Background: How BAF250a regulates nucleosome configuration in ES cells is not clear.
Results: BAF250a regulates nucleosome occupancy and H3K27me3 to control gene expression during ES cell differentiation.
Conclusion: BAF250a plays a key role in poised chromatin regulation.
Significance: Understanding the mechanisms of chromatin remodeling in poised chromatin regulation provides epigenetic insights into ES cell differentiation.
Keywords: chromatin remodeling, differentiation, embryonic stem cell, epigenetics, histone modification, BAF250a, nucleosome occupancy
Abstract
The unique chromatin signature of ES cells is fundamental to the pluripotency and differentiation of ES cells. One key feature is the poised chromatin state of master developmental genes that are transcriptionally repressed in ES cells but ready to be activated in response to differentiation signals. Poised chromatin in ES cells contains both H3 Lys-4 trimethylation (H3K4me3) and H3 Lys-27 trimethylation (H3K27me3) methylation, indicating activating and repressing potential. However, the contribution of non-covalent chromatin structure to the poised state is not well understood. To address whether remodeling of nucleosomes is important to the poised state, we characterized the function of BAF250a, a key regulatory subunit of the ES cell ATP-dependent Brahma-associated factor (BAF) chromatin remodeling complex (esBAF). Acute deletion of BAF250a disrupted the differentiation potential of ES cells by altering the expression timing of key developmental genes and pluripotent genes. Our genome-wide nucleosome and histone modification analyses indicated that the disruption of gene expression timing was largely due to changes of chromatin structures at poised genes, particularly those key developmental genes mediated by BAF250a. Specifically, BAF250a deletion caused a nucleosome occupancy increase at H3K4me3- and/or H3K27me3-associated promoters. Moreover, H3K27me3 levels and the number of bivalent promoter genes were reduced in BAF250a KO ES cells. We revealed that BAF250a ablation led to elevated Brg1 but reduced Suz12 recruitment at nucleosome occupancy-increased regions, indicating an unexpected and complicated role of BAF250a in regulating esBAF and Polycomb repressive complex (PRC) activities. Together, our studies identified that BAF250a mediates esBAF and PRC functions to establish the poised chromatin configuration in ES cells, which is essential for the proper differentiation of ES cells.
Introduction
Embryonic stem cells are capable of self-renewal and differentiation into all somatic and germ line cell types. A comprehensive network of transcription and epigenetic factors regulates proper ES cell function (1). Recent studies have shown that chromatin structure plays an essential role in ES cell regulation (2). One unique feature of the ES cell genome is the presence of an open chromatin structure compared with somatic cells (3), with higher chromatin accessibility, as characterized by higher levels of the active histone modification H3K4me3, lower occupancy of nucleosomes, and less heterochromatin (4). During ES cell differentiation, developmental genes and signals are rapidly activated to direct lineage specification. In ES cells, most key developmental transcription factors are in bivalent state which is characterized by the presence of both H3K4me3 and H3K27me3. During differentiation, these developmental genes quickly lose the repressive histone mark H3K27me3 and become activated (5). However, despite being a major component of chromatin, the role of nucleosome configuration in maintaining the poised chromatin structure and regulating rapid gene activation during differentiation is not clear.
Nucleosome configuration is regulated through ATP-dependent chromatin remodeling complexes, which use the energy of ATP hydrolysis to slide, remodel, displace, and replace the nucleosomes (6–8). In mammals, BAF2 complexes, are also called SWItch/Sucrose NonFermentable (SWI/SNF) complexes, are one of the major classes of ATP-dependent chromatin remodeling complexes. BAF complexes play important roles during various biological processes with different assemblies of the complexes (9, 10). In ES cells, the BAF complex is composed of unique subunit components termed esBAF (11). Disruption of esBAF components, such as BAF250a, BRG1, and BAF155, leads to ES cell pluripotency defects (12, 13). However, the chromatin remodeling activity of esBAF in regulating nucleosome dynamics in ES cells has not been explored.
In this study, we sought to address how esBAF complexes maintain specific chromatin architecture, in particular the nucleosomes, and attempted to connect the nucleosome and related chromatin structures to ES cell maintenance and rapid differentiation. To achieve this goal, we generated tamoxifen-inducible BAF250a KO ES cells. We then applied genome-wide nucleosome mapping by MNase-sequencing and histone modifications, including H3K4me3 and H3K27me3, by ChIP-sequencing to study the function of esBAF in regulating key chromatin structures in ES cells. We further identified the regulatory roles of BAF250a in recruiting esBAF and PRC2, a complex that deposits H3K27 trimethylation. Our result showed that BAF250a mediates the proper nucleosome occupancy and H3K27me3 modification at ES cells by regulating esBAF and PRCs, and disruption of these epigenetic regulations caused the differentiation defects in BAF250a KO ES cells.
Experimental Procedures
Derivation of Mouse Tamoxifen-inducible BAF250a KO ES Cells
E3.5 blastocysts were collected by crossing CreErt2;BAF250af/f with BAF250af/f mice. Blastocysts were washed with M2 medium (M7167), followed by treatment of acidic Tyrode's solution to remove the zonae pellucidae. Blastocysts were cultured for 7 days for outgrowth, and ES cell were generated by dissociating the outgrowth as described previously (14). Inducible BAF250a KO ES cells with the Rosa26-LacZ reporter were also generated using E3.5 blastocysts from CreErt2;BAF250af/f with BAF250af/f;Rosa26-LacZ mice.
