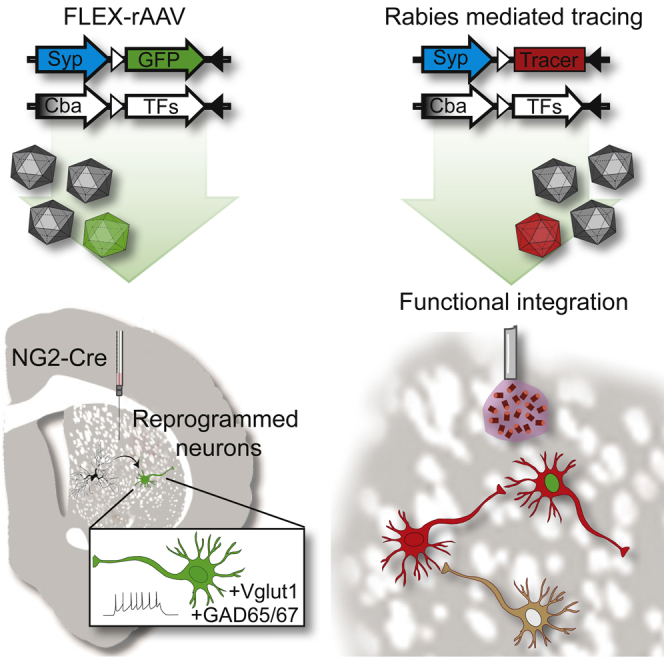
Summary
The possibility of directly converting non-neuronal cells into neurons in situ in the brain would open therapeutic avenues aimed at repairing the brain after injury or degenerative disease. We have developed an adeno-associated virus (AAV)-based reporter system that allows selective GFP labeling of reprogrammed neurons. In this system, GFP is turned on only in reprogrammed neurons where it is stable and maintained for long time periods, allowing for histological and functional characterization of mature neurons. When combined with a modified rabies virus-based trans-synaptic tracing methodology, the system allows mapping of 3D circuitry integration into local and distal brain regions and shows that the newly reprogrammed neurons are integrated into host brain.
Graphical Abstract
Highlights
-
•
Ascl1, Lmx1a, and Nurr1 convert striatal NG2 glia into functional neurons
-
•
Cre-inducible AAV conversion vectors result in efficient neural conversion in vivo
-
•
A neuron-specific reporter allows for long-term phenotypic and functional analysis
-
•
Rabies-based tracing show that converted neurons integrate into local circuitry
Torper et al. have developed an AAV-based system for in vivo neural conversion with neuron-specific reporters allowing for long-term histological and functional characterization. The in-vivo-converted neurons are shown to be functional and integrate into host neural circuits, as assessed with rabies-virus-based tracing.
Introduction
Lineage conversion, where one terminally differentiated cell is converted into another terminally differentiated cell type via viral-mediated expression of transcription factors, has recently been achieved for many cell types (Vierbuchen and Wernig, 2011), including cells of the CNS such as neurons, astrocytes, oligodendrocytes, and neural progenitor cells (Caiazzo et al., 2015; Thier et al., 2012; Vierbuchen et al., 2010; Yang et al., 2013). In the brain, resident glia and transiently appearing proliferating progenitors have been successfully converted into neurons or neural progenitor cells in situ using defined combinations of transcription factors (Guo et al., 2014; Torper et al., 2013), and in some cases, functional properties such as action potentials and/or postsynaptic currents have been observed (Heinrich et al., 2014; Niu et al., 2013). However, to date, it is not clear if the new neurons can integrate into the host neuronal circuitry, and studies of this are partly hampered by lack of good experimental systems.
In this study, we present an adeno-associated virus (AAV)-based conversion system that can be used to specifically target and convert resident glia at high efficiency. When combined with a neuron-specific reporter system, it allows for long-term monitoring and functional analysis of the reprogrammed neurons. Using this system, we show that the striatal NG2 glia can be converted to GABAergic and glutamatergic neurons that remain stable over a long period of time and display electrophysiological properties of functional neurons. By combining the reporter system with virus-based tracing methodology, we show that the new neurons integrate into local circuitry in an efficient manner.
Results
To specifically target NG2-expressing cells, we developed Cre-recombinase-dependent AAV vectors. In these flip-excision (FLEX) vectors, the conversion factors and reporter genes are inserted in an antisense direction, flanked by two pairs of antiparallel, heterotypic loxP sites (Atasoy et al., 2008). Each of the three conversion factors, Ascl1, Lmx1a, and Nurr1 (ALN), are placed under the control of the ubiquitous chicken β-actin (cba) promoter on individual vectors (Figure S1A). To construct a reporter system that restricts GFP expression to reprogrammed neurons, we placed GFP under the control of the neuron-specific synapsin promoter in a FLEX vector (Figure S1A) so that GFP would expressed only in neurons originating from a Cre-expressing cell.
