Abstract
Cartilage-hair hypoplasia (CHH) is an autosomal recessive disorder which is characterized by bone metaphysis anomalies with manifestations that include short stature, defective cellular immunity, and predisposition to several cancers. It is caused by mutations in RMRP, which is transcribed as an RNA component of the mitochondrial RNA-processing ribonuclease. We report the clinical and molecular data of a Moroccan patient with CHH. Sequencing of RMRP identified 2 mutations in the patient: the known mutation g.97G>A and the variation g.27G>C, which has not been reported previously. Given the high mutational heterogeneity, the high frequency of variations in the region, and the fact that RMRP is a non-coding gene, assigning the pathogenicity to RMRP mutations remains a difficult task. Therefore, we compared the characteristics of the primary and secondary structures of mutated RMRP sequences. The location of our mutations within the secondary structure of the RMRP molecule revealed that the novel g.27G>C mutation causes a disruption in the Watson-Crick base pairing, which results in an impairment of a highly conserved P3 domain. Our work prompts considering the consequences of novel RMRP nucleotide variations on conserved RNA structures to gain insights into the pathogenicity of mutations.
Key Words: Cartilage-hair hypoplasia, Mutational analysis, RNA folding , RNase P/MRP
Cartilage-hair hypoplasia (CHH; MIM#250250) is a form of short-limbed dwarfism due to skeletal dysplasia. It was first recognized in the Old Order Amish in the USA, a religious isolate, where the carrier frequency was 1/19 [McKusick, 1964; McKusick et al., 1965]. Later, it has been reported in other populations; however, there is no precise measure of its incidence worldwide.
CHH is an autosomal recessive disorder characterized by skeletal involvement, short stature, variable features like blond fine sparse hair, and defective cellular immunity affecting T-cell-mediated responses [Makitie et al., 1995]. Patients may have a severe combined immunodeficiency requiring bone marrow transplantation or they may be asymptomatic [Castigli et al., 1995; Makitie et al., 1998]. CHH is caused by mutations in the RMRP gene (RNA component of mitochondrial RNA-processing endoribonuclease; MIM#157660) [Ridanpää et al., 2001], an untranslated gene with only 267 nucleotides that encode an RNA subunit of an RNase-MRP complex which is involved in multiple cellular and mitochondrial functions [Martin and Li, 2007]. RNase MRP RNA is the first nuclear-encoded RNA in which mutations have been found to lead to human diseases.
More than 66 mutations in families with CHH have been reported in a variety of ethnic groups, mainly Europeans [Bonafé et al., 2002; Nakashima et al., 2003; Thiel et al., 2005; Thiel and Rauch, 2011]. Up to now, no RMRP mutations have been reported in the Moroccan population. RMRP is characterized by a very high density of SNPs and several pathogenic variants. Subsequently, molecular diagnosis remains problematic.
Here, we studied RMRP variations in a patient with CHH and identified 1 novel mutation. Based on a previous extensive conservation study in eukaryotes [Li et al., 2002], we report the structural analysis of the identified mutations as a mean to assign their potential pathogenicity.
Case Report
A 6-year-old Moroccan girl was referred to our department for karyotyping to evaluate her statural delay. She was the third of 3 children, and although here parents originated from the same region, they were not consanguineous. There was no family history of CHH or immunodeficiency, and neonatal measurements were normal. The pregnancy was not medically followed, but it was reported without complications. At birth, the girl weighed 2,800 g and had a length of 52 cm and a head circumference of 36 cm. Her postnatal growth was slow. Clinical examination at the age of 6 years showed a delay in growth development with a weight of 15 kg (-3 SD), height of 93 cm (-4 SD), and a head circumference of 52 cm (mean). The child had very fine and sparse hair, and the lower limbs appeared disproportionately shorter than the upper limbs. Her hands were short and broad, and her cognitive development was normal.
Growth hormone deficiency and other endocrinopathies were eliminated. Repeated failure to trigger cell division in response to treating her lymphoblasts with phytohaemagglutinin for chromosomal analysis induced an immune work-up which revealed marked lymphopenia and hypogammaglobulinemia. Immunological evaluation of the patient showed a deficit of cellular immunity in T8 cells with a value of 0.24 × 103/µl (normal range: 0.37-1.1 × 103/µl) as well as hypogammaglobulinemia with an IgA and IgM deficit and a normal IgG value.