X-Gal Staining of Chimeras
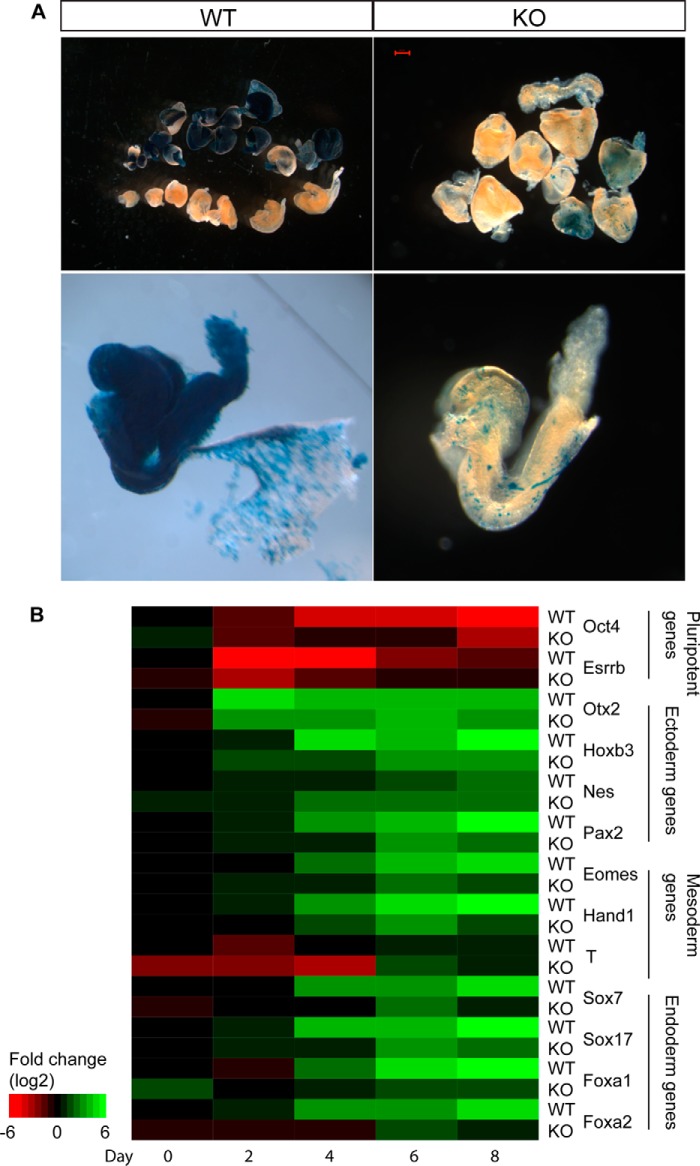
WT and BAF250a KO ES cells with Rosa26-LacZ reporter were injected into WT embryos and harvested at E9.5. The embryos were then fixed and stained with X-gal. Embryonic tissue derived from BAF250a-inducible KO ES cells were identified by X-gal staining (blue). In principle, normal ES cells contribute to all embryo lineages.
Cell Culture and Purification of Nuclei
ES cells were cultured under standard conditions (DMEM, 2 mm glutamine, sodium pyruvate, non-essential amino acids, 15% FBS, 0.1 mm 2-mercaptoethanol, and 1000 units/ml leukemia inhibitory factor) on mouse embryonic fibroblast feeders. ES cells were treated with 4-hydroxytamoxifen for 36 h and recovered for 36 h to generate BAF250a KO ES cells.
ES cells were fixed in 1% formaldehyde for 10 min at room temperature, and the fixation was stopped in 0.125 m glycine for 5 min. ES cells were rinsed with cold PBS and collected by scraper. 2 × 107 cells were washed with cold PBS twice and resuspended in nuclear lysis buffer (5 mm PIPES (pH 8.0), 85 mm KCl, and 0.5% Nonidet P-40) and rocked at 4 °C for 10 min. The nuclei pellet was collected by centrifugation at 2000 rpm for 5 min.
Embryoid Body (EB) Differentiation
Mouse ES cells were dissociated and resuspended in 15% FBS/DMEM at a concentration of 3.3 × 104 cells/ml. 15 μl of cell suspension was then added to the dish and aggregated to form EBs using the hanging drop method (15). EBs were collected every other day for gene expression analysis.
Mononucleosome Preparation and MNase-seq 2 × 107
Nuclei were digested with three concentrations of MNase for 15 min at room temperature. MNase-digested nuclei were decross-linked at 55 °C overnight with proteinase K digestion. DNAs were harvested via phenol-chloroform extraction. Nucleosomal DNAs were run on 1% agarose gels, and the mononucleosomal DNAs (150 bp) were gel-purified. Mononucleosomal DNAs from three digestions were pooled and prepared for sequencing (16).
Nucleosome Mapping Analysis
Sequencing reads were aligned to mouse genome data (mm9) using Bowtie (17) with the following options: -M 1 -q -v 2 -k 1 -X 2000 -best -strata. The distribution of mononucleosome fragment sizes was calculated. MNase-seq fragments with different fragment size distribution will lead to extensive false positives in the analysis (18). The fragment sizes determined by paired-end MNase-seq were calculated to confirm that the fragment sizes of different samples were comparable. Nucleosome occupancy was analyzed by DANPOS (18) with default parameters. Briefly, different replicates were pooled, and the nucleosome occupancy was calculated at base pair resolution. The nucleosome occupancy was then normalized with quantile normalization. The nucleosome coverage was calculated by read per million reads. All aligned plots of nucleosome occupancy were calculated with 10-bp binning, and the midpoints of fragments were considered nucleosome centers. The coordinates of transcription start site (TSS) of each gene cluster were retrieved from the UCSC genome browser. The binding site coordinates of Suz12 were obtained from GSE11431 (19).
Chromatin Immunoprecipitation
Nuclei were pelleted and digested with MNase in MNase digestion buffer for 15 min at 37 °C. Chromatins were collected by brief sonication and incubated with Mangabeads and antibodies against H3K4me3 (Millipore, catalog no. 07-473), H3K27me3 (Millipore, catalog no. 07-449), and Suz12 (Abcam, catalog no. ab12073) overnight. The chromatin solutions were washed four times with LiCl wash buffer and eluded with elution buffer at 65 °C. DNAs were purified using phenol:chloroform extraction. Enrichment of immunoprecipitation DNA was then validated by quantitative PCR.
ChIP-seq Library Preparation
Approximately 100 ng of ChIPed DNAs were used to prepared the library using the NEBNext kit (E6240) following the protocol of the manufacturer.
