLE 3.
Summary for detection of PNAG synthesis by immunofluorescence in indicated strain
The following symbols are used: −, no immunofluorescence detected; +, 25–50% of WT levels of PNAG observed; ++, 51–80% of WT levels of PNAG observed; +++, >80% of WT levels of PNAG observed.
| Strain | Complemented with | Control mAb | mAb to PNAG |
|---|---|---|---|
| E. coli K1 WT | − | +++ | |
| E. coli K1 ΔpgaA | − | − | |
| E. coli K1 ΔpgaB | − | − | |
| E. coli K1 ΔpgaC | − | − | |
| E. coli K1 ΔpgaD | − | − | |
| E. coli K1 ΔpgaA | (pUCP18Tc) | − | − |
| E. coli K1 ΔpgaB | − | − | |
| E. coli K1 ΔpgaC | − | − | |
| E. coli K1 ΔpgaD | − | − | |
| E. coli K1 ΔpgaA | (pUCP18Tc-pgaA) | − | +++ |
| E. coli K1 ΔpgaB | (pUCP18Tc-pgaB) | − | +++ |
| E. coli K1 ΔpgaC | (pUCP18Tc-pgaC) | − | +++ |
| E. coli K1 ΔpgaD | (pUCP18Tc-pgaD) | − | +++ |
| E. coli K1 ΔpgaA | (pUCP18Tc-epsH) | − | +++ |
| E. coli K1 ΔpgaA | (pUCP18Tc-epsJ) | − | + |
| E. coli K1 ΔpgaB | (pUCP18Tc-epsI) | − | +++ |
| E. coli K1 ΔpgaC | (pUCP18Tc-epsH) | − | + |
| E. coli K1 ΔpgaC | (pUCP18Tc-epsJ) | − | +++ |
| E. coli K1 ΔpgaD | (pUCP18Tc-epsK) | − | +++ |
| E. coli K1 ΔpgaABCD | (pUCP18Tc-epsHIJK) | − | +++ |
| K. pneumoniae Δpga | (pUCP18Tc) | − | − |
| K. pneumoniae Δpga | (pUCP18Tc-epsHIJK) | − | +++ |
We also analyzed the B. subtilis DS991 WT and ΔepsK strains as well as WT E. coli for the presence of intracellular PNAG. WT E. coli had a low, but detectable, level of intracellular PNAG (Table 4). Both WT and ΔepsK B. subtilis had detectable intracellular PNAG (Table 4), with the B. subtilis ΔepsK strain having about 60% more intracellular PNAG, indicating that in the absence of the predicted EpsK transporter, there was accumulation of the PNAG polysaccharide inside the B. subtilis ΔepsK cells.
TABLE 4.
Levels of intracellular PNAG in WT and B. subtilis ΔepsK strain
| Strain | ng/108 CFU (mean ± S.D.) |
|---|---|
| B. subtilis DS991 (WT) | 152 ± 52 |
| B. subtilis ΔepsK | 242 ± 64 |
| E. coli E11 (WT) | 6 ± 1 |
Finally, we found that the predicted MATE function of the B. subtilis EpsK protein had similarity to PelG and PslK in P. aeruginosa used for the synthesis of the PEL and PSL polysaccharides, respectively. P. aeruginosa, unlike E. coli and K. pneumoniae, does not normally synthesize PNAG, so we introduced the pUCP18Tc-epsHIJK plasmid into WT and algF-interrupted (Tn::algF) P. aeruginosa strain FRD1 to ascertain whether PNAG could be synthesized. We did not detect PNAG synthesis in either the WT P. aeruginosa strain or the strain carrying pUCP18Tc-epsHIJK, but we did show alginate production by virtue of binding of mAb F429 to these bacterial cells (Fig. 4). We hypothesized that it might be problematic to produce or detect low level PNAG production in WT FRD1 P. aeruginosa if this organism is expressing both the positively charged PNAG and the negatively charged alginate in the same cell. We thus cloned pUCP18Tc-epsHIJK into P. aeruginosa FRD1 (Tn501::algF), wherein the Tn (transposon) insertion has a polar effect on the alginate biosynthetic locus, leading to loss of alginate production, as verified by loss of binding of mAb F429 to cells of this strain (Fig. 4). Addition of the pUCP18Tc-epsHIJK plasmid resulted in expression of PNAG on the recombinant P. aeruginosa FRD1 (Tn501::algF) cells as detected by immunofluorescence analysis of binding of mAb F598 to PNAG. This binding was lost after treatment of the recombinant P. aeruginosa strains with dispersin B and periodate, indicative of PNAG synthesis in alginate-negative P. aeruginosa by proteins encoded by the B. subtilis epsHIJK genes.
