Background: Peripheral membrane proteins can interact with zwitterionic phospholipid headgroups or lipid tails.
Results: Incorporation of fluorinated Phe or Tyr differentiates side chain insertion from PC-cation-π complexes.
Conclusion: Enhanced binding of a fluorinated protein implies side chain insertion; weakened binding represents a cation-π complex.
Significance: Fluorinated Tyr or Phe incorporation can elucidate membrane binding modes.
Keywords: crystal structure; lipid-protein interaction; phosphatidylcholine; phospholipase C; site-directed mutagenesis; 3,5-difluorotyrosine; cation-π interactions; pentafluorophenylalanine
Abstract
Cation-π interactions, where protein aromatic residues supply π systems while a positive-charged portion of phospholipid head groups are the cations, have been suggested as important binding modes for peripheral membrane proteins. However, aromatic amino acids can also insert into membranes and hydrophobically interact with lipid tails. Heretofore there has been no facile way to differentiate these two types of interactions. We show that specific incorporation of fluorinated amino acids into proteins can experimentally distinguish cation-π interactions from membrane insertion of the aromatic side chains. Fluorinated aromatic amino acids destabilize the cation-π interactions by altering electrostatics of the aromatic ring, whereas their increased hydrophobicity enhances membrane insertion. Incorporation of pentafluorophenylalanine or difluorotyrosine into a Staphylococcus aureus phosphatidylinositol-specific phospholipase C variant engineered to contain a specific PC-binding site demonstrates the effectiveness of this methodology. Applying this methodology to the plethora of tyrosine residues in Bacillus thuringiensis phosphatidylinositol-specific phospholipase C definitively identifies those involved in cation-π interactions with phosphatidylcholine. This powerful method can easily be used to determine the roles of aromatic residues in other peripheral membrane proteins and in integral membrane proteins.
Introduction
Membrane binding by peripheral membrane proteins is often mediated by specific recognition of a variety of anionic phospholipids (e.g. phosphatidylserine (1), phosphoinositides (2, 3), phosphatidic acid (4)). Although phosphatidylcholine (PC),2 a major component of eukaryotic membranes, is usually considered a structural lipid, it can also serve as a specific anchor for peripheral membrane proteins. For example, the extracellular Bacillus thuringiensis virulence factor phosphatidylinositol-specific phospholipase C binds poorly to vesicles composed of anionic phospholipids at physiological salt concentrations but shows increasing avidity for vesicles containing PC (5). The targets of this enzyme are glycosylphosphatidylinositol-anchored proteins in the external leaflet of the plasma membrane, and affinity for PC would aid in directing the enzyme to its mammalian cell target. Mutagenesis (6), NMR (7), and molecular dynamics (MD) (6) simulations are consistent with choline cation-tyrosine π complexes providing a significant amount of the binding energy for B. thuringiensis PI-PLC. However, none of these experiments absolutely distinguishes nonspecific side-chain insertion from very specific formation of a cation-π complex with PC. The cation-π complex is formed above the hydrocarbon boundary in the hydrated part of the phospholipid, whereas membrane insertion places the side chain into the hydrocarbon portion. Substitution of alanine in place of tyrosine produces a large change in the side chain volume and, therefore, cannot identify the source of lost binding affinity. NMR experiments can localize long-lived PC binding sites but cannot define the binding mechanism. In contrast, such complexes can be identified in MD simulations, but many are transient in nature and may or may not be populated when the protein interacts with membranes. Clearly, a more direct experimental method is needed to demonstrate the existence of such interactions.
We have developed a method to separate the two membrane binding modes using site-specific incorporation of fluorinated amino acids, in this case pentafluorophenylalanine (F-F5) and 3,5-difluorotyrosine (Y-F2). The fluorinated amino acids are more hydrophobic (8) and are, therefore, likely to enhance binding by insertion. However, the altered electrostatics of the fluorinated aromatic ring should destabilize cation-π interactions (9).
The method is first tested with the extracellular Staphylococcus aureus PI-PLC, which has little affinity for PC and binds preferentially to membranes rich in anionic phospholipids (10, 11). However, the introduction of two tyrosines at Asn-254 and His-258 (N254Y/H258Y), to mimic the Bacillus enzyme, leads to PC binding specificity, presumably due to the cation-π complexes observed in a crystal structure of N254Y/H258Y with a dibutyroyl-PC bound (12). In contrast, a phenylalanine (Phe-249) in a surface loop around the S. aureus PI-PLC active site is thought to contribute to binding by inserting into the membrane (9, 10). A fluorinated Tyr at residue 258 weakens protein binding to PC-rich vesicles, confirming the importance of choline cation-Tyr π interactions at this site, whereas a fluorinated Phe at 249 enhances vesicle binding for all vesicle compositions indicating nonspecific hydrophobic interactions with lipid tails. With this confirmation that the methodology works as predicted, the many surface Tyr of B. thuringiensis PI-PLC were replaced with 3,5-difluotoryrosine to determine which Tyr residues are involved in cation-π interactions with PC. This method for determining how aromatic residues interact with phospholipids is readily applicable to other peripheral membrane proteins.
EXPERMENTAL PROCEDURES
Chemicals
F-F5 was prepared by one-step deprotection of the commercially available t-butoxycarbonyl-l-pentafluorophenylalanine. Y-F2 was synthesized following established methods (13).