ChIP-seq Data Analysis
The ChIP-seq reads were aligned to mouse genome data (mm9) using Bowtie with the following options: -M 1 -q -v 2 -k 1 -X 2000 -best -strata. Peak calling was performed using SCIER 1.1 (20) with the following parameters: window size, 200; gap size, 600; false discovery rate, 0.05. The input DNAs were used as a control. The ChIP-seq reads at each base pair were normalized to read per million reads. The coverage of the ChIP-seq signal across TSS was calculated by averaging the ChIP-seq signals at 10-bp binning.
Purification of the BAF Complex
The cell lysates were prepared from cells as described previously (21). Briefly, 10 million cells were harvested and washed with PBS twice. Cell pellets were then subjected to lysis buffer. The cell lysate were immunoprecipitated with Brg1 antibody and protein G beads at 4 °C overnight. The protein G beads/proteins were washed three times with wash buffer and eluted with elution buffer. The precipitated proteins were identified with specific BAF component antibody by Western blot analyses.
Expression Array
Total RNA was isolated from WT and inducible BAF250a KO ES cells using TRIzol according to the protocol of the manufacturer, followed by quality assessment using Bioanalyzer 2100 (Agilent). RNA was then subjected to in vitro transcription and hybridized to Affymetrix Mouse 2.0 ST arrays.
Generation of V5-tagged Brg1 Mouse ES Cells
The V5 sequence was inserted into the C terminus of Brg1 using CRISPR/Cas9 technology (22). In brief, guide RNA (5′-CCGCTCAGGAAGTGGCAGTG-3′), the V5 sequence with a 50-bp 5′ and 3′ arm (single-stranded DNA synthesized from IDT), and the Cas9 plasmid were electroporated into 1 million ES cells. Two thousand cells were then seeded in a 10-cm dish, andV5 knockin ES cells clones were picked and identified by PCR.
Results
Acute Deletion of BAF250a Disrupted the Differentiation Potential of ES Cells
We have shown previously that BAF250a is required for the maintenance of ES cell pluripotency (12). Because long-term culture of BAF250a KO ES cells without feeder cells could lead to secondary events (12), we generated tamoxifen-inducible BAF250a KO ES cells to investigate the immediate effect of BAF250a deletion. In these conditional KO ES cells, BAF250a was excised rapidly, and BAF250a protein was depleted 72 h after tamoxifen treatment (data not shown). Interestingly, these cells continued to abundantly express pluripotent genes such as OCT4, SOX2, and NANOG (data not shown). To study the effect of BAF250a on ES cell pluripotency and differentiation, we first performed chimera analyses by injecting both WT and BAF250a KO ES cells into E3.5 blastocysts. About 60% of WT and BAF250a KO ES cells could contribute to form chimeras (WT, 15 chimeras in 22 embryos; KO, 6 chimeras in 10 embryos). However, WT ES cells could contribute to almost all cell lineages, whereas BAF250a KO ES cells displayed remarkable defects in contributing to three germ layers of embryos (Fig. 1A), suggesting that BAF250a is required for potency of ES cells.
FIGURE 1.
BAF250 deletion disrupted ES cell differentiation potential. A, WT ES cells contributed to the whole chimera embryo after blastocysts injection (left panel). BAF250a KO ES cells rarely contributed to the chimera embryos (right panel). Contributions of ES cells to embryos are indicated by X-gal staining (blue). B, quantitative PCR analysis of Oct4, Esrrb, Otx2, Hoxb3, Nestin, Pax2, Eomes, Hand1, T, Sox7, Sox17, Foxa1, and Foxa2 during differentiation. Expression -fold changes of relative mRNA expression of each gene were calculated by comparison with the expression at WT day 0.
To study the regulation of BAF250a on developmental genes during differentiation, we performed an in vitro EB differentiation assay and analyzed gene expression by quantitative PCR. We found that, during differentiation, the expression of developmental genes such as Otx2, Nes, Hoxb3, Pax2, Eomes, T, Hand1, Sox7, Sox17, Foxa1, and Foxa2 were aberrant. In particular the mesoderm and endoderm genes were delayed dramatically in BAF250a KO cells (Fig. 1B). Importantly, pluripotent genes such as Oct4 and Esrrb were not shut down properly during differentiation (Fig. 1B). These results suggest that BAF250a-mediated epigenetic mechanisms are essential for gene expression plasticity during ES cell differentiation.
BAF250a Ablation Causes an Increase of Nucleosome Occupancy at TSS Regions
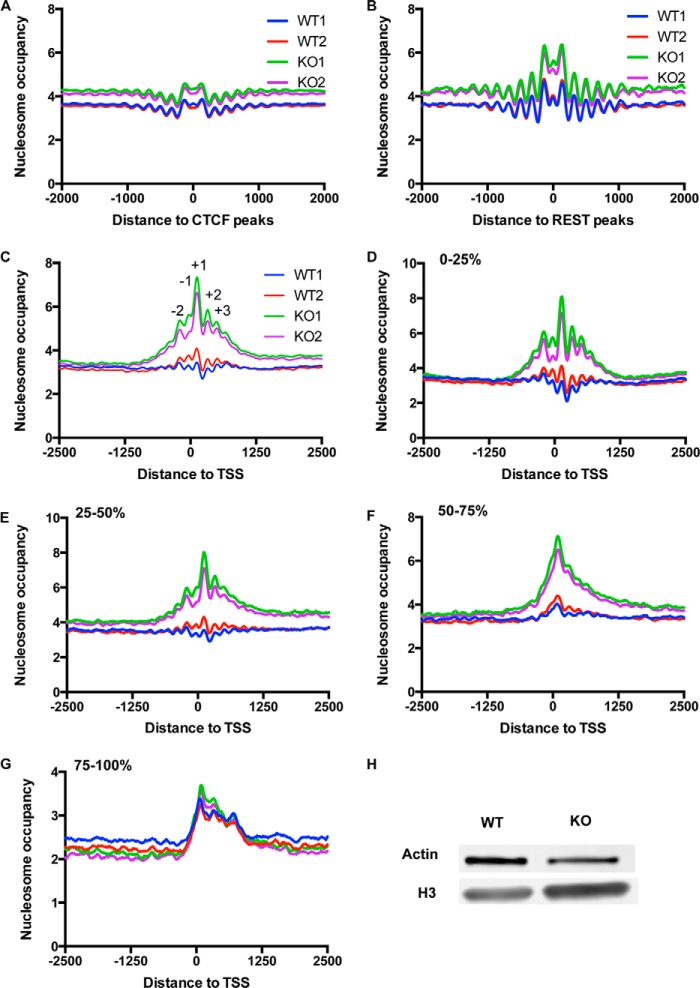
Given the nucleosome remodeling function of the esBAF complex, it is likely that BAF250a regulates ES cell function through mediate nucleosome structures. Therefore, we examined the genome-wide nucleosome profile in both WT and BAF250a-inducible KO ES cells using mild micrococcal nuclease (MNase) digestion coupled with high-throughput sequencing of mononucleosome-protected DNA fragments, as described previously (23, 24). To validate the MNase-seq results, we analyzed the fragment distribution of MNase-seq. The size distribution of all samples was similar, and the average fragment size in WT and KO cells was 153/154 and 159/161, respectively, indicating that they had undergone a similar degree of MNase digestion. We then analyzed the nucleosome profile at the binding sites of chromatin insulators such as CTCF and REST. Consistent with previous reports (25, 26), nucleosomes were well positioned at the binding sites of CTCF and the REST repressor protein in both WT and BAF250a KO ES cells (Fig. 2, A and B), indicating successful mapping of nucleosomes.