FIGURE 4.
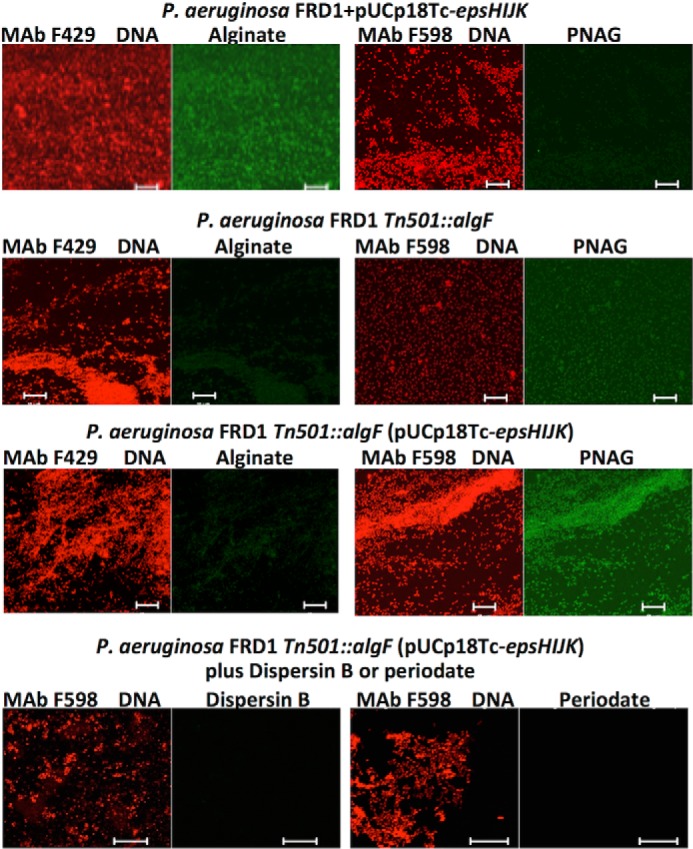
Production of PNAG in alginate-deficient P. aeruginosa FRD1 in the presence of the B. subtilis epsHIJK locus. WT P. aeruginosa FRD1 produces alginate and binds Alexa Fluor 48-labeled mAb F429 but not PNAG-specific mAb F598. A polar Tn-insertion in the algF gene eliminated alginate production and insertion of the B. subtilis epsHIJK locus leads to PNAG production. Immunoreactive PNAG in this latter strain is lost after treatment with dispersin B or periodate. White bars, 10 μm.
Detection of PNAG by WT and Recombinant B. subtilis and E. coli Strains
To confirm that PNAG was produced by WT B. subtilis but not B. subtilis ΔepsH or ΔepsJ, and by WT E. coli E11 and E. coli E11 (Δpga) (pUCP18Tc-epsHIJK) but not E. coli (Δpga), we extracted surface material from cells as described (14) and used slot blots for immunologic detection of extracted antigen. As shown in Fig. 5, WT B. subtilis as well as WT E. coli E11 produced immunoreactive PNAG, whereas the B. subtilis ΔepsH and ΔepsJ strains did not, nor did E. coli (Δpga). Complementation of the PNAG-deficient E. coli (Δpga) strain with the B. subtilis epsHIJK genes restored detectable PNAG production. Confirmation that the immunoreactive material was likely PNAG was obtained by treating extracts with dispersin B or periodate, both of which destroy PNAG. Such treated materials had no reactivity with the mAb to PNAG in a slot-blot assay (Fig. 5). Finally, chemical analysis of the extracts indicated hexosamine was only detectable in the PNAG-positive extracts. Attempts to further purify PNAG to a degree sufficient for analysis by NMR were unsuccessful. This is consistent with almost all prior publications indicating that methods have not yet been developed to purify PNAG sufficiently for NMR analysis in the absence of hyper-expression of the biosynthetic proteins in organisms like S. aureus, E. coli, or A. baumannii (13, 14, 45, 46). In organisms such as S. epidermidis (8) and Vibrio parahaemolyticus (47), isolation of PNAG-related small molecular weight fragments has been achieved only by use of natural hyper-producers of biofilms as sources of the initial extracts.