To validate the vectors, we first transfected T293 cells and transduced the cultures with a lentiviral vector expressing Cre. In the absence of Cre, there is no transgene expression. In the presence of Cre, the transgene is excised and recombined stably in a correctly reinserted orientation, allowing the transgene to be transcribed and protein to be expressed (Figure S1B). Next, we injected our vectors into the striatum of transgenic mice aged 12–16 weeks (Figure 1A) that express Cre under the control of the mouse NG2 (Cspg4) promoter/enhancer (Zhu et al., 2008). When injecting a ubiquitous reporter vector (cba-GFP, n = 5; Figure S1A) that expresses GFP in all Cre-positive cells, we found that GFP was exclusively expressed in NG2 glia and co-localized with NG2 (Figure 1C), SOX10, and PDGFRα (Figures S1F and S1G). When we injected our new neuron-specific reporter (syn-GFP) into the striatum of NG2-cre-expressing mice aged 12–16 weeks, we did not detect any GFP-expressing cells (Figure 1B), with the exception of 1 animal out of 11 analyzed, where we observed a total of 18 GFP+ cells. We also injected the cba-GFP and syn-GFP vectors into wild-type mice and never detected any GFP expression (Figure S1C; n = 10 for each reporter). Taken together, these data confirm a specific Cre-dependent control of gene expression only in NG2-expressing cells in vivo and establish that the neuron-specific reporter is not active in these cells. All subsequent experiments were performed in mice aged 12–16 weeks; we included control animals injected with syn-GFP only in each experiment and never observed any conversion in these animals (Figures 1G and 1I).
Figure 1.
Ascl1, Lmx1a, and Nurr1 Efficiently Convert NG2 Glia into Neurons
(A) Schematic of experimental procedures.
(B) The syn-GFP reporter constructs are not expressed in NG2 glia.
(C) Cre-inducible GFP reporter under the ubiquitously expressed promoter cba (green) co-labels with NG2 (red).
(D–G) Converted cells appear after 1 week and increase in number and morphological maturity by 12 weeks.
(H and I) NG2 glia can be converted into neurons by Ascl1 alone, albeit in lower numbers than when using ALN for conversion.
Scale bars represent 50 μm in (C)–(F) and 10 μm in (D′)–(F′). A, Ascl1; L, Lmx1a; N, Nurr1. Error bars represent SEM. See also Figure S1.
To analyze the potential for neural conversion of striatal NG2 glia using ALN as conversion factors, we injected a mix of ALN and syn-GFP (1:1:1:1) into the striatum of NG2-Cre mice and also injected syn-GFP alone as control (n ≥ 4 for each time point and vector combination). Animals were sacrificed 2 days and 1, 4, and 12 weeks after vector injections. In the ALN-injected group, no GFP-positive cells could be detected when analyzed after 2 days of injection (n = 4, not shown), and only a few GFP-positive cells could be detected when analyzed 1 week after injection (n = 6; Figures 1D and 1D′). The number of GFP-positive cells increased by 4 and 12 weeks (n = 4 and 7 for each time point, respectively; Figures 1E, 1E′, 1F, and 1F′). No GFP-expressing cells could be detected in the control-injected animals (Figures 1G, S1D, and S1E). Quantifications showed an increasing number of converted GFP+ cells over time, reaching 6,912 ± 3,052 per animal at 4 weeks (n = 3). As Ascl1 alone can convert somatic cells into neurons whereas Lmx1a and Nurr1 cannot (Chanda et al., 2014, Pfisterer et al., 2011), we next performed injections with Ascl1 only and compared these mice with ALN-injected and control animals. We found some converted neurons in the Ascl1-only group (Figure 1H), albeit in much lower numbers compared to ALN-injected animals (Figure 1I). The control animals injected with syn-GFP only did not contain any reprogrammed neurons (Figure 1I).
To establish a neuronal phenotype of the ALN-converted cells, we stained for the neuronal marker NeuN. The GFP+ cells co-expressed NeuN (Figures 2A–2A′′), and the proportion of converted cells co-expressing GFP and NeuN increased from 20.8% ± 5.9% (n = 3) at 4 weeks to 46.8% ± 2.9% (n = 3) after 12 weeks (Figure 2B). The reprogrammed cells also co-expressed MAP2, and confocal analysis confirmed the presence of cells positive for syn-GFP, NeuN, and MAP2, further confirming a mature neuronal phenotype of the converted cells (Figures 2C–2C′′′). Staining for NG2 and GFP confirmed that no cells co-expressed GFP and NG2, suggesting a rapid loss of glial properties in the converted cells (Figures 2D–2D′′).