FIGURE 2.
BAF250a deletion in ES cells led to a nucleosome occupancy increase around TSS. A and B, nucleosomes were well positioned around the CTCF and REST binding sites. The distance to CTCF and REST binding sites was plotted at the x axis. C, MNase-seq revealed that BAF250a deletion resulted in a nucleosome occupancy increase at TSS regions of all genes 72 h after BAF250a deletion. D—G, the nucleosome position underwent a progressive disorganization with the gene expression level reduced. The nucleosome occupancy increase level was high in first quantile-expressed (0–25%) genes (D), second quantile-expressed (25–50%) genes (E), and third quantile-expressed (50–75%) genes (F) but low at fourth quantile-expressed (75–100%) genes (G). TSSs are indicated in the x axes at 0. Shown in the y axes are the counts of nucleosome tags. H, Western blot analysis of Histone H3 in WT and KO ES cells.
To study the nucleosome changes in BAF250a KO ES cells, we studied the nucleosome profiles at TSS regions. We observed a well organized nucleosome positioning at TSS (Fig. 2C). However, in our study, we found a nucleosome located at nucleosome-free regions. These nucleosomes were also found in other limited digestion studies (27–29). The observation of these nucleosomes indicates that these nucleosomes are most accessible and can be further digested with higher MNase concentrations. To investigate the relationship of TSS nucleosome occupancy and transcription, we clustered the gene promoters into four groups on the basis of the gene expression level and examined the nucleosome profile in each group. Nucleosomes were well positioned at highest expressed genes (Fig. 2D), whereas nucleosomes were not positioned around TSSs of the lowest expressed genes (Fig. 2G). In second and third quantile-expressed genes, nucleosomes became less organized (Fig. 2, E and F), suggesting a correlation between nucleosome phrasing at TSS and gene expression.
When comparing the nucleosome occupancy at WT and KO TSS, we observed an overall nucleosome occupancy increase in BAF250a KO ES cells across TSS regions (Fig. 2C) but not other regions. Western blot analysis of H3 protein showed that there was an increase of chromatin H3 protein after BAF250a KO (Fig. 2H), suggesting an increase of nucleosome in BAF250a KO ES cells. Moreover, the unstable nucleosomes at nucleosome-free regions were less evident in BAF250a KO ES cells (Fig. 2C), suggesting that BAF250a may have an important function in stabilizing these nucleosomes. Finally, we found that the nucleosome occupancy increase became less pronounced when gene expression decreased (Fig. 2, D–G), suggesting that BAF250a-regulated nucleosome density is positively correlated with gene expression in general.
esBAF Regulated Nucleosome Occupancy on Promoters Associated with H3K4me3 and/or H3K27me3
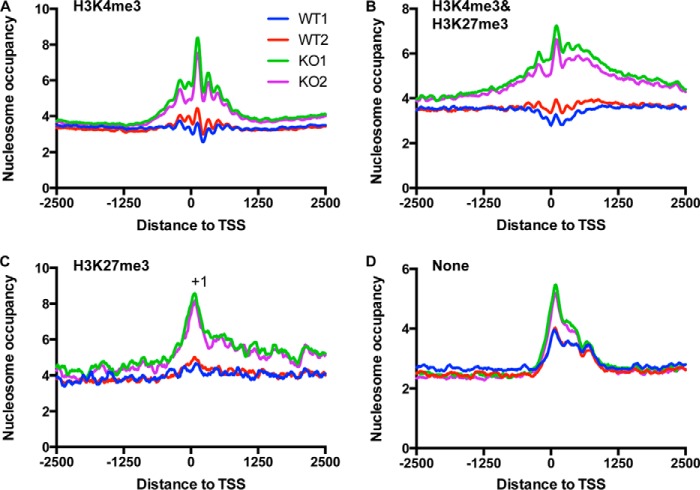
To test how BAF250a-regulated nucleosome changes correlate with promoter chromatin status, we compared the nucleosome profiles at promoters associated with H3K4me3 alone, both H3K4me3 and H3K27me3, H3K27me3 alone, and neither H3K4me3 or H3K27me3 (none) (30) in WT and BAF250a-inducible KO cells. Similar to high expression gene promoters, we observed well positioned nucleosomes at H3K4me3 promoters, whereas only +1 nucleosomes were positioned in H3K27me3 promoters and promoters with no histone modifications (Fig. 3, A–D). Notably, the bivalent promoters exhibited relatively good nucleosome phrasing (Fig. 3B), although these genes were silenced. These observations suggested that the open chromatin structure characterized by well positioned nucleosomes at bivalent promoters may be related to rapid activation of these genes during differentiation. Moreover, the increase of nucleosome occupancy in BAF250a KO ES cells was prominent in promoters associated with H3K4me3 or H3K27me3 but not in the no histone modification promoters. These results suggest that BAF250a-mediated nucleosome remodeling may contribute to the open chromatin structure at promoters associated with histone modifications in ES cells.