Radiography of the hands showed a delay in bone aging (bone age of 4 years for a chronologic age of 5 years) and a cone-shaped epiphysis in the first phalanx of the first finger and in the second phalanx of the other fingers. The other skeletal radiographs showed widening, cupping, and defective mineralization of the metaphyses. As the diagnosis of CHH was suspected because of sparse hair, an immunoglobulin anomaly, and cone-shaped epiphyses of the phalanges, an analysis of the RMRP gene was proposed. The family was convoyed for genetic testing and was informed about the study, for which consent was signed by the parents.
Materials and Methods
Detection of RMRP Mutations
The nucleotide sequence of RMRP was retrieved from GenBank (accession No. M29916 in Homo sapiens). The secondary structure was retrieved from the Rfam database (accession No. RF00030), and the transcription initiation site was designated as + 1. Genomic DNA was isolated from peripheral leukocytes according to standard procedures [Miller et al., 1988]. The entire transcribed region and ∼115 bp of the promoter region of the human RMRP were amplified by PCR and screened for mutations by direct sequencing. PCR primers were RMRP1F (5′-ttagaaagttatgcccgaaaacg-3′), RMRP1R (5′-ctagagggagctgacggatg-3′), RMRP2F (5′-aagtccgccaagaagcgtat-3′) and RMRP2R (5′-aggtggactgatcgcttgac-3′). Afterwards, products were sequenced for both strands using an ABI PRISM™ 310 genetic analyzer (Applied Biosystems, Foster City, Calif., USA).
RNA Structure Model
Although much of the sequence dependence of RNA thermodynamics is unknown, for RNA sequences of <700 nucleotides it is possible to correctly predict roughly 70% of the secondary structure from thermodynamics alone [Mathews, 2004]. Based on this percentage, which suggests that thermodynamics is a major determinant of the secondary structure and thus of evolution of structured RNAs, we used (a) a rapid search of a possible secondary structure in the primary structure using the GArna package [Titov et al., 2000] and (b) a 2-step recursion and dynamic programming to find the one structure which has the minimum free energy [McCaskill, 1990]. A multiple sequence alignment was constructed based on sequence information alone, and then the lowest free energy structure was predicted that was common to all or most sequences [Hofacker et al., 2002; Bernhart et al., 2008]. Calculations were improved by also providing free energy change bonuses for base pair formation at sites of covariation, where the structure was conserved but the sequence was not. Algorithms were used according to their Web availability [Brodskiĭ et al., 1995; Bernhart et al., 2008; Smith et al., 2010].
Results and Discussion
Identification of Novel RMRP Variants
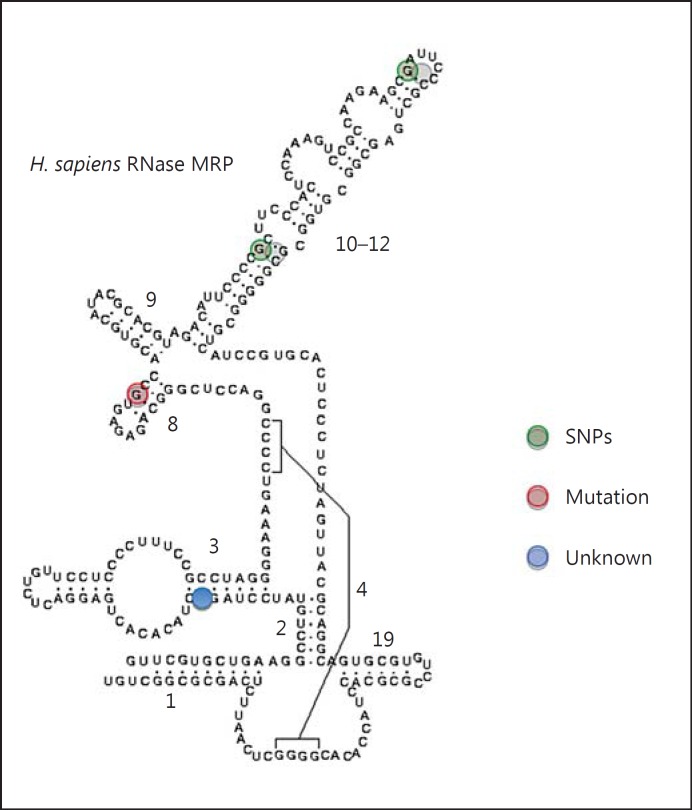
We identified several heterozygous single nucleotide variations in the patient (g.-48C>A, g.-13A>C, g.-6G>A, g.27G>C, g.97G>A, g.127G>C, and g.156G>C). Except for the variation g.97G>A which was inherited from the mother, all other variations were passed on from the father. Whilst only the g.97G>A variation was previously described as a causative mutation [Martin and Li, 2007], the other variations were either reported as polymorphisms (i.e. g.-48C>A, g.-6G>A, g.127G>C, and g.156G>C) [Hermanns et al., 2005; Martin and Li, 2007] or had never been described before (i.e. g.-13A>C and g.27G>C; fig. 1).