Figure 2.
In-Vivo-Converted Neurons Express Pan-neuronal Markers and Downregulate NG2
(A and B) NeuN (blue) is expressed in 20.8% ± 5.9% (n = 3) of re-programmed cells at 4 weeks after injection and 46.8% ± 2.9% (n = 3) of re-programmed cells at 12 weeks after injection.
(C––D′′) Converted neurons co-express GFP (green), MAP2 (red), and NeuN (blue) (C–C′′′) but lose their NG2 expression (D–D′′).
(E and F) The converted neurons express vGlut1 (red) (E) and GAD65/67 (red) (F).
Scale bars represent 25 μm in (A)–(C′′′), 50 μm in (D)–(E), and 10 μm in (F). Error bars represent SEM. See also Figure S2.
Next, we co-stained GFP with a panel of subtype-specific markers: tyrosine hydroxylase (TH) for dopaminergic neurons, GAD65/67 for GABAergic neurons, vGlut1 for glutamatergic neurons, DARPP32 for striatal projection neurons, and paravalbumin (PA), calbindin (CALB), and choline acetyltransferase (Chat) for striatal interneurons. The gene combination used in this study (ALN) has been shown to convert mouse fibroblasts into functional neurons, including dopamine neurons, in vitro (Caiazzo et al., 2011). However, we did not detect any converted neurons expressing TH, even though ALN was used to drive neural conversion in vivo (Figure S2F). Given that all factors were delivered on separate vectors, the lack of TH could potentially be explained by lack of co-expression of the dopamine fate determinants in the reprogrammed neurons. However, when analyzing this, we found a large overlap in expression when staining for GFP and NURR1 as well as for GFP and LMX1A (Figures S1H–S1I′′). Quantifications showed that 79% ± 8% and 83.7% ± 6% of the GFP+ neurons co-expressed LMX1A and NURR1, respectively. To exclude that the lack of a dopamine-like phenotype is due to the cell of origin, we repeated the experiment using astrocytes instead of NG2 glia as a starter population for neural conversion. In these studies, we used a GFAP-Cre mouse to specifically target the resident astrocyte population. We injected syn-ALN and syn-GFP into the striatum of GFAP-Cre mice as well as syn-GFP only (n = 5 for each group). 6 weeks after vector injections, we could detect a high level of neural conversion as assessed by GFP expression (Figure S2F) in the animals injected with ALN, but not in animals injected with syp-GFP alone, matching what we have reported from reprogramming GFAP-Cre glia using Ascl1, Brn2a, and Mytl1 (Torper et al., 2013). Co-staining with TH showed that the ALN-converted neurons originating from GFAP+ astrocytes, like the ALN-converted neurons originating from NG2+ glia, do not express TH (Figure S2F). 16% of the reprogrammed neurons expressed vGlut1 (Figure 2E), and 68% expressed GAD65/67 (Figures 2F and S2A), while none were found to co-express DARPP32, PA, CALB, or CHAT (Figures S2B–S2E).
Twelve weeks after conversion, the reprogrammed neurons had acquired a mature neuronal morphology, and dendritic spine-like protrusions could be observed (Figure 3A). At this time point, we used patch-clamp electrophysiology to investigate the functional properties of the neurons obtained (Figure 3B). All the GFP-expressing neurons recorded showed functional electrophysiological properties of neurons, with a resting membrane potential (V0) of −61.4 ± 9.7 mV (n = 5) and the ability to evoke repetitive action potentials after current injections, with a mean amplitude of 33.5 ± 2.29 mV and an action potential threshold of 25 ± 7.19 pA (Figure 3C; Table S1; n = 6). Furthermore, all the converted cells recorded from showed fast-inactivated inward and outward currents characteristic of sodium and delayed-rectifier potassium currents, with a peak amplitude of INa at 625 ± 131 pA and IK at 2,020 ± 290 pA (Figure 3D; n = 6). Both action potentials and inward currents could be blocked with the neurotoxin tetrodotoxin (TTX) (Figure 3E). We could also detect post-synaptic events with a mean frequency of 3.67 ± 1.93 Hz and a peak amplitude of 5.03 ± 0.64 pA, indicative of formation of a functional postsynaptic compartment and possible establishment of synaptic networks (Figure 3F; Table S1).
Figure 3.