FIGURE 3.
MNase-seq revealed distinct nucleosome profiles at promoters associated with different histone modifications. A and B, nucleosomes at active promoters (A) and poised promoters (B) had well positioned nucleosomes. C and D, in contrast, only +1 nucleosome was positioned at repressed promoters (C), and no organized nucleosomes were at no histone modification promoters (D). BAF250a deletion led to a high nucleosome occupancy increase at promoters with either H3K4me3 or H3K27me3 but less at promoters without histone modifications. TSSs are indicated in the x axes at 0. Shown in the y axes are the counts of nucleosome tags.
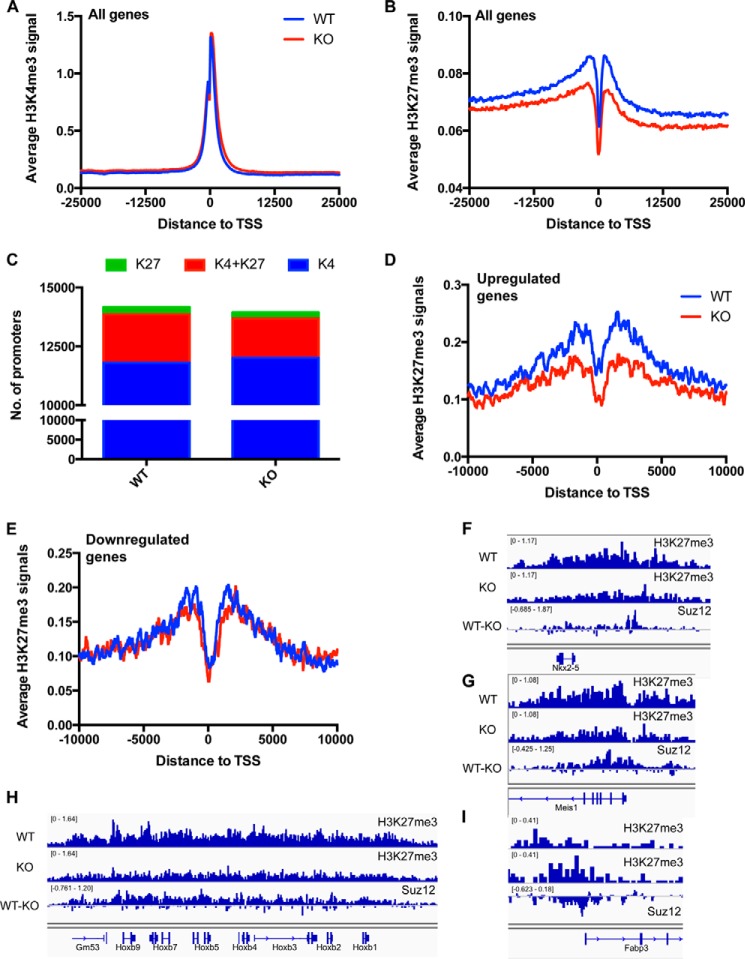
Loss of BAF250a Resulted in Disruption of Bivalent Chromatin Modifications
The changing of nucleosome occupancy in BAF250a KO ES cells may lead to disruption of deposition of histone modifications and, therefore, poised chromatin signatures. To address this possibility, we examined the global modification of H3K4me3 and H3K27me3 by ChIP-seq. We found that H3K4me3 did not show significant changes at TSS regions (Fig. 4A), whereas a prominent reduction of H3K27me3 binding was found at TSS in BAF250a KO ES cells (Fig. 4B). Reduction of H3K27me3 also led to a loss of bivalent promoters in BAF250a KO ES cells (Fig. 4C), indicating that the poised chromatin structure was affected upon BAF250a deletion. However, loss of BAF250a did not lead to a decrease of H3K27me3 at all genes. The H3K27me3 level decreased at the up-regulated genes, whereas it remained unchanged in down-regulated genes (Fig. 4, D and E). For example, there was an H3K27me3 reduction at Hoxb gene clusters and several developmental genes such as Nkx2.5 and Meis1, but the Fabp3 promoter showed an increased H3K27me3 level in BAF250a KO ES cells (Fig. 4, F–I). Changes in H3K27me3 prompted us to examine the recruitment of the PRC complex. We performed a genome-wide ChIP-seq assay using antibodies against Suz12, a core component of PRC2 complex. Suz12 binding was reduced at Hoxb gene regions and developmental genes such as Nkx2.5 and Meis1 (Fig. 4, F–H). Notably, Suz12 binding was increased at H3K27me3-elevated regions such as Fabp3 (Fig. 4I), suggesting a possible regulation of BAF250a on H3K27me3 through modulation of Suz12 binding. Together, these results showed that BAF250a functioned synergistically with PcG proteins and contributed to a repressive role of esBAF in ES cells.
FIGURE 4.
Deletion of BAF250a caused bivalent gene defects. A and B, no change of H3K4me3 modifications (A) and decreased H3K27m3 modifications (B) in BAF250a KO ES cells, as shown by average ChIP-seq tag densities of H3K4me3 and H3K27me3 at TSS regions of all genes. C, loss of bivalent genes in BAF250a KO ES cells 72 h after BAF250a deletion. D, decrease of H3K27me3 at TSS of up-regulated genes in BAF250a KO ES cells. E, no changes of H3K27me3 at down-regulated genes in BAF250a KO ES cells. F—I, ChIP-seq signals of H3K27me3 and Suz12 in both WT and KO at regions of Nkx2.5 (F), Meis1 (G), the HoxB cluster (H), and Fabp3 (I). The WT and KO lanes represent the signals of H3K27me3 in BAF250a WT and KO ES cells, respectively. The WT-KO lane represents the differential signals of Suz12 in WT ES cells and KO ES cells. Signals higher than 0 indicate decreased binding of Suz12 in KO ES cells, and signals less than 0 indicated increased binding of Suz12 in KO ES cells.