Preparation and Characterization of Fluorinated PI-PLC Enzymes
To site-specifically incorporate these two unnatural amino acids into S. aureus PI-PLC, pEVOL-pylT-PylRS and pEVOL-F2YRS (14) were used. Although broadly similar to the system to incorporate Y-F2, the full characterization of pEVOL-pylT-PylRS for F-F5 incorporation will be described in a separate report. These plasmids each contain a tRNA-tRNA synthetase pair evolved for the incorporation of F-F5 or Y-F2, respectively. An amber stop codon was substituted at specific sites in S. aureus PI-PLC gene for incorporation of the fluorinated amino acid at the desired position. To express fluorinated mutants F249F-F5, H258F-F5, and N254Y/H258Y-F2, the mutant plasmids and the appropriate pEVOL (15) plasmid were cotransformed into Escherichia coli BL21(DE3) (10). Proteins were purified by chromatography using a nickel-nitrilotriacetic acid affinity resin and then an anion exchange resin, as described previously for this protein and its variants (11, 12).
A previously described B. thuringiensis PI-PLC plasmid (16) was subcloned into the pET-23b vector to create a C-terminal His tag. A similar expression and purification procedure was used for incorporation of Y-F2 into B. thuringiensis PI-PLC to generate Y86Y-F2, Y88Y-F2, Y204Y-F2, Y246Y-F2, Y247Y-F2, Y248Y-F2, and Y251Y-F2. However, the introduction of these fluorinated amino acids was done using N168C as the control. Fluorophores can be easily attached to this single Cys and have been shown to have minimal effect on enzyme activity or binding (5).
Mass Spectrometry
Samples were analyzed using instrumentation at the University of Massachusetts Mass Spectrometry Center. Samples were dissolved initially in water and then diluted to 1–10 μm in acetonitrile/water/acetic acid (50:47:3 v/v/v). Intact protein molecular weights were determined with a MicrOTOF II ESI-TOF mass spectrometer (Bruker Daltonics, Inc., Billerica MA) using electrospray ionization in positive mode. MS/MS data were obtained using a SolariX 7T FT-ICR (Bruker Daltonics) using collision-induced dissociation in the storage quadrupole (∼5 V collision energy). The most intense charge state ion [M+43H]43+ was selected for isolation. Although complete top-down fragmentation data were not obtained, representative fragments confirmed that fluorinated amino acids were correctly incorporated at the amber codon sites. Interestingly, the fluorinated analogs tended to fragment with a significantly lower (2–3 V) collision energy resulting in a quite different fragmentation pattern from the non-fluorinated version. This phenomenon will be the subject of future investigation.
MS analysis of tryptic peptides was required to identify the position of the Y-F2 in S. aureus N254Y/H258Y-F2. Tryptic peptides were also used to confirm the position of the fluorinated amino acid in H258F-F5.
Enzyme Activity Assays
Enzymatic activities for all the S. aureus PI-PLC variants, determined toward 4 mm PI in small unilamellar vesicles (SUVs) with various amounts of PC, were measured by 31P NMR spectroscopy as described previously (11, 12). The total phospholipid concentration of vesicles varied from 4 mm (mole fraction PC, XPC = 0) to 20 mm (XPC = 0.8). The assay buffer was 50 mm MES, pH 6.5.
Vesicle Binding Assays
Fluorescence correlation spectroscopy (FCS) was used to measure binding of the control proteins and their fluorinated counterparts to PG/PC SUVs. The S. aureus proteins were labeled on the N-terminal amino groups with Alexa Fluor 488 succinimidyl ester (11, 12). Vesicle binding of these proteins (3.5 nm) to SUVs composed of PG (the substrate analog) and PC, prepared by sonication, was measured at pH 6.5 (in 50 mm MES containing 1 mg/ml BSA), the optimum pH for enzyme activity (11). For FCS experiments with the B. thuringiensis PI-PLC, all Y-Y2 mutants also contained N168C, which was covalently labeled with Alexa Fluor 488 maleimide (5). B. thuringiensis PI-PLC proteins at 10 nm were in phosphate-buffered saline, pH 7.4, with 1 mg/ml BSA for titrations with the PG/PC SUVs.
The FCS measurements were carried out at 22 °C using a home-built confocal setup based on an IX-70 inverted microscope. Details of the experimental set-up and data analysis have been previously described (6, 17). Proteins were titrated with SUVs, and the fraction of protein bound to vesicles was determined from two species fits to the autocorrelations (obtained in cross-correlation mode), G(τ) = Apgp(τ) + Avgv(τ), where p and v denote free protein and SUVs that are fluorescent due to PI-PLC binding, respectively, and A is the amplitude of species p and v (18–20). The dependence of the fraction protein bound on total phospholipid concentration was used to extract an apparent Kd.
Crystallization
Purified PI-PLC variants were concentrated to 20 mg/ml and then combined with myo-inositol (100 mm). The protein solution was then diluted to 10 mg/ml using deionized water and incubated on ice for a minimum of 2 h before setting up crystallization trays. The PI-PLC samples were crystallized at 4 °C by vapor diffusion using hanging drops of 3 μl at a final protein concentration of 10 mg/ml. For both mutants, the protein crystallized in 150 mm ammonium acetate, 100 mm sodium acetate, pH 4.6, with 10 mm magnesium nitrate and 20–24% PEG 4000.
Crystallography
Data were collected at 100 K using a Rigaku MicroMax-07 HF high intensity microfocus rotating copper anode x-ray generator coupled with Osmic VariMax Optics and a R-Axis IV++ image plate area detector. Data were indexed and reduced using d*TREK (21). Both structures were solved by molecular replacement in PHENIX (22) using PHASER (23) in PHENIX with the S. aureus structure 3V18 as a model. All models were refined with Phenix.refine, with manual model building in COOT (24). Maps were generated in PHENIX. Structural comparisons were made using SSM superposition (25) in COOT and alignment in PyMOL (26).