Reprogrammed Neurons Show Electrophysiological Properties of Functional Neurons
(A) Converted neurons acquire a mature neuronal morphology and exhibit dendrite spine-like protrusions.
(B) Example of a cell selected for patch-clamp recordings 12 weeks after conversion.
(C and D) Reprogrammed cells evoke action potentials (AP) upon depolarizing current injection (C) and inward sodium (Na+) and outward potassium (K+) current upon depolarization in artificial cerebrospinal fluid (ACSF) (D).
(E) Action potentials and inward outward current could be blocked by adding the neurotoxin TTX to the buffer.
(F) Examples of spontaneous post-synaptic currents.
See also Table S1.
Functional neuronal properties, including post-synaptic events, have previously been described for in-vivo-reprogrammed striatal astrocytes and for reprogrammed cortical NG2 glia (Heinrich et al., 2014; Niu et al., 2013), but these types of recordings cannot distinguish between functional integration with host neurons and communication among reprogrammed neurons. To confirm circuitry integration, and to map what neurons form synaptic inputs to the newly reprogrammed cells, we used modified rabies virus (ΔG-rabies) to trace connectivity of the reprogrammed neurons. ΔG-rabies is a deletion mutant virus where the gene coding for glycoprotein (GP; necessary for transsynaptic spread) is replaced by the gene coding for mCherry (Wickersham et al., 2007a, 2007b) and pseudotyped to infect cells only via the avian tva receptor (Barnard et al., 2006). To selectively trace connectivity of our in-vivo-reprogrammed neurons, we constructed a FLEX tracing vector with a tricistronic construct encoding for nuclear GFP (ncGFP), tva receptor, and rabies GP under the synapsin promoter (Figure S3A). Expression of this vector is exclusive to the reprogrammed neurons and renders them infectable with Δrabies via the tva receptor. Since the reprogrammed neurons also express GP from the tracing construct, the rabies can assemble into infectious particles in the reprogrammed neurons and transmit mCherry retrogradely across one synapse to host neurons that provide efferent input. Thus, targeted reprogrammed neurons (starter neurons) will appear red and green, while traced host neurons that synapse onto the starter neurons appear red only (schematic in Figure 4A). As a positive control, we used injection of unflexed tracing vectors containing the same synapsin-driven tricistronic construct with ncGFP, tva, and GP (Figure S3B). Because it is not FLEXed, this vector infects host neurons and thus traces the connectivity of endogenous striatal cells. We also injected animals with a vector containing ncGFP and tva but lacking GP (Figure S3B). This control vector allows for Δrabies infection via the tva receptor but no transsynaptic spreading, as the cell lacks GP (Wickersham et al., 2007a).
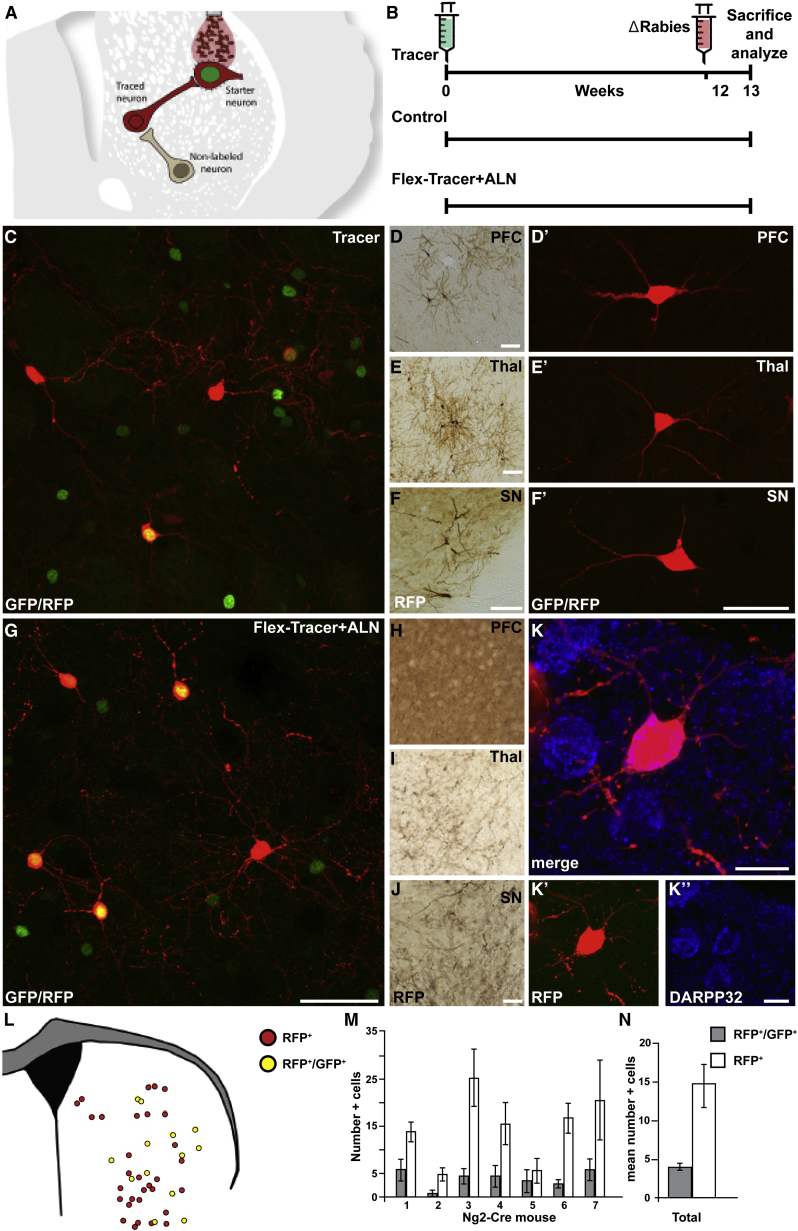
Figure 4.