BAF250a Deletion Led to Increased Brg1 Binding and Nucleosome Occupancy and Reduced H3K27me3 Density at Genes Essential for ES Cell Differentiation
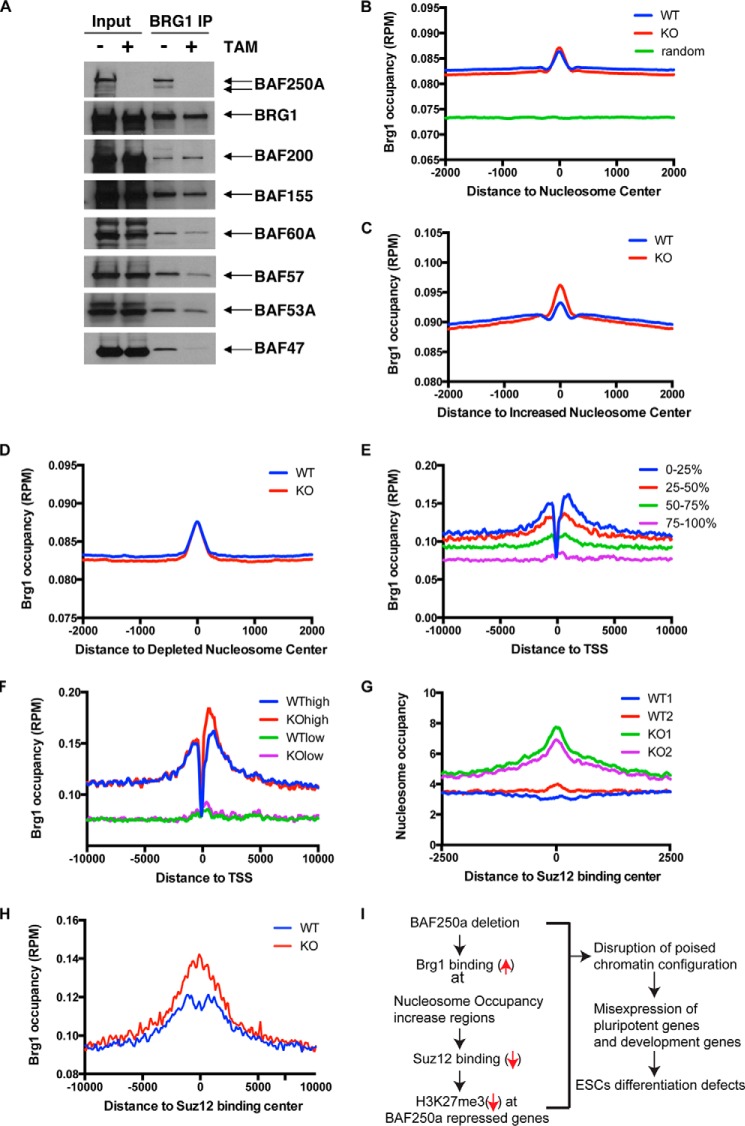
To identify the underlying molecular mechanism, we first examined the effect of BAF250a deletion on BAF complex assembly by coimmunoprecipitation. We found that the interaction of Brg1 with other core BAF subunits, such as BAF155, was not affected, whereas the interaction of Brg1 with BAF60a, BAF57, and BAF53a was diminished, and the interaction of BAF47 with Brg1 was almost abolished (Fig. 5A). Because components of the BAF complex contribute to the specificity of the BAF complex, these results indicate that Brg1 recruitment may be affected in BAF250a KO ES cells. To further examine this hypothesis, we performed ChIP-seq analyses against the catalytic subunit of esBAF Brg1 to study the possible change of Brg1 occupancy. We knocked in a V5 tag to the C terminus of Brg1 that allowed us to successfully study genome-wide Brg1 binding. Subsequently, we examined whether Brg1 was involved in the process of modulation of nucleosome density in BAF250a KO ES cells. We found that Brg1 was enriched in the nucleosome regions compared with random regions (Fig. 5B). In addition, Brg1 occupancy was increased in regions that exhibited elevated nucleosome density in BAF250a KO ES cells, whereas there was no increase of Brg1 occupancy in regions with a reduced density of nucleosomes (Fig. 5, C and D). We found that Brg1 bound to promoter regions and was positively correlated with gene expression (Fig. 5E), which is consistent with what has been described previously (11). Increased binding of Brg1 in BAF250a KO cells was less profound in low expression genes (Fig. 5F). These data suggest that BAF250a may be required for proper recruitment of Brg1 to poised and active genes to maintain proper nucleosome configuration for gene expression in ES cells.
FIGURE 5.
The nucleosome occupancy increase in BAF250a KO ES cells was associated with Brg1 recruitment. A, protein interaction of Brg1 with the BAF complex subunits BAF200, BAF155, BAF60a, BAF57, BAF53a, and BAF47 in WT and BAF250a KO ES cells 72 h after BAF250a deletion. B, Brg1 was enriched at the nucleosome center. C, elevated Brg1 binding at increased nucleosome occupancy regions at BAF250a KO ES cells. D, Brg1 binding was not changed at depleted nucleosome regions at BAF250a KO ES cells. E, the Brg1 binding level was positively correlated with gene expression. F, the Brg1 binding increase was more profound at high expression gene promoters. WThigh/Kohigh, the top 0–25% expressed genes in WT/KO cells; WTlow/Kolow, 75–100% expression genes in WT/KO cells. G and H, increased nucleosome occupancy (G) and brg1 binding (H) in Suz12 binding sites. I, model of BAF250a regulating ES cell differentiation.