FIGURE 5.

Detection of PNAG in extracts of E. coli and B. subtilis strains. Extracts of the indicated E. coli or B. subtilis strain were immobilized on membranes and probed with mAb F598 to PNAG followed by anti-human IgG conjugated to HRP. Both WT E. coli and E. coli Δpga carrying the cloned B. subtilis epsHIJK genes expressed PNAG, while the Δpga strain did not contain detectable PNAG. Similarly, extracts of cells of B. subtilis DS991 had detectable PNAG, but none was present in the strains lacking either the epsH or epsJ genes. Extracts with detectable PNAG lost reactivity with mAb F598 following dispersin B or periodate treatment and contained detectable hexosamine. N/A, not applicable; Pos, positive; Neg, negative.
Opsonic Killing Mediated by Antibody to PNAG
The susceptibility of the B. subtilis and E. coli strains to OPK was tested in an assay using antibodies raised in either a rabbit or goat to the 9GlcNH2-TT vaccine (36). Antibodies in both of these antisera readily mediated OPK of WT B. subtilis and WT E. coli (Fig. 6). Deletion of the pga locus in E. coli resulted in no effect of antibody to PNAG on cell survival in an OPK assay, whereas introduction of the pUCP18Tc-epsHIJK plasmid into pga-deleted E. coli restored the susceptibility to OPK. Thus, B. subtilis EpsH–K proteins produced an antigen in E. coli functionally equivalent to native PNAG from this organism.
FIGURE 6.
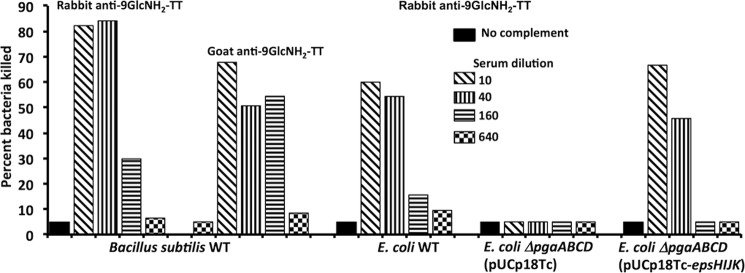
Opsonophagocytic killing of indicated E. coli or B. subtilis strains by antibodies to PNAG raised by immunization with a conjugate vaccine of 9GlcNH2-TT. Bars indicate mean percentage of bacteria killed in duplicate compared with those in the corresponding dilution of nonimmune goat or rabbit antiserum to PNAG.
Discussion
B. subtilis has served as one of the major model organisms for scrutinizing biofilm formation and multicellular activity in bacterial communities (18, 48). Many of the factors, genes, and conditions needed for producing and modulating formation of biofilms by this organism are well studied (49). Components of the B. subtilis biofilm include EPS and polymeric substances such as poly-dl-glutamic acid and proteins, including TapA, TasA, and BslA (48). However, the chemistry of the biofilm EPS constituents are not defined, and no definitive polysaccharide structures have previously been identified. Here, we found that within the 15-gene eps cluster of B. subtilis there was a 4-gene locus encompassing epsHIJK that encodes proteins that can synthesize either the conserved bacterial surface polysaccharide PNAG, a common component of microbial biofilms, or an antigenically cross-reactive material. In the B. subtilis biofilm, PNAG, or a related structure, likely serves as a scaffold as well as an anchoring substrate for the other components in the biofilm, which requires gene products from the eps locus other than epsHIJK for full matrix formation, as many of the eps gene products are needed to establish the biofilm phenotype (48).
To synthesize PNAG using the B. subtilis EpsH–K proteins, we speculate that EpsH is an undecaprenyl priming transferase that makes undecaprenyl-3-O-acyl N-acetylglucosamine. EpsI could either be modifying this first GlcNAc or possibly adding on another sugar monomer and also providing a deacetylase function. EpsJ is potentially the poly-GlcNAc transferase that is needed for long chain extension to the lipid linker of the UDP-N-acetylglucosamine precursor to synthesize the PNAG-like molecule, and EpsK either facilitates the activity of EpsJ and/or transports the polysaccharide out of the cell.