Results and Discussion
Incorporation of F-F5 and Y-F2 into S. aureus PI-PLC
ESI-TOF MS analysis was used to show that the fluorinated amino acids were properly incorporated into S. aureus PI-PLC F249F-F5 and H258F-F5. F249F-F5 gave an observed average mass of 36,807 Da, and H258F-F5 gave an average mass of 36,819 Da, whereas wild type had an observed mass of 36,719 Da (Fig. 1). These results are in agreement with the calculated mass. MS/MS analysis also confirmed the fluorinated modification at these desired sites (Fig. 1B). Thus, the ESI-MS and MS/MS together confirmed successful incorporation of F-F5 at sites 249 and 258 using the cloning and expression strategies described. The N254Y/H258Y-F2 variant showed a high avidity for salt, making it difficult to desalt the sample sufficiently for MS analysis, precluding successful analysis by ESI-TOF. Instead, the protein containing Y-F2 was trypsinized, and the mixture was examined for fluorinated peptides to determine the site of fluoro-amino acid incorporation. Fig. 1C shows the ions corresponding to fragments of the tryptic peptide containing Y-F2 at residue 258 in S. aureus PI-PLC. The MS/MS fragments detected for relevant peptides in both N254Y/H258Y-F2 and H258F-F5 are provided in Tables 1 and 2, confirming the successful incorporation of the fluorinated amino acids.
FIGURE 1.
MS confirmation of fluorinated amino acid incorporation in S. aureus PI-PLC. A, intact MS analysis of wild type, F249F-F5, and H258F-F5 S. aureus PI-PLC variants. B, MS/MS analysis of S. aureus PI-PLC, F249F-F5, and H258F-F5. C, MS/MS identification of the tryptic peptide fragments containing the fluorinated tyrosine in N254Y/H258Y-F2. Red and blue indicate the b (N-terminal) and y (C-terminal) ions, respectively. The peptide amino acid sequence is shown in the upper right with the mutations N254Y and H258Y-F2 indicated in red.
TABLE 1.
MS/MS fragments of the tryptic peptide containing the N254Y/H258Y-F2 mutation with intensities >900
The full peptide sequence is 226AKTDS NKDNVYVNFL SVASGGSAFN STYYYAS Y-F 2IN PEIAK265 and the MS/MS data, including all identified peptides, are shown in Fig. 1C.
| Fragment sequence | Predicted mass | Measured mass | |
|---|---|---|---|
| Da | Da | ||
| C-terminal fragment | |||
| y6+ | 260N PEIAK265 | 671.37 | 671.60 |
| y7+ | 259IN PEIAK265 | 784.46 | 784.60 |
| y8+ | 258 Y-F2 IN PEIAK265 | 983.50 | 983.60 |
| y9+ | 257SY-F2IN PEIAK265 | 1070.53 | 1070.73 |
| y10+ | 256AS Y-F2IN PEIAK265 | 1141.57 | 1141.87 |
| y11+ | 255YASY-F2IN PEIAK265 | 1304.63 | 1304.87 |
| y12+ | 254YYASY-F2IN PEIAK265 | 1467.70 | 1467.87 |
| y213+ | 245GGSAFN STYYYASY-F2IN PEIAK265 | 784.69 | 784.60 |
| y242+ | 242VASGGSAFN STYYYASY-F2IN PEIAK265 | 1305.10 | 1304.87 |
| y273+ | 239FLSVASGGSAFNSTYYYASY-F2IN PEIAK265 | 986.13 | 985.80 |
| N-terminal fragment | |||
| b182+ | 226AKTDS NKDNVYVNFL SVA242 | 983.99 | 983.93 |
| b282+ | 226AKTDSNKDNVYVNFLSVASGGSAFN STY253 | 1469.69 | 1469.80 |
| b313+ | 226AKTDS NKDNVYVNFL SVASGGSAFN STYYYA256 | 1112.52 | 1112.20 |
| b323+ | 226AKTDS NKDNVYVNFL SVASGGSAFN STYYYAS257 | 1141.53 | 1141.87 |
| b333+ | 226AKTDS NKDNVYVNFL SVASGGSAFN STYYYAS Y-F2258 | 1207.88 | 1208.27 |
| b383+ | 226AKTDS NKDNVYVNFL SVASGGSAFNSTYY YAS Y-F2IN PEI263 | 1396.64 | 1396.87 |
TABLE 2.
MS/MS fragments of the tryptic peptide containing the H258F-F5 mutation
The full peptide sequence is 226AKTDS NKDNVYVNFL SVASGGSAFN STYNYASF-F5IN PEIAK265, and the MS/MS data are shown in Fig. 1.