Fig. 1.
Nucleotide variations and RNase MRP structure. SNPs, the mutation, and the unknown variation that were identified in a Moroccan patient with CHH are shown. The secondary structure was issued from a Sanger annotation of RMRP in the Rfam database (RF00030).
Therefore, the mutation analysis of RMRP led us to consider that the patient is compound heterozygous for RMRP mutations. The mutation g.97G>A was located on the maternal allele, similarly to what has been previously described [Martin and Li, 2007], while the novel variations g.-13A>C and g.27G>C were paternally inherited.
Structural Analyses of Folded RMRP Variants
Mutations leading to CHH are predominantly found in both the transcribed region and the promoter region (from the TATA box to the transcription initiation site) of the RMRP gene. The most frequently found mutation in CHH patients is the g.70A>G point mutation [Ridanpää et al., 2003] with a founder effect in Finland [Makitie et al., 1995]. RMRP is a noncoding gene, which highlights the challenge to explore the consequences of mutations. Correct segregation of mutations in the families and absence of mutations in controls are 2 commonly used criteria. Other suggested criteria are evolutionary conservation of nucleotides involved in CHH-associated mutations and involvement in base-pairing in the secondary structure model [Bonafé et al., 2002].
Several mutations that co-segregate with the disease in multiple unrelated families with CHH do not map in known base-pairing positions. In their report of 20 novel RMRP mutations in patients with CHH, Bonafé et al. [2005] summarized all the putative pathogenic mutations and SNPs and indicated that putative pathogenic mutations are located in highly conserved nucleotides, whereas SNPs are located in non-conserved positions. They also aligned the promoter regions of the corresponding genes from various mammalian species and found a similar pattern: CHH-associated mutations occur in regions of strongly conserved nucleotide sequences, whereas non-pathogenic SNPs mainly occupy areas of nonconserved nucleotides of the promoter region.
The variation g.-13A>C is located in a poorly conserved region, which so far has been affected by pathogenic duplications or insertions but not by single base substitutions. Moreover, a population of 50 healthy Moroccan individuals was unsuccessfully screened for the g.-13A>C variation. Based on these assumptions, we consider that g.-13A>C may be a rare and nonpathogenic variation.
The other variation, g.27G>C, is located in the RMRP transcript. To assess its potential pathogenicity, a rapid search into the primary structure for a possible secondary structure was performed. As illustrated in online supplementary figure 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000430970), this nucleotide variation led to a lowered predicted significance of the first 70 nucleotides. Consistently, mapping the nucleotide variation to the RMRP structure (fig. 1) revealed its location in a domain previously referred to as the P3 helix in lower eukaryotes [Li et al., 2002; Perederina et al., 2010]. We used a simulation strategy of RNA folding to analyze the mutational consequences in the structuration of this small 46-bp-long hairpin (fig. 2). Interestingly, the g.27G>C substitution compromised the formation of this hairpin structure. Likewise, 2 other mutations, which were previously described as segregating with CHH in affected families, i.e. g.63C>T and g.64T>A, resulted in a similar impairment [Bonafé et al., 2005]. Furthermore, we did not find the variant g.27G>C in 50 healthy Moroccan individuals that were assessed as controls. Altogether, these findings lead us to refer to the variation g.27G>C as a mutation.