In regard to EpsK, it is unclear how an α-helical membrane transporter from a Gram-positive organism would function in PNAG synthesis and transport in E. coli. Using BLAST analysis we noted that EpsK is a member of the Wzx family of proteins (Table 2), which show overall little conservation in their primary amino acid sequences, but Wzx proteins can interchangeably export different polysaccharides containing N-acetylglucosamine or N-acetylgalactosamine as the initiating sugar (50). Therefore, as a member of the Wzx family of proteins, it is entirely plausible that EpsK is transporting the PNAG polymer out of the cell in both B. subtilis and E. coli.
These structural inferences of the EpsHIJK proteins are consistent with the predictions that synthesis of PNAG, alginate, and cellulose, all components of microbial biofilms, is mediated by proteins with similar hallmarks and functions but not necessarily with conserved architectures (41, 51). Moreover, it appears that some of the proteins, despite little overall amino acid sequence identity, nonetheless maintain sufficiently conserved and interchangeable functions, as documented here for the B. subtilis and E. coli PNAG biosynthetic proteins.
We found that deletion of these genes in B. subtilis disrupted PNAG production as detected by loss of immunoreactivity with mAb F598. Cloning either the epsHIJK genes individually into E. coli strains with in-frame deletions in the endogenous pgaA–D genes, or as a 4-gene cluster into pgaABCD-deleted E. coli or K. pneumoniae, resulted in production of immunochemically detectable PNAG, as did cloning the epsHIJK genes into alginate-deficient P. aeruginosa. Expression of epsHIJK in E. coli mutants deficient in pga genes could result in either production of authentic PNAG or a material that is a component of another E. coli factor, such as an LPS or a glycoprotein. We also found that the epsH and epsJ genes can both complement the E. coli pgaA and the pgaC genes, yet in both of these organisms loss of any one of these genes results in a PNAG-negative phenotype. It appears there is no cross-complementation within the host strain. Although the structural similarity of EpsH and EpsJ explains, in part, the ability of each to provide some cross-complementation, it seems that they likely have some additional nonshared specific functions within B. subtilis that are provided by other proteins when they are expressed in E. coli.
Although PNAG itself, or a structure containing a PNAG component, appears to be an EPS element involved in B subtilis biofilm formation, it is likely that other EPS molecules are also needed for full pellicle formation. Studies of the eps locus indicate that deletion of numerous other genes also disrupts biofilm formation, and some of these appear to be classic EPS biosynthetic genes (17, 18, 48). The EpsM–O proteins are predicted to be an acetyltransferase (EpsM), an aminotransferase/sugar dehydratase (EpsN), and a GT with a GT-B fold (EpsO). The EpsE protein is involved in both motility and biofilm formation (21) acting as a clutch of the flagellar cellular motility apparatus to inhibit movement, while also having a predicted GT structure.
Recently, Elsholz et al. (52) reported that the B. subtilis EPS serves as a positive regulator of its own synthesis by binding to the extracellular portion of a receptor encoded by the epsA gene that interacts with a tyrosine kinase encoded by epsB and inhibits EpsB autophosphorylation. Dephosphorylated EpsB is associated with enhanced EPS production. In an epsH mutant unable to produce biofilm and, as shown here, PNAG, a His-tagged recombinant EpsB protein was highly phosphorylated due to lack of EPS production. We found neither EpsA nor EpsB was needed for synthesis of immunoreactive PNAG in B. subtilis. This is consistent with the finding from Elsholz et al. (52) that PNAG-related material prepared using a modified method from Mack et al. (8) that yields small molecular weight polysaccharide intercellular adhesin had no effect on EpsB autophosphorylation. Given that the epsH mutant was not producing a factor binding to the EpsA extracellular domain, it appears that other EPS components either depend on EpsH for synthesis or PNAG is needed to facilitate the inhibition of EpsB autophosphorylation. However, the actual factors regulating B. subtilis EPS production via EpsA and EpsB are currently uncharacterized in regard to specific chemical properties.