| C-terminal fragment | Fragment sequence | Predicted mass | Measured mass |
|---|---|---|---|
| Da | Da | ||
| y5+ | 261PEIAK265 | 557.33 | 557.33 |
| y6+ | 260N PEIAK265 | 671.37 | 671.40 |
| y7+ | 259IN PEIAK265 | 784.46 | 784.47 |
| y8+ | 258F-F5IN PEIAK265 | 1021.48 | 1021.47 |
| y9+ | 257SF-F5IN PEIAK265 | 1108.51 | 1108.60 |
| y10+ | 256ASF-F5IN PEIAK265 | 1179.55 | 1179.60 |
| y11+ | 255YASF-F5IN PEIAK265 | 1342.61 | 1342.60 |
| y12+ | 254NYASF-F5IN PEIAK265 | 1456.65 | 1456.60 |
| y13+ | 253YNYASF-F5IN PEIAK265 | 1619.72 | 1619.73 |
| y15+ | 251STYNYASF-F5IN PEIAK265 | 1807.80 | 1807.40 |
| y16+ | 250N STYNYASF-F5IN PEIAK265 | 1921.84 | 1921.40 |
Crystal Structure of S. aureus-fluorinated Proteins
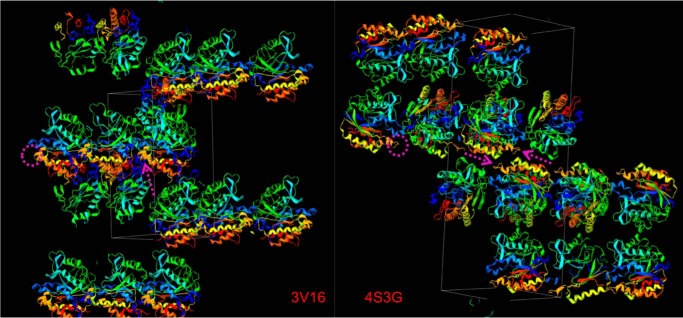
Replacement of Phe with the fluorinated analog F-F5 should destabilize cation-π interactions. To test this we took advantage of the observation that an intramolecular cation-π interaction between Phe-249 and His-258 is seen in wild type S. aureus PI-PLC crystallized under acidic conditions (10). The structure of F249F-F5 was solved and compared with the structure for wild type obtained under acidic pH conditions (structure statistics are presented in Table 3). Electron density is contoured to 1σ for the aromatic residues near the mutation in Fig. 2A to show the quality of the data. The cation-π interaction between Phe-249 and the cationic His-258 has a planar stacking distance of 3.5–4.3 Å between the aromatic rings (10). With the F-F5 introduced at position 249, the cation-π complex can no longer form (Fig. 2C), and overall F249F-F5 adopts a conformation similar to found in wild type crystallized at basic pH, where His-258 is no longer protonated (10). However, the mobile loop preceding helix G is in a slightly different orientation. The exact disposition of this loop, variable in many of the S. aureus mutant crystal structures (10–12), is near protein contacts in the F249F-F5 crystal. This specific orientation of the loop for F249F-F5 likely aids in stabilizing the more hydrophobic F-F5 side chain in the crystal (Fig. 3).
TABLE 3.
Full refinement and model statistics for F249F-F5 and H258F-F5structures
r.m.s.d., root mean square deviation.
| F249F-F5 (PDB 4S3G) | H258F-F5 (PDB 4RV3) | |
|---|---|---|
| Diffraction data | ||
| Resolution range (Å) | 42.26-2.5 | 49.87-2.0 |
| No. of reflections | 12,823 | 22,307 |
| No. of reflections in free set | 623 | 1139 |
| Space group | P43212 | P43212 |
| Unit cell | ||
| a (Å) | 60.05 | 59.85 |
| b (Å) | 60.05 | 59.85 |
| c (Å) | 191.33 | 180.41 |
| Completeness | 99.7 | 96.6 |
| Rmerge | 0.053 | 0.056 |
| Refinement | ||
| Rcrysta | 0.212 | 0.180 |
| Rfreeb | 0.291 | 0.238 |
| No. of residues | 301 | 302 |
| No. of non-hydrogen protein atoms | 2,408 | 2,433 |
| No. of H2O molecules | 54 | 161 |
| No. of non-hydrogen inositol atoms | 12 | 12 |
| No. of ions | 1 | 1 |
| r.m.s.d. bonds (Å) | 0.010 | 0.007 |
| r.m.s.d. bonds angles (degree) | 1.337 | 1.020 |
| Average B-factor (Å2) | 49.0 | 29.0 |
a Rwork = {|‖Fo| − | Fc‖)/| Fo|}, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
b Rfree is calculated from a set of 5% randomly selected reflections that were excluded from the refinement.
FIGURE 2.
Representative electron densities of part of the rim loop in the mutant F249F-F5 (A) and part of the helix G in the mutant H258F-F5 (B) structures are shown in gray and contoured at 1σ. Overlay of the pH 4.6 structures for wild type (yellow, PDB code 3V16) and F249F-F5 (red, PDB code 4S3G) (C) and H258Y (magenta, PDB code 3V1H) and H258F-F5 (cyan, PDB code 4RV3) S. aureus PI-PLC (D). Key residues are labeled. myo-inositol is depicted in space-filling representation.
FIGURE 3.
Crystal packing for wild type (PDB code 3V16) and the F249F-F5 variant (PDB code 4S3G) PI-PLC enzymes. The rim loop region containing residue Phe or F-F5 at position 249 is circled in magenta and indicated with arrows in the respective unit cells. Notice that the hydrophobic residue in the rim loop affects monomer packing in the crystal.
H258F-F5 was also crystallized under acidic conditions; Fig. 2B provides an example of the electron density. Because His-258 has been replaced, the intramolecular cation-π interaction between residues 258 and Phe-249 cannot form. Therefore, in this variant the rim loop containing Phe-249 adopts a conformation close to that seen for H258Y (10) (Fig. 2D) that also cannot form the intramolecular cation-π complex. More importantly, the Tyr at residue 258 is nearly superimposable with the F-F5, indicating that introduction of the fluorinated amino acid has virtually no effect on helix G. These structures show that the electrostatics of the fluorinated Phe preclude formation of cation-π complexes and have little effect on the well defined secondary structure in which they may reside.
S. aureus Fluorinated PI-PLC Binding to Vesicles
Binding of S. aureus PI-PLC variants to PG/PC SUVs was monitored by FCS at pH 6.5 where S. aureus PI-PLC shows maximal activity. Wild type protein has weak affinity for PC, and the apparent Kd increases substantially as XPC increases (Fig. 4A, open circles). Replacing Phe-249 with F-F5 enhances binding for all vesicle compositions (Fig. 4A, filled circles). This enhancement is emphasized by plotting the ratio of the apparent Kd for F249F-F5 relative to that of wild type (Fig. 4D). The increased affinity of F249F-F5 for all vesicle compositions correlates with partitioning of the more hydrophobic F-F5 into the bilayer.