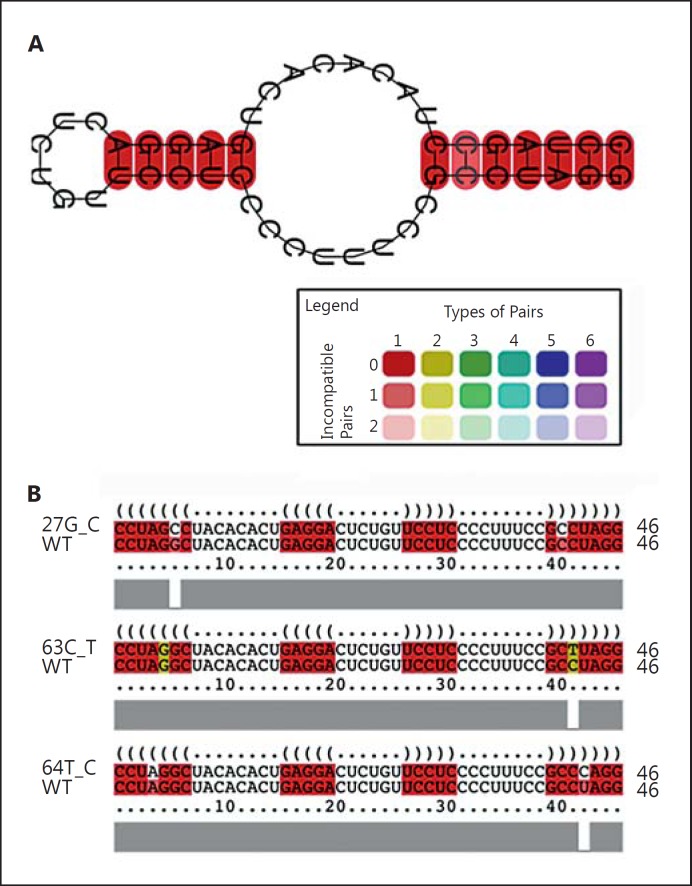
Fig. 2.
Characterization of mutations in the P3 domain. The representations were generated using RNAalifold of the Vienna Package [Bernhart et al., 2008]. A The disruptive mutation g.27G>C is shown on the consensus structure of the wild-type 2D sequence of the P3 helix. The base pairs are colored using the color code shown in the legend in a way that hue shows sequence conservation and saturation decreases with the number of incompatible base pairs. B Disruptive effects of mutations concerning the P3 domain. Alignment of each mutation located in the P3 helix is annotated with the consensus wild-type structure and its secondary structure as predicted by RNAalifold. The consensus structure is printed as a string of dots and brackets on top of the alignment. The string is well bracketed in a way that base pairs within the helix structure are indicated by corresponding opening and closing brackets. As in A, compatible base pairs are colored, and the hue uses the same color code. The mutations g.63C>T and g.64T>A, which were previously described as segregating with CHH patients, resulted in a similar impairment.