One final aspect to consider is that the polysaccharide synthesized by the B. subtilis EpsH–K proteins may not be PNAG but a molecular entity with sufficient β-1–6-linked N-acetylglucosamine in it to make it immunoreactive with antibodies to PNAG but also containing other components. mAb F598 binding to PNAG requires a minimum of seven β-1–6-linked N-acetylglucosamine residues (36, 53), indicating a minimal PNAG-related constituent present in the material synthesized by the EpsH–K proteins. We do note that every microbial strain wherein we or others have identified a dispersin B and periodate-sensitive, hexosamine-containing antigen that reacts with antibodies to native PNAG, and wherein the reactive material has been isolated, turns out to be chemically verified PNAG (14, 45, 54–56). Furthermore, only 1–6-linked hexosamines, and no other possible amino-sugar linkages, are sensitive to periodate, and only β-1–6-linked N-acetylglucosamine can be digested by dispersin B (22). Also, for every microbe for which genes encoding PNAG biosynthetic proteins have been identified and deleted, there is loss of antibody reactivity with material on the cell surface due to gene loss. Thus, we consider it highly unlikely that the material produced by the EpsH–K proteins is not PNAG, but at a minimum, it is at least a PNAG-containing molecular structure.
Overall, our results show PNAG, or a closely related entity, is a component of the B. subtilis biofilm matrix synthesized by the EpsH–K proteins. These proteins can also be expressed and are functional in Gram-negative organisms, including E. coli, K. pneumoniae, and P. aeruginosa. However, it is possible that the EpsH–K proteins are also required for producing other polysaccharide components of the biofilm matrix, inasmuch as PNAG does not appear to be the only carbohydrate entity in this organism's biofilm (48). However, no carbohydrate component other than PNAG identified here has been fully characterized as being synthesized by proteins encoded by genes within the B. subtilis eps locus, leaving open the question as to the chemical composition of additional eps-dependent polymeric carbohydrates present in this structure. Analysis of EPS composition would undoubtedly advance the understanding of the B. subtilis biofilm formation process, but it should be noted that published methods used to isolate EPS (52) would contain very little native PNAG, as this molecule is poorly soluble at neutral pH, particularly after alcohol precipitation (14, 45). Thus, in the absence of an appropriate method, researchers have not been able to purify PNAG to obtain an appreciable yield for detailed chemical structural determination. We did find, however, that extracts from the WT B. subtilis cells and recombinant E. coli cells carrying the pUCP18Tc-epsHIJK plasmid contained immunoreactive dispersin B and the periodate-sensitive hexosamine-containing material present. Thus, the analysis of the products of the B. subtilis epsHIJK locus described here are fully consistent with the production by WT B. subtilis of PNAG, which is likely a necessary component of this organism's biofilm based on the genetic data showing the essentiality of epsH–K gene products for production of this structure (19).
Author Contributions
D. R. performed experiments, analyzed data, contributed to the study concept, and wrote the manuscript. C. C. B., Y. F. Z., S. P., M. K., D. B. K., D. J. L., P. L. H., and D. S. performed experiments, analyzed data, provided reagents, edited the manuscript, and contributed to the study concept. G. B. P. supervised the project, developed the study concept, analyzed data, and wrote the manuscript.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants EY016104 and AI057159 (a component of Award U54A1057159) (to G. B. P.) and Grant GM093030 (to D. B. K.). This work was also supported by the AXA Research Fund and the Fondation pour la Recherche Médicale (to D. R.), awards from the William Randolph Hearst Fund (to D. R.) and the Hood Foundation (to D. S.) and the Seedlings Foundation (to D. S. and C. C.-B.), and Grant 43998 from the Canadian Institutes of Health Research (to P. L. H.). G. B. P. is an inventor of intellectual properties (human monoclonal antibody to PNAG and PNAG vaccines) that are licensed by Brigham and Women's Hospital to Alopexx Vaccine, LLC, and Alopexx Pharmaceuticals, LLC, entities in which G. B. P. also holds equity. As an inventor of intellectual properties, G. B. P. also has the right to receive a share of licensing-related income (royalties, fees) through Brigham and Women's Hospital from Alopexx Pharmaceuticals, LLC, and Alopexx Vaccine, LLC. G. B. P.'s interests were reviewed and are managed by the Brigham and Women's Hospital and Partners Healthcare in accordance with their conflict of interest policies. C. C.-B. and D. S. are inventors of intellectual properties (use of human monoclonal antibody to PNAG and use of PNAG vaccines) that are licensed by Brigham and Women's Hospital to Alopexx Vaccine, LLC, and Alopexx Pharmaceuticals, LLC. As inventors of intellectual properties, C. C.-B. and D. S. also have the right to receive a share of licensing-related income (royalties, fees) through Brigham and Women's Hospital from Alopexx Pharmaceuticals, LLC, and Alopexx Vaccine, LLC.