FIGURE 4.
Effects of the fluorinated aromatic amino acids on binding to PG/PC SUVs. Apparent dissociation constants for S. aureus PI-PLC variants containing fluorinated aromatic amino acids as a function of mole percent PC (XPC) for F249F-F5 (●) compared with wild type (○) (A), H258F-F5 (▴) compared with H258F (▵) (B), and N254Y/H258Y-F2 (■) compared with N254Y/H258Y (□) (C). D, the ratio of the Kd for the fluorinated protein to the Kd for the relevant control, shown in the previous panels, as a function of XPC; symbols are the same as in A–C.
The affinity of H258F for the SUVs is comparable with that of the wild type protein (within a factor of two across the range of phospholipid compositions). The binding of H258F-F5 compared with H258Y (Fig. 4B) is similar for XPC ≤ 0.5. However, at high XPC, H258F-F5 binding is 5-fold weaker than H258F. At XPC = 0.9, the affinity of H258F-F5 for SUVs was too low to reliably measure by FCS. Thus, although wild type has low affinity for PC-rich membranes, its affinity is further reduced by F-F5 incorporation in helix G. Because structurally H258F-F5 is very similar to H258Y, the loss of affinity at high XPC suggests that there is some tendency for a Phe residue introduced at position 258 in the membrane binding interface to interact with choline headgroups. Formation of such a weak complex would require significant PC in the bilayer.
N254Y/H258Y, as reported previously, shows a dramatic increase in affinity for PC-rich SUVs indicative of the introduction of a PC site with these mutations (12). As the PC content increases above XPC = 0.5, tight binding of N254Y/H258Y is observed (Fig. 4C, open circles). The shape of the curve is consistent with a PC cation/Tyr-π interaction contributing to binding in the PC-rich region and electrostatic interactions dominating binding to more anionic PG-rich vesicles. Rather than introduce F-F5 at this position, we chose to incorporate Y-F2. However, substituting Y-F2 for a surface tyrosine can also alter the electrostatic distribution because the Y-F2 hydroxyl pKa is shifted to ∼7.5 (27). S. aureus PI-PLC activity is optimal at pH 6.5, and the binding was measured at that pH. Therefore, deprotonation of the hydroxyl should not interfere significantly with the binding. For the most anionic SUVs, where introduction of a negative charge on the protein should have the strongest effect on binding (XPC = 0 and 0.2), the Kd for N254Y/H258Y-F2 is the same as that for N254Y/H258Y (Fig. 4C).
As SUVs become enriched in PC, N254Y/H258Y-F2 loses much of the binding affinity compared with N254Y/H258Y. This is easily seen from plotting the ratio of Kd for N254Y/H258Y-F2 to that of its control N254Y/H258Y (Fig. 4D). From this ratio we can estimate the free energy lost when substituting Y-F2 for Tyr. At XPC = 0.65, 0.8, and 0.9, ΔΔG values are +6.1, +5.7, and +7.6 kJ/mol, respectively. This very dramatic loss of affinity is consistent with the role of Tyr-258 in cation-π complexes with choline headgroups (12).
Interestingly, for pure PC SUVs, ΔΔG is +2.7 kJ/mol when the Tyr is fluorinated. The lower ΔΔG and the fact that N254Y/H258Y-F2 binding could still be measured toward pure PC vesicles might suggest that the cation-π interactions have been weakened. Perhaps this is not surprising as only two fluorine atoms were incorporated into the ring. However, another interpretation is that the large ΔΔG values for N254Y/H258Y-F2 binding to SUVs with some PG indicate that the cation-π complex requires PG binding, likely in the active site as it is a competitive inhibitor, for optimal formation.
Enzyme Activity of Fluorinated S. aureus PI-PLC
With the exception of the pure PI SUVs, enzymatic activities for all the S. aureus PI-PLC variants were measured under conditions where the total amount of phospholipid is significantly above the apparent Kd measured by FCS for PG/PC SUVs. Thus, nearly all the proteins should partition onto the vesicles. For all of the variants specific activities increase with increasing PC content (Fig. 5). For wild type this is not due to a specific PC interaction but rather because anionic lipids antagonize dimerization, and S. aureus PI-PLC dimers appear to have increased activity (11). For this PI-PLC, zwitterionic phospholipids reduce the surface concentration of PI, and this in turn prevents anionic phospholipids from binding to a cationic site adjacent to the active site that antagonizes dimerization (11). Therefore, all variants exhibited higher activities with increasing XPC.
FIGURE 5.
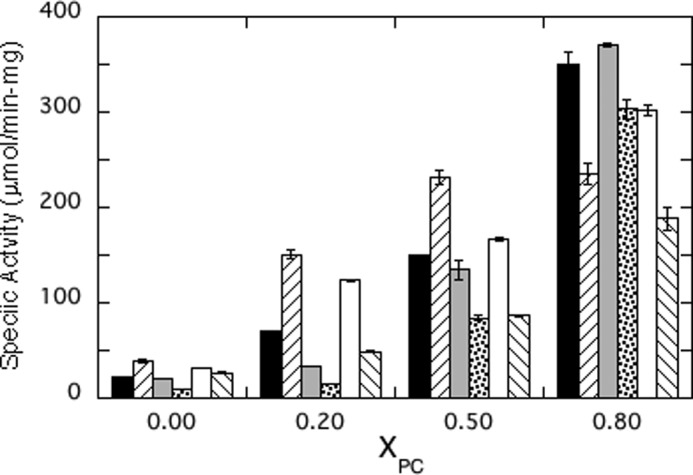
Specific activities of S. aureus PI-PLC mutants toward PI/PC SUVs as a function of mole fraction PC (XPC): wild type (■), F249F-F5 (/), H258F (light gray bars), H258F-F5 (·), N254Y/H258Y (□), and N254Y/H258Y-F2 (\). PI was fixed at 4 mm with increasing amounts of PC. Protein concentrations were adjusted to 0.3–4 μg/ml to ensure <20% cleavage of the PI (4 mm).