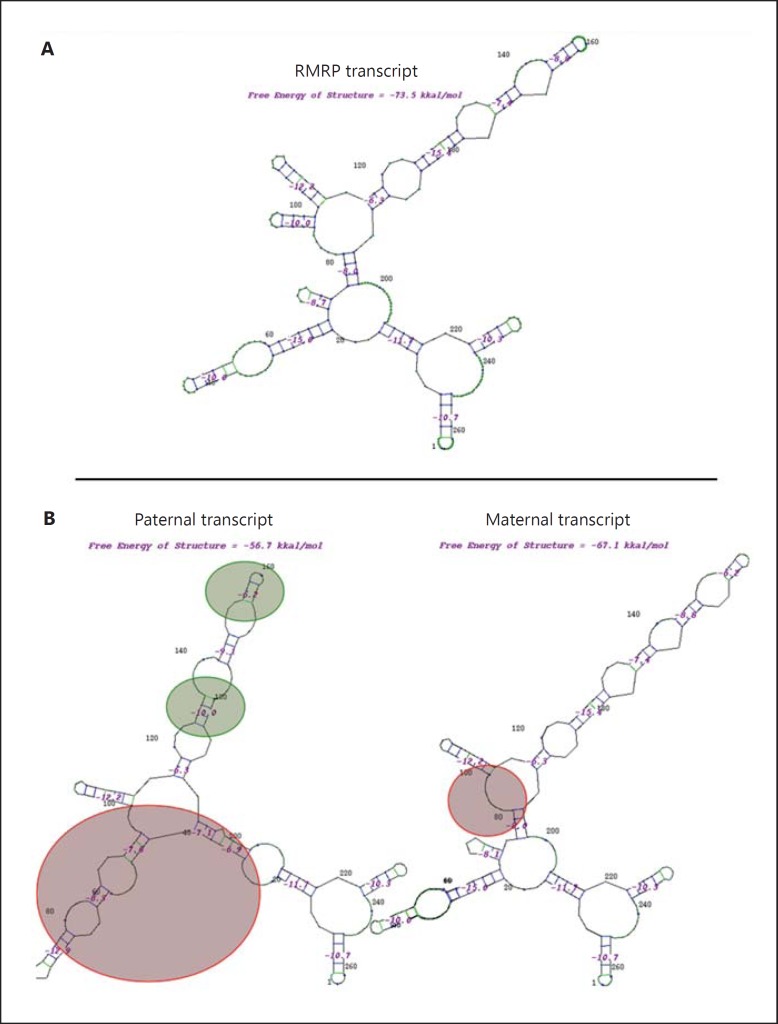
We then assessed how our hypotheses on this mutation could induce a predicted disruption of RMRP functionality. To this end, we studied the RNA folding of RMRP (fig. 3A) and the effect of all variants whether paternally or maternally inherited on the predicted RMRP structure (fig. 3B). We observed a dramatic effect of the g.27G>C mutation which, by disrupting the Watson-Crick base pairing in the P3 domain, globally impaired the overall structure of RMRP, while SNPs locally lowered the thermodynamic stability without disrupting the structure (fig. 3B). Likewise, we visualized the mutational consequence of g.97G>A in altering the RMRP structure of the maternal transcript (fig. 3B).
Fig. 3.
Mutational analyses of the 2D structure of RMRP. Representations were generated using the GeneBee RNA secondary structure prediction software [Brodskiĭ et al, 1995]. The free energy is indicated for each structure. A Secondary structure of the human RMRP transcript. A multiple alignment of human (267 bp, M29916), murine (275 bp, NR001460), and bovine (279 bp, NR036646) sequences was first carried out to retrieve the most fitted folding of human RMRP. B Mutational analyses of paternally and maternally inherited RMRP transcripts on the secondary structure. The analyzed transcript sequences comprised polymorphic variations as well. Encircled substructures indicate the structural effect of sites of variations, whether polymorphism (green) or mutation (red), on the RMRP structure.
In eukaryotes, the P3 domain has an important function. The RNase P lineage has split into 2, giving rise to a closely related enzyme, RNase MRP, which has similar components but has evolved to have different specificities. The eukaryotic RNases P/MRP have acquired an essential helix-loop-helix protein-binding RNA domain P3 that has an important function in eukaryotic enzymes and distinguishes them from bacterial and archaeal RNases P.
Altogether, these findings are indicative of the pathogenicity of this novel g.27G>C mutation, while the g.-13A>C variation may rather be considered as a rare polymorphism. Importantly, they also present a way of assessing mutations in the context of polymorphic variants on the disruption of RMRP functionality.
In conclusion, we have characterized a Moroccan patient with CHH and described a novel mutation in the RMRP gene (g.27G>C). We believe that in the future identification of RMRP mutations and their deduced consequences on the RMRP secondary structure may help to clarify their pathogenicity. This should prove an invaluable clue to delineate the phenotypic spectrum of CHH and help classify metaphyseal chondrodysplasia. At last, the ability to rapidly model the RMRP structure may in turn pave the way to the discovery of therapeutics that target RNA.