This article contains supplemental Table S1.
- EPS
- exopolysaccharides
- PNAG
- poly-N-acetylglucosamine
- GT
- glycosyltransferase
- MMG
- minimal medium glucose
- LB
- lysogeny broth
- OPK
- opsonophagocytic killing
- MATE
- multiple antimicrobial extrusion protein
- PDB
- Protein Data Bank
- TT
- tetanus toxoid.
References
- 1. Hall-Stoodley L., Stoodley P. (2009) Evolving concepts in biofilm infections. Cell Microbiol. 11, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 2. Epstein A. K., Pokroy B., Seminara A., Aizenberg J. (2011) Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc. Natl. Acad. Sci. U.S.A. 108, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McDougald D., Rice S. A., Barraud N., Steinberg P. D., Kjelleberg S. (2012) Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10, 39–50 [DOI] [PubMed] [Google Scholar]
- 4. Harriott M. M., Noverr M. C. (2011) Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 19, 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bakaletz L. O. (2012) Bacterial biofilms in the upper airway–evidence for role in pathology and implications for treatment of otitis media. Paediatr. Respir. Rev. 13, 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mack D., Davies A. P., Harris L. G., Knobloch J. K., Rohde H. (2009) Staphylococcus epidermidis biofilms: functional molecules, relation to virulence, and vaccine potential. Top. Curr. Chem. 288, 157–182 [DOI] [PubMed] [Google Scholar]
- 7. Cywes-Bentley C., Skurnik D., Zaidi T., Roux D., Deoliveira R. B., Garrett W. S., Lu X., O'Malley J., Kinzel K., Zaidi T., Rey A., Perrin C., Fichorova R. N., Kayatani A. K., Maira-Litràn T., et al. (2013) Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, E2209–E2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mack D., Fischer W., Krokotsch A., Leopold K., Hartmann R., Egge H., Laufs R. (1996) The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKenney D., Pouliot K. L., Wang Y., Murthy V., Ulrich M., Döring G., Lee J. C., Goldmann D. A., Pier G. B. (1999) Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284, 1523–1527 [DOI] [PubMed] [Google Scholar]
- 10. Cramton S. E., Gerke C., Schnell N. F., Nichols W. W., Götz F. (1999) The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heilmann C., Schweitzer O., Gerke C., Vanittanakom N., Mack D., Götz F. (1996) Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 12. Foulston L., Elsholz A. K., DeFrancesco A. S., Losick R. (2014) The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 5, e01667–01614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X., Preston J. F., 3rd, Romeo T. (2004) The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi A. H., Slamti L., Avci F. Y., Pier G. B., Maira-Litrán T. (2009) The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1–6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skurnik D., Davis M. R., Jr., Benedetti D., Moravec K. L., Cywes-Bentley C., Roux D., Traficante D. C., Walsh R. L., Maira-Litràn T., Cassidy S. K., Hermos C. R., Martin T. R., Thakkallapalli E. L., Vargas S. O., McAdam A. J., et al. (2012) Targeting pan-resistant bacteria with antibodies to a broadly conserved surface polysaccharide expressed during infection. J. Infect. Dis. 205, 1709–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Branda S. S., Chu F., Kearns D. B., Losick R., Kolter R. (2006) A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238 [DOI] [PubMed] [Google Scholar]
- 17. Marvasi M., Visscher P. T., Casillas Martinez L. (2010) Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol. Lett. 313, 1–9 [DOI] [PubMed] [Google Scholar]
- 18. Shank E. A., Kolter R. (2011) Extracellular signaling and multicellularity in Bacillus subtilis. Curr. Opin. Microbiol. 14, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagorska K., Ostrowski A., Hinc K., Holland I. B., Obuchowski M. (2010) Importance of eps genes from Bacillus subtilis in biofilm formation and swarming. J. Appl. Genet. 51, 369–381 [DOI] [PubMed] [Google Scholar]
- 20. Blair K. M., Turner L., Winkelman J. T., Berg H. C., Kearns D. B. (2008) A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320, 1636–1638 [DOI] [PubMed] [Google Scholar]