To determine how BAF250a regulates H3K27me3 density in ES cells, we analyzed the nucleosomes occupancy and Brg1 binding to target loci of Suz12, a core component in PRC2. There was pronounced nucleosome occupancy increase at Suz12 binding sites (Fig. 5G), accompanied by elevated Brg1 binding in BAF250a KO ES cells (Fig. 5H). For example, we found that Brg1 binding was increased at the Hox cluster region with decreased H3K27me3 modification. Consistent with decreased H3K27me3, Suz12 binding was reduced in the HoxB cluster region (Fig. 4H). Previous studies have shown that Suz12 binds to low nucleosome occupancy regions during ES cell differentiation to regulate H3K27me3 modifications (31, 32). Therefore it is likely that the nucleosome occupancy increase in BAF250a KO ES cells led to lower binding of Suz12. Collectively, BAF250a deletion resulted in elevated Brg1 binding at Suz12 binding regions and a subsequent nucleosome occupancy increase. The nucleosome occupancy increase also contributed to reduced Suz12 binding and abnormal modification of H3K27me3 in BAF250a KO ES cells (Fig. 5I). The H3K27me3 defects in ES cells caused by the nucleosome occupancy increase may also explain the inefficient shutdown of pluripotent genes during ES cell differentiation (Fig. 1B).
Discussion
In this study, we demonstrate that BAF250a, a regulatory component of esBAF, controls key aspects of chromatin structure at the level of nucleosome position and modification. Deletion of BAF250a in ES cells significantly decreased the potential of ES cells to contribute to differentiated lineages, leading us to propose that the ability of esBAF to appropriately modify nucleosomes is critical to differentiation. We found that BAF250a differentially regulates nucleosome structure in promoters associated with different gene expression levels and histone modifications. Importantly, our results show that BAF250a mediates the level of Brg1 binding, particularly at BAF-repressed genes, to control proper nucleosome occupancy at ES cells. Moreover, BAF250a deletion led to reduced Suz12 recruitment and decreased H3K27me3 modifications at developmental genes in ES cells. Collectively, these epigenetic disruptions may cause incomplete shutdown of pluripotent genes and aberrant expression of developmental genes and contribute to the differentiation defects of BAF250a KO ES cells.
Our results show that BAF250a-mediated promoter nucleosome occupancy increase at promoters correlates with histone marks H3K4me3 and H3K27me3, suggesting a potential role for nucleosome positioning in the poised chromatin signature. In this study, we found that nucleosomes are well positioned in the promoters of highly expressed genes and that they become disorganized in the promoters of lower expressed genes, suggesting that a well positioned nucleosome is a feature of open chromatin. Moreover, we found that nucleosomes are well organized in H3K4me3 promoters and less organized in bivalent H3K4me3 and H3K27me3 promoters and that only +1 nucleosomes are positioned at promoters decorated with H3K27me3 and promoters that lacked these marks. Noticeably, there is a more prominent nucleosome occupancy increase in promoters associated with either H3K3me4 alone or both H3K4me3 and H3K27me3 modifications. Specifically, the binding level of Brg1 descends from H3K4me3 promoters, bivalent promoters, H3K27me3 promoters, and promoters lacking these histone modifications. Importantly, Brg1 occupancy increases in regions with elevated nucleosome occupancy in BAF250a KO ES cells. These results suggest that BAF250a directs esBAF to remodel the nucleosome structure into a poised chromatin configuration at poised genes in ES cells.
Furthermore, our results show that BAF250a not only mediates nucleosome density on poised gene promoters but also regulates bivalent marks in ES cells. We found that global H3K27me3 decreased in BAF250a KO ES cells. In particular, we observed a decreased H3K27me3 level in BAF250a-repressed genes but no significant change of H3K27me3 in BAF250a-activated genes. The alteration of H3K27me3 levels may indicate a specific repressive role of BAF250a in ES cells. Although Brg1 deletion does not result in global H3K27me3 and H3K4me3 changes, H3K27me3 density decreased in Brg1-repressed genes and increased in Brg1-activated genes in Brg1 KO ES cells (33). These differences in histone modification changes in BAF250a KO and Brg1 KO ES cells reflect distinct functions exercised by esBAF components on histone modification regulations. Therefore, it seems that the BAF250a-containing esBAF complex has a specific function in gene repression via regulating the H3K27me3 modification in ES cells.
A comparison of our data to a study in mouse embryonic fibroblasts suggests that BAF complexes regulate nucleosomes in a cell type-specific manner. BAF complexes have different composition assemblies in different cell type. Therefore, BAF complexes may achieve a cell-specific function by different assemblies of the BAF complex (9). Indeed, deletion of Brg1 or BAF47 in mouse embryonic fibroblast leads to an overall nucleosome occupancy decrease at TSS regions (34), a finding that differs from the nucleosome increase we observed in BAF250a KO ES cells. These results likely reflect the distinct functions of BAF complexes in different cell types. It has been shown that ES cells have less heterochromatin than differentiated cells (3). ES cells have more abundant active histone modifications such as H3K4me3 and more active transcription with lower nucleosome occupancy. The nucleosome occupancy increase in BAF250a KO ES cells may indicate that the KO ES cell chromatin is transitioning into a more differentiated status. The tendency to differentiation observed in prolonged culture of BAF250a KO ES cells also suggests that esBAF is essential for ES cell-specific chromatin structure maintenance. In contrast, BAF mutations in somatic cells may lead to more open chromatin structure changes. Indeed, many of these mutations cause aberrant cell growth and cancer (35). Therefore, a BAF-mediated, cell type-specific nucleosome structure is essential for proper cell function.
In summary, our results show that BAF250a is a key player in regulating nucleosome occupancy and bivalent histone modifications in ES cells. The dynamic nucleosome and histone modifications mediated by BAF250a ensure that ES cells respond to differentiation signals in a timely manner. Ablation of BAF250a leads to altered nucleosome structures that severely hamper the plasticity of ES cell chromatin, which, in turn, contributes to a general delay in the expression of developmental genes and continued expression of pluripotent genes during differentiation. The aberrant expression of both developmental and pluripotent genes by BAF250a ablation causes the early germ layer developmental defects in vivo and various lineage differentiation defects in EB-based differentiation in vitro. Our studies further indicate that BAF250a is required for proper Brg1 recruitment and also regulates Suz12 recruitment at nucleosome occupancy-increased regions, indicating an intricate role of BAF250a in regulating esBAF and PRC activities. Future studies of the BAF complex and its involvement in high-order chromatin structures will elucidate the unique role of the esBAF chromatin remodeling complex in ES cell differentiation and development.