Supplementary Material
Supplementary data
References
- 1.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonafé L, Schmitt K, Eich G, Giedion A, Superti-Furga A. RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin Genet. 2002;61:146–151. doi: 10.1034/j.1399-0004.2002.610210.x. [DOI] [PubMed] [Google Scholar]
- 3.Bonafé L, Dermitzakis ET, Unger S, Greenberg CR, Campos-Xavier BA, et al. Evolutionary comparison provides evidence for pathogenicity of RMRP mutations. PloS Genet. 2005;1:444–454. doi: 10.1371/journal.pgen.0010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodskiĭ LI, Ivanov VV, Kalaĭdzidis IaL, Leontovich AM, Nikolaev VK, et al. GeneBee-NET: an internet based server for biopolymer structure analysis. Biokhimiia. 1995;60:1221–1230. [PubMed] [Google Scholar]
- 5.Castigli E, Irani AM, Geha RS, Chatila T. Defective expression of early activation genes in cartilage-hair hypoplasia (CHH) with severe combined immunodeficiency (SCID) Clin Exp Immunol. 1995;102:6–10. doi: 10.1111/j.1365-2249.1995.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, et al. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum Mol Genet. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 7.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Frank DN, Pace N, Zengel JM, Lindahl L. Phylogenetic analysis of the structure of RNase MRP in yeast. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makitie O, Sulisalo T, de la Chapelle A, Kaitila I. Cartilage-hair hypoplasia. J Med Genet. 1995;32:39–43. doi: 10.1136/jmg.32.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makitie O, Kaitila I, Savilahti E. Susceptibility to infections and in vitro immune functions in cartilage-hair hypoplasia. Eur J Pediatr. 1998;157:816–820. doi: 10.1007/s004310050943. [DOI] [PubMed] [Google Scholar]
- 11.Martin AN, Li Y. RNase MRP RNA and human genetic diseases. Cell Res. 2007;17:219–226. doi: 10.1038/sj.cr.7310120. [DOI] [PubMed] [Google Scholar]
- 12.Mathews D. Predicting the secondary structure common to two RNA sequences with Dynalign. Curr Protoc Bioinformatics. 2004;12:Unit 12.4. doi: 10.1002/0471250953.bi1204s08. [DOI] [PubMed] [Google Scholar]
- 13.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 14.McKusick VA. Metaphyseal dysostosis and thin hair; a ‘new’ recessively inherited syndrome? Lancet. 1964;1:832–833. doi: 10.1016/s0140-6736(64)93029-6. [DOI] [PubMed] [Google Scholar]
- 15.McKusick VA, Eldridge R, Hostetler JA, Ruangwit U, Egeland JA. Dwarfism in the Amish. II. Cartilage-hair hypoplasia. Bull Johns Hopkins Hosp. 1965;116:285–326. [PubMed] [Google Scholar]
- 16.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakashima E, Mabuchi A, Kashimada K, Onishi T, Zhang J, et al. RMRP mutations in Japanese patients with cartilage-hair hypoplasia. Am J Med Genet. 2003;123A:253–256. doi: 10.1002/ajmg.a.20281. [DOI] [PubMed] [Google Scholar]
- 18.Perederina A, Esakova O, Quan C, Khanova E, Krasilnikov AS. Eukaryotic ribonucleases P/MRP: the crystal structure of the P3 domain. EMBO J. 2010;29:761–769. doi: 10.1038/emboj.2009.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 20.Ridanpää M, Jain P, McKusick VA, et al. The major mutation in the RMRP gene causing CHH among the Amish is the same as that found in most Finnish cases. Am J Med Genet C Semin Med Genet. 2003;121:81–83. doi: 10.1002/ajmg.c.20006. [DOI] [PubMed] [Google Scholar]
- 21.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiel CT, Rauch A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best Pract Res Clin Endocrinol Metab. 2011;25:131–142. doi: 10.1016/j.beem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, et al. Severely incapaciting mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Titov II, Ivanisenko VA, Kolchanov NA. FITness – a WWW-resource for RNA folding simulation based on genetic algorithm with local minimization. Comput Techn. 2000;5:48–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data