- 21. Guttenplan S. B., Blair K. M., Kearns D. B. (2010) The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6, e1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramasubbu N., Thomas L. M., Ragunath C., Kaplan J. B. (2005) Structural analysis of dispersin B, a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans. J. Mol. Biol. 349, 475–486 [DOI] [PubMed] [Google Scholar]
- 23. Martínez A., Ramírez O. T., Valle F. (1997) Improvement of culture conditions to overproduce β-galactosidase from Escherichia coli in Bacillus subtilis. Appl. Microbiol. Biotechnol. 47, 40–45 [DOI] [PubMed] [Google Scholar]
- 24. Bertani G. (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 26. Hsieh P.-F., Lin T.-L., Yang F.-L., Wu M.-C., Pan Y.-J., Wu S.-H., Wang J.-T. (2012) Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS ONE 7, e33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu X., Skurnik D., Pozzi C., Roux D., Cywes-Bentley C., Ritchie J. M., Munera D., Gening M. L., Tsvetkov Y. E., Nifantiev N. E., Waldor M. K., Pier G. B. (2014) A poly-N-acetylglucosamine-Shiga toxin broad-spectrum conjugate vaccine for Shiga toxin-producing Escherichia coli. MBio 5, e00974–00914-e00974–00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chuang C. H., Wang Y. H., Chang H. J., Chen H. L., Huang Y. C., Lin T. Y., Ozer E. A., Allen J. P., Hauser A. R., Chiu C. H. (2014) Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut 63, 736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cherepanov P. P., Wackernagel W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 [DOI] [PubMed] [Google Scholar]
- 30. Yasbin R. E., Young F. E. (1974) Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 14, 1343–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franklin M. J., Ohman D. E. (1993) Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175, 5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skurnik D., Roux D., Cattoir V., Danilchanka O., Lu X., Yoder-Himes D. R., Han K., Guillard T., Jiang D., Gaultier C., Guerin F., Aschard H., Leclercq R., Mekalanos J. J., Lory S., Pier G. B. (2013) Enhanced in vivo fitness of Pseudomonas aeruginosa carbapenem-resistant oprD mutants revealed through In-seq analysis. Proc. Natl. Acad. Sci. U.S.A. 110, 20747–20752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith R. L., Gilkerson E. (1979) Quantitation of glycosaminoglycan hexosamine using 3-methyl-2-benzothiazolone hydrazone hydrochloride. Anal. Biochem. 98, 478–480 [DOI] [PubMed] [Google Scholar]
- 34. Maira-Litrán T., Kropec A., Goldmann D. A., Pier G. B. (2005) Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl-β-(1–6)-glucosamine. Infect. Immun. 73, 6752–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romero-Steiner S., Frasch C. E., Carlone G., Fleck R. A., Goldblatt D., Nahm M. H. (2006) Use of opsonophagocytosis for serological evaluation of pneumococcal vaccines. Clin. Vaccine Immunol. 13, 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gening M. L., Maira-Litrán T., Kropec A., Skurnik D., Grout M., Tsvetkov Y. E., Nifantiev N. E., Pier G. B. (2010) Synthetic β-(1→6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect. Immun. 78, 764–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valvano M. A. (2011) Common themes in glycoconjugate assembly using the biogenesis of O-antigen lipopolysaccharide as a model system. Biochemistry 76, 729–735 [DOI] [PubMed] [Google Scholar]
- 38. Willis L. M., Whitfield C. (2013) Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44 [DOI] [PubMed] [Google Scholar]
- 39. Whitfield C., Paiment A. (2003) Biosynthesis and assembly of Group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 40. Gerke C., Kraft A., Süssmuth R., Schweitzer O., Götz F. (1998) Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273, 18586–18593 [DOI] [PubMed] [Google Scholar]
- 41. Morgan J. L., Strumillo J., Zimmer J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alibert-Franco S., Pradines B., Mahamoud A., Davin-Regli A., Pagès J. M. (2009) Efflux mechanism, an attractive target to combat multidrug resistant Plasmodium falciparum and Pseudomonas aeruginosa. Curr. Med. Chem. 16, 301–317 [DOI] [PubMed] [Google Scholar]