However, superimposed on this behavior are some interesting trends. F249F-F5 had higher activity than wild type toward PI in vesicles with XPC ≤ 0.5 (Fig. 5). This increased activity for F249F-F5 correlates with increasing the hydrophobicity of a side chain that partitions into the membrane, perhaps enhancing processive catalysis on these vesicles. It should be noted that the specific activity decreased at XPC = 0.8. Although the exact cause of this is not known, it parallels what is observed for B. thuringiensis PI-PLC, where the specific activity drops at high mole fraction of PC where the vesicle binding is tight (5). The affinity of the substrate for the active site depends on the mole fraction of substrate in the interface, its ability to diffuse to the bound enzyme active site, and also the ability of the substrate to dissociate from one vesicle to find another (on the time scale of these assays, the enzyme has to sample many vesicles for ∼10% hydrolysis). The tighter binding fluorinated Phe could perturb any of these steps.
With one exception (H258F-F5, which has very weak binding at XPC = 0.8), the total phospholipid concentration used at each XPC was above the apparent Kd for binding to PG/PC SUVs. The fraction of fluorinated protein bound to SUVs can be compared with that of its control protein at each XPC. That ratio is 1.00 ± 0.10. In contrast, the specific activities for these paired enzymes toward vesicles containing PC show larger changes (excluding F249F-F5 and H258F-F5 at XPC = 0.8). The average ratio of smaller to larger specific activity value is 0.55 ± 0.13. The bigger change for enzyme activities suggests that under conditions where the bulk of the protein is partitioned onto the vesicles, PC cation-protein π interactions in H258F and N254Y/H258Y must also enhance either substrate access or hydrophobic product exit from the active site of the anchored protein. Similarly, increasing the hydrophobic interaction with F249F-F5 increases the specific activity, and this must reflect a step once the protein is partitioned onto the vesicles.
Y-F2 in B. thuringiensis PI-PLC Identifies PC Cation-Tyr π Interactions
Given the success in using fluorinated aromatic amino acid incorporation to distinguish side chain partitioning into a bilayer from PC cation-π interactions, we extended this methodology to B. thuringiensis PI-PLC. This PI-PLC has a large number of Tyr residues in rim loops as well as in helix G that could interact with the bilayer (Fig. 6). Recent MD simulations have suggested transient cation-π complexes exist when the protein is bound to a PC bilayer (6). Experimental tests to see which Tyr residues were critical for membrane binding substituted the Tyr with Ala. This substitution impaired binding if the mutated Tyr partitioned into the bilayer. However, the Ala mutations only indicate which residues are important for membrane binding, not the mode of binding.
FIGURE 6.
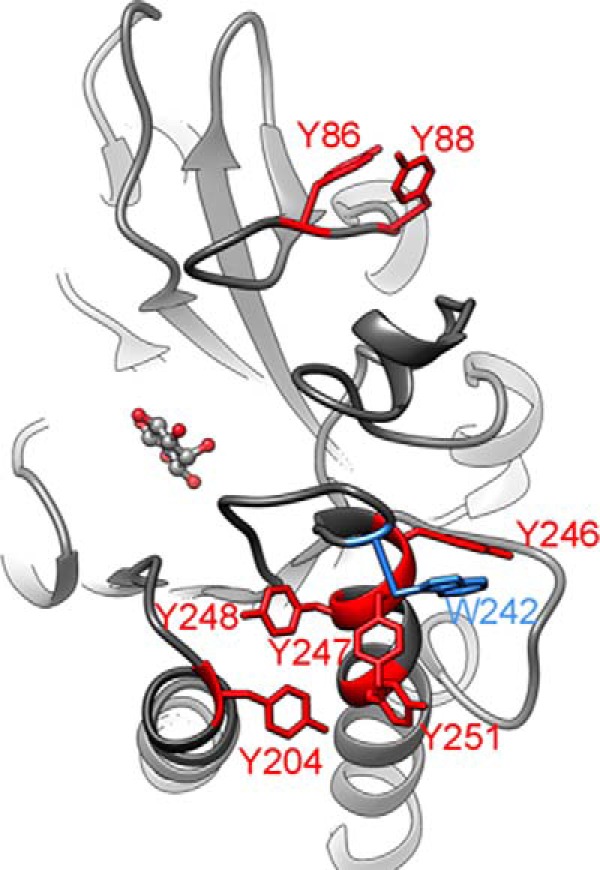
View of B. cereus PI-PLC (PDB 1ptg (30) showing the aromatic residues in the area that interacts with the membrane. Blue highlights the loop with Trp-242, which is critical for membrane insertion. Red highlights all the tyrosine residues in this region that may be involved in PC cation-aromatic π interactions. An inositol molecule is shown occupying the active site. The Bacillus cereus and B. thuringiensis PI-PLC sequences are 97% identical, and the membrane binding interface is totally conserved. There is no existing structure for wild type B. thuringiensis PI-PLC.
To investigate how B. thuringiensis PI-PLC Tyr residues interact with phospholipids, we constructed seven Y-F2 variants (Y86Y-F2, Y88Y-F2, Y204Y-F2, Y246Y-F2, Y247Y-F2, Y248Y-F2, and Y251Y-F2) and examined their binding to PG and PC SUVs at pH 7.4, the optimal pH for the activity of this enzyme. At this pH the hydroxyl group of each surface Y-F2 will be ∼50% ionized introducing a partial negative charge in the vicinity of the bilayer. That is likely to affect the affinity of the proteins for PG surfaces (quantified by the Kd for pure PG SUVs) but have only a modest effect on binding to a zwitterionic bilayer (pure PC SUVs). For the latter, a reduction in Kd would indicate nonspecific partitioning of that Tyr into the bilayer, i.e. interactions with lipid tails, whereas an increase in Kd would indicate participation in PC cation-π binding. A summary of the apparent Kd values obtained by FCS are shown in Table 4.