Author Contributions
I. L. designed the experiments, collected and analyzed data, and wrote the manuscript. J. W. designed the experiments and collected data. Z. Y., X. G., P. F., J. D., and L. G. collected and/or assembled data. W. W. and R. K. provided financial support and interpreted data. Z. W. conceived and designed the study, wrote the manuscript writing, and approved the final version of the manuscript.
Note Added in Proof
Nucleosome positioning results were missing from the version of Fig. 2 that was published on June 12, 2015 as a Paper in Press. These results are now included in the corrected Fig. 2, A and B. Fig. 2, C—H, has been relabeled accordingly. These changes do not affect the interpretation of the results or the conclusions.
This work was supported, in whole or in part, by National Institutes of Health Grant HL109054 (to W. Z.) and Intramural Research Program of the NIA/National Institutes of Health Grant AG000650-10 (to W. W.). This work was also supported by the Samuel and Jean Frankel Cardiovascular Center, University of Michigan (Inaugural Fund) (to W. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
The normalized microarray data reported in this paper have been submitted to the Gene Expression Omnibus Repository with accession number GSE 59082.
- BAF
- Brahma-associated factor
- EB
- embryoid body
- E
- embryonic day
- MNase
- micrococcal nuclease
- H3K4me3
- H3 Lys-4 trimethylation
- H3K27me3
- H3 Lys-27 trimethylation
- esBAF
- ES cell specific ATP-dependent BAF complex
- PRC
- polycomb repressive complex
- MNase-seq
- MNase-sequencing
- ChIP-seq
- ChIP-sequencing
- TSS
- transcription start site.
References
- 1. Surani M. A., Hayashi K., Hajkova P. (2007) Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762 [DOI] [PubMed] [Google Scholar]
- 2. Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M. (2011) Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mattout A., Meshorer E. (2010) Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 22, 334–341 [DOI] [PubMed] [Google Scholar]
- 4. Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 6. Lorch Y., Maier-Davis B., Kornberg R. D. (2010) Mechanism of chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narlikar G. J., Sundaramoorthy R., Owen-Hughes T. (2013) Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154, 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitehouse I., Flaus A., Cairns B. R., White M. F., Workman J. L., Owen-Hughes T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400, 784–787 [DOI] [PubMed] [Google Scholar]
- 9. Wu J. I., Lessard J., Crabtree G. R. (2009) Understanding the words of chromatin regulation. Cell 136, 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ho L., Crabtree G. R. (2010) Chromatin remodelling during development. Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106, 5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao X., Tate P., Hu P., Tjian R., Skarnes W. C., Wang Z. (2008) ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U.S.A. 105, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagy A., Marina G., Kristina V., Richard B. (2003) Manipulating the Mouse Embryo, pp. 359–398, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 15. Takahashi T., Lord B., Schulze P. C., Fryer R. M., Sarang S. S., Gullans S. R., Lee R. T. (2003) Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107, 1912–1916 [DOI] [PubMed] [Google Scholar]
- 16. Bowman S. K., Simon M. D., Deaton A. M., Tolstorukov M., Borowsky M. L., Kingston R. E. (2013) Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics 14, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J., Dent S., He X., Li W. (2013) DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 23, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y.-H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W.-K., Clarke N. D., Wei C.-L., Ng H.-H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 20. Zang C., Schones D. E., Zeng C., Cui K., Zhao K., Peng W. (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan Z., Wang Z., Sharova L., Sharov A. A., Ling C., Piao Y., Aiba K., Matoba R., Wang W., Ko M. S. (2008) BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells 26, 1155–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dennis J. H., Fan H.-Y., Reynolds S. M., Yuan G., Meldrim J. C., Richter D. J., Peterson D. G., Rando O. J., Noble W. S., Kingston R. E. (2007) Independent and complementary methods for large-scale structural analysis of mammalian chromatin. Genome Res. 17, 928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kharchenko P. V., Woo C. J., Tolstorukov M. Y., Kingston R. E., Park P. J. (2008) Nucleosome positioning in human HOX gene clusters. Genome Res. 18, 1554–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu G., Schones D. E., Cui K., Ybarra R., Northrup D., Tang Q., Gattinoni L., Restifo N. P., Huang S., Zhao K. (2011) Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valouev A., Johnson S. M., Boyd S. D., Smith C. L., Fire A. Z., Sidow A. (2011) Determinants of nucleosome organization in primary human cells. Nature 474, 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen K., Wilson M. A., Hirsch C., Watson A., Liang S., Lu Y., Li W., Dent S. Y. (2013) Stabilization of the promoter nucleosomes in nucleosome-free regions by the yeast Cyc8-Tup1 corepressor. Genome Res. 23, 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deng T., Zhu Z. I., Zhang S., Leng F., Cherukuri S., Hansen L., Mariño-Ramírez L., Meshorer E., Landsman D., Bustin M. (2013) HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Mol. Cell. Biol. 33, 3377–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu G., Cui K., Northrup D., Liu C., Wang C., Tang Q., Ge K., Levens D., Crane-Robinson C., Zhao K. (2013) H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woo C. J., Kharchenko P. V., Daheron L., Park P. J., Kingston R. E. (2010) A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Woo C. J., Kharchenko P. V., Daheron L., Park P. J., Kingston R. E. (2013) Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol. Cell. Biol. 33, 3274–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ho L., Miller E. L., Ronan J. L., Ho W. Q., Jothi R., Crabtree G. R. (2011) esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 13, 903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tolstorukov M. Y., Sansam C. G., Lu P., Koellhoffer E. C., Helming K. C., Alver B. H., Tillman E. J., Evans J. A., Wilson B. G., Park P. J., Roberts C. W. (2013) Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc. Natl. Acad. Sci. U.S.A. 110, 10165–10170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kadoch C., Hargreaves D. C., Hodges C., Elias L., Ho L., Ranish J., Crabtree G. R. (2013) Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]