- 43. Little D. J., Bamford N. C., Pokrovskaya V., Robinson H., Nitz M., Howell P. L. (2014) Structural basis for the de-N-acetylation of poly-β-1,6-N-acetyl-d-glucosamine in Gram-positive bacteria. J. Biol. Chem. 289, 35907–35917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Little D. J., Poloczek J., Whitney J. C., Robinson H., Nitz M., Howell P. L. (2012) The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-d-glucosamine. J. Biol. Chem. 287, 31126–31137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maira-Litrán T., Kropec A., Abeygunawardana C., Joyce J., Mark G., 3rd, Goldmann D. A., Pier G. B. (2002) Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70, 4433–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Conover M. S., Sloan G. P., Love C. F., Sukumar N., Deora R. (2010) The Bps polysaccharide of Bordetella pertussis promotes colonization and biofilm formation in the nose by functioning as an adhesin. Mol. Microbiol. 77, 1439–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ye L., Zheng X., Zheng H. (2014) Effect of sypQ gene on poly-N-acetylglucosamine biosynthesis in Vibrio parahaemolyticus and its role in infection process. Glycobiology 24, 351–358 [DOI] [PubMed] [Google Scholar]
- 48. Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. (2013) Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Romero D. (2013) Bacterial determinants of the social behavior of Bacillus subtilis. Res. Microbiol. 164, 788–798 [DOI] [PubMed] [Google Scholar]
- 50. Marolda C. L., Vicarioli J., Valvano M. A. (2004) Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150, 4095–4105 [DOI] [PubMed] [Google Scholar]
- 51. Whitney J. C., Howell P. L. (2013) Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 21, 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Elsholz A. K., Wacker S. A., Losick R. (2014) Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 28, 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gening M. L., Tsvetkov Y. E., Pier G. B., Nifantiev N. E. (2007) Synthesis of β-(1→6)-linked glucosamine oligosaccharides corresponding to fragments of the bacterial surface polysaccharide poly-N-acetylglucosamine. Carbohydr. Res. 342, 567–575 [DOI] [PubMed] [Google Scholar]
- 54. Kaplan J. B., Velliyagounder K., Ragunath C., Rohde H., Mack D., Knobloch J. K., Ramasubbu N. (2004) Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186, 8213–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Izano E. A., Sadovskaya I., Vinogradov E., Mulks M. H., Velliyagounder K., Ragunath C., Kher W. B., Ramasubbu N., Jabbouri S., Perry M. B., Kaplan J. B. (2007) Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb. Pathog. 43, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bobrov A. G., Kirillina O., Forman S., Mack D., Perry R. D. (2008) Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 10, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 57. Simon R., O'Connell M., Labes M., Pühler A. (1986) Plasmid vectors for the genetic analysis and manipulation of rhizobia and other Gram-negative bacteria. Methods Enzymol. 118, 640–659 [DOI] [PubMed] [Google Scholar]
- 58. Kearns D. B., Losick R. (2003) Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49, 581–590 [DOI] [PubMed] [Google Scholar]
- 59. Pozsgai E. R., Blair K. M., Kearns D. B. (2012) Modified mariner transposons for random inducible-expression insertions and transcriptional reporter fusion insertions in Bacillus subtilis. Appl. Environ. Microbiol. 78, 778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Konkol M. A., Blair K. M., Kearns D. B. (2013) Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195, 4085–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Muyrers J. P., Zhang Y., Testa G., Stewart A. F. (1999) Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27, 1555–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doublet B., Douard G., Targant H., Meunier D., Madec J. Y., Cloeckaert A. (2008) Antibiotic marker modifications of λRed and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 75, 359–361 [DOI] [PubMed] [Google Scholar]
- 63. Urban T. A., Griffith A., Torok A. M., Smolkin M. E., Burns J. L., Goldberg J. B. (2004) Contribution of Burkholderia cenocepacia flagella to infectivity and inflammation. Infect. Immun. 72, 5126–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Patrick J. E., Kearns D. B. (2008) MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70, 1166–1179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.