TABLE 4.
Apparent Kd value for B. thuringiensis PI-PLC Y→Y-F2 mutants and extrapolated ΔΔG for loss of binding
| PI-PLC | PG SUVs Kd | ΔΔG (PG)a | PC SUVs Kd | ΔΔG (PC)a | % Occupancy cation-πb | PC SUVS, Y→Ab Kd |
|---|---|---|---|---|---|---|
| mm | mm | mm | ||||
| WT | 5.60 ± 0.04 | 0.030 ± 0.006 | ||||
| Y86Y-F2 | 41.2 ± 1.2 | 4.9 | 0.62 ± 0.03 | 7.4 | 22 | 0.072 ± 0.005 |
| Y88Y-F2 | 30.3 ± 1.5 | 3.6 | 0.78 ± 0.07 | 8.0 | 96 | 1.2 ± 0.3 |
| Y204Y-F2 | 36.9 ± 5.5 | 4.7 | 1.04 ± 0.06 | 8.7 | 26 | 0.87 ± 0.58 |
| Y246Y-F2 | 34.5 ± 10.1 | 4.5 | 1.09 ± 0.02 | 8.8 | 77 | 1.2 ± 0.3 |
| Y247Y-F2 | 28.2 ± 1.4 | 4.0 | 0.053 ± 0.004 | 1.4 | 8.9 | 0.082 ± 0.007 |
| Y248Y-F2 | 40.2 ± 9.4 | 4.9 | 2.31 ± 0.07 | 10.6 | 1.9 | 2.5 ± 0.5 |
| Y251Y-F2 | 28.7 ± 8.6 | 4.0 | 0.69 ± 0.46 | 7.7 | 36 | 0.8 ± 0.4 |
a Extrapolated from the ratio of Kd(mutant)/Kd(WT) at 22 °C.
b Y→A is the mutant PI-PLC with alanine in the place of the Tyr at that position. These binding data are from Grauffel et al. (6).
Successful incorporation of Y-F2 into all variants was confirmed by fluorine NMR (data not shown). All Y-F2 variants show impaired binding to PG SUVs with the apparent Kd increasing from 5.6 mm for wild type to 30–40 mm for the fluorinated proteins. The average ΔΔG for these Y-F2 variants binding to PG SUVs, +4.4 ± 0.5 kJ/mol, represents the electrostatic cost of placing a fluorinated ring (and partial negative charge) at an anionic interface.
In contrast to the uniform behavior of these variants with a PG vesicle, the apparent Kd values for pure PC SUVs covered a much wider range. The Y247Y-F2 Kd was <2-fold higher than that of wild type B. thuringiensis PI-PLC, indicating as suggested from previous experiments and MD simulations (6) that Tyr247 does not contribute significantly to cation-π interactions. The largest increase in the apparent Kd for PC containing vesicles was observed for Y248Y-F2 (from 0.03 to 2.3 mm). However, the MD simulations strongly suggest this is not because of cation-π interactions but rather arises from intramolecular interactions. Tyr-248 in the rim loop helps properly position Trp-242 (6), and this Trp is very important for membrane binding. All mutations to Tyr-248 appear to disrupt this interaction.
Y204Y-F2 and Y246Y-F2 show the next largest increases in Kd followed by Y86Y-F2, Y88Y-F2, and Y251Y-F2. In the MD simulations, all of these Tyr residues contribute to cation-π interactions but with different occupancies. The apparent Kd values for the Y-F2 variants also correlate well with those for the alanine variants (Table 4). For both there is a qualitative correlation of the loss of binding affinity with cation-π occupancy observed in the MD simulations.
The important observation is that none of the Kd values for Y-F2 variants binding to PC SUVs decreased. Therefore, for this PI-PLC, the surface Tyr residues do not partition into the membrane but rather engage in cation-π interactions to different degrees. For PC-rich bilayers, cation-π interactions are ideal for transiently anchoring these proteins long enough to search an initial area for their glycosylphosphatidylinositol-anchored substrates to cleave ∼10 substrates followed by hopping to a new region on the PC-rich outer plasma membrane (28).
This approach using fluorinated amino acids to distinguish the type of membrane interaction of a particular aromatic side chain should have broad applicability to many other systems. Integral membrane proteins have aromatic residues at the membrane surface, and these may also engage in cation-π interactions with the membrane. For example, PC modulation of an aspartate transporter from Pyrococcus horikoshii is postulated to occur through a cation-π interaction with a surface Tyr and the lipid headgroup. In that case, the interaction influences conformational flexibility of the oligomerization domain, which in turn modulates the rate of transport (29). Use of Y-F2 in such a system could provide direct evidence for such a cation-π interaction.
Summary
The incorporation of fluorinated amino acids into these two amphitropic proteins has allowed us to assess directly how specific aromatic amino acids interact with membranes. This approach should be useful in providing a direct test of predictions from MD simulations and in designing membrane binding interfaces based on PC cation-protein π interactions. Thus, this methodology is broadly applicable to proteins that interact with membranes and can provide new insight into the roles of specific aromatic amino acids in membrane-protein interactions.
Author Contributions
T. H. created and characterized all S. aureus and B. thuringiensis variants. S. E. carried out the MS experiments. Y.-J. L. and W. L. generated the pEVOL-pylT-PylRS system for incorporation of F-F5. J. W. generated the pEVOL-F2YRS system for incorporation of Y-F2 into proteins. J. G. helped with the synthesis of Y-F2 and interpretation of the binding data. T. H. and M. R. planned experiments and wrote the manuscript along with A. G., who also supervised FCS experiments. All authors reviewed the results and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 60418 (to M. F. R.).
The atomic coordinates and structure factors (codes 4S3G and 4RV3) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- F-F5
- pentafluorophenylalanine
- Y-F2
- 3,5-difluorotyrosine
- PI-PLC
- phosphatidylinositol (PI)-specific phospholipase C
- SUV
- small unilamellar vesicle
- FCS
- fluorescence correlation spectroscopy
- ESI
- electrospray ionization
- XPC
- mole fraction PC.
References
- 1. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 2. Kutateladze T. G. (2010) Translation of the phosphoinositide code by PI effectors. Nat. Chem. Biol. 6, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krick R., Busse R. A., Scacioc A., Stephan M., Janshoff A., Thumm M., Kühnel K. (2012) Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family. Proc. Natl. Acad. Sci. U.S.A. 109, E2042–E2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karathanassis D., Stahelin R. V., Bravo J., Perisic O., Pacold C. M., Cho W., Williams R. L. (2002) Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 21, 5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pu M., Roberts M. F., Gershenson A. (2009) Fluorescence correlation spectroscopy of phosphatidylinositol-specific phospholipase C monitors the interplay of substrate and activator lipid binding. Biochemistry 48, 6835–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grauffel C., Yang B., He T., Roberts M. F., Gershenson A., Reuter N. (2013) Cation-π interactions as lipid-specific anchors for phosphatidylinositol-specific phospholipase C. J. Am. Chem. Soc. 135, 5740–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pu M., Orr A., Redfield A. G., Roberts M. F. (2010) Defining specific lipid binding sites for a peripheral membrane protein in situ using subtesla field-cycling NMR. J. Biol. Chem. 285, 26916–26922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pace C. J., Gao J. (2013) Exploring and exploiting polar-π interactions with fluorinated aromatic amino acids. Acc. Chem. Res. 46, 907–915 [DOI] [PubMed] [Google Scholar]
- 9. Dougherty D. A. (2013) The cation-π interaction. Acc. Chem. Res. 46, 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldstein R., Cheng J., Stec B., Roberts M. F. (2012) Structure of the S. aureus PI-specific phospholipase C reveals modulation of active site access by a titratable π-cation latched loop. Biochemistry 51, 2579–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng J., Goldstein R., Stec B., Gershenson A., Roberts M. F. (2012) Competition between anion binding and dimerization modulates Staphylococcus aureus phosphatidylinositol-specific phospholipase C enzymatic activity. J. Biol. Chem. 287, 40317–40327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng J., Goldstein R., Gershenson A., Stec B., Roberts M. F. (2013) The cation-π box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 288, 14863–14873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seyedsayamdost M. R., Yee C. S., Stubbe J. (2007) Site-specific incorporation of fluorotyrosines into the R2 subunit of E. coli ribonucleotide reductase by expressed protein ligation. Nat. Protoc. 2, 1225–1235 [DOI] [PubMed] [Google Scholar]
- 14. Li F., Shi P., Li J., Yang F., Wang T., Zhang W., Gao F., Ding W., Li D., Li J., Xiong Y., Sun J., Gong W., Tian C., Wang J. (2013) A genetically encoded 19F NMR probe for tyrosine phosphorylation. Angew. Chem. Int. Ed. Engl. 52, 3958–3962 [DOI] [PubMed] [Google Scholar]
- 15. Young T. S., Ahmad I., Yin J. A., Schultz P. G. (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 395, 361–374 [DOI] [PubMed] [Google Scholar]
- 16. Feng J., Wehbi H., Roberts M. F. (2002) Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 277, 19867–19875 [DOI] [PubMed] [Google Scholar]
- 17. Cheng J., Karri S., Grauffel C., Wang F., Reuter N., Roberts M. F., Wintrode P. L., Gershenson A. (2013) Does changing the predicted dynamics of a phospholipase C alter activity and membrane binding? Biophys. J. 104, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Middleton E. R., Rhoades E. (2010) Effects of curvature and composition on α-synuclein binding to lipid vesicles. Biophys. J. 99, 2279–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rusu L., Gambhir A., McLaughlin S., Rädler J. (2004) Fluorescence correlation spectroscopy studies of Peptide and protein binding to phospholipid vesicles. Biophys. J. 87, 1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson N. L. (1991) In Topics in Fluorescence Spectroscopy (Lakowicz J. R., ed.) p. 337, Plenum Press, New York [Google Scholar]
- 21. Pflugrath J. W. (1999) The finer things in x-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 26. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3 Schrodinger, LLC, New York [Google Scholar]
- 27. Ravichandran K. R., Liang L., Stubbe J., Tommos C. (2013) Formal reduction potential of 3,5-difluorotyrosine in a structured protein: insight into multistep radical transfer. Biochemistry 52, 8907–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang B., Pu M., Khan H. M., Friedman L., Reuter N., Roberts M. F., Gershenson A. (2015) quantifying transient interactions between Bacillus phosphatidylinositol-specific phospholipase C and phosphatdoylcholine0rich vesicles. J. Am. Chem. Soc. 137, 14–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McIlwain B. C., Vandenberg R. J., Ryan R. M. (2015) Transport rates of a glutamate transporter homologue are influenced by the lipid bilayer. J. Biol. Chem. 290, 9780–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heinz D. W., Ryan M., Bullock T. L., Griffith O. H. (1995) Crystal structure of the phosphatidylinositol-specific phospho-lipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 14, 3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]