Reprogrammed Neurons Are Innervated by the Pre-existing Local Neural Circuitry
(A) Schematic of modified-rabies-mediated monosynaptic tracing. Reprogrammed cells infected by modified rabies appear with a green nucleus and a red cytoplasm, whereas any neuron providing synaptic input to the reprogrammed neurons appears as red only.
(B) Experimental timeline.
(C) Injection of unflexed tracer serves as positive methodological control. Targeted host neurons (green and red) receive synaptic input from local striatal neurons (red only).
(D–F′) Traced neurons (DAB and red) are also detected in the motor cortex (PFC), thalamus (Thal), and substantia nigra (SN).
(G) Tracing of reprogrammed neurons using FLEX tracer + ALN (red and green) shows traced host neurons (red).
(H–J) The absence of traced neurons in PFC, Thal, and SN suggest that the reprogrammed cells integrate only locally.
(K–K′′) Co-labeling of mCherry and DARRP32 (blue) shows that the traced neurons are of a medium spiny neuron subtype.
(L) Distribution of the reprogrammed neurons (yellow) and traced neurons (red) in a representative animal.
(M and N) Quantifications of reprogrammed (gray bars) and traced (white bars) neurons (n = 7).
Scale bars represent 50 μm in (C) and (D′)–(G), 100 μm in (D)–(F) and (H)–(J), and 10 μm in (K)–(K′′). Error bars represent SEM. See also Figure S3.
Three groups of animals were injected with (1) unflexed tracing vector as a positive control (tracer-control), (2) flexed tracing vector together with ALN to trace connectivity of reprogrammed cells (FLEX tracer + ALN), and (3) control vector lacking GP. 12 weeks after injection of these tracing and control vectors, Δrabies was injected in the same area, and 1 week later, the animals were perfused and used for histology (Figure 4B). In animals injected with tracer control (that traces from endogenous neurons), we could detect targeted host starter cells expressing GFP and mCherry at the injection site (Figure 4C), as well as traced-red-only cells (i.e., striatal neurons that have made a synaptic contact with the targeted cells) (Figure 4C). Outside the striatum, we could also detect traced mCherry+/GFP− neurons in structures that normally innervate the striatum, such as the motor cortex (Figure 4D), thalamus (Figure 4E), and substantia nigra (Figure 4F). In the animals injected with control vector lacking GP, no tracing was observed (Figure S3C), confirming the transsynaptic spread of mCherry.
To trace connectivity of the reprogrammed neurons, animals were injected with the FLEX tracer vector in combination with ALN (Figure 4B). In these animals, we detected targeted reprogrammed starter cells expressing GFP and mCherry at the injection site (Figure 4G). Also in this group, red-only-traced cells representing endogenous host neurons that have made a synaptic contact with the reprogrammed neurons could be detected (Figure 4G). Co-staining of mCherry with DARPP32 showed that host cells that connect with the reprogrammed neurons are of the medium spiny neuron subtype (Figures 4K–4K′′). Interestingly, no mCherry-positive neurons could be detected outside the striatum in this group, indicating that the new neurons preferentially integrate into local circuitry (Figures 4H–4J).
The starter neurons and the traced neurons were evenly distributed within the striatum (Figure 4L). Quantifications of the reprogrammed starter neurons and the traced host neurons showed that each animal contained more traced host neurons than reprogrammed starter neurons (Figure 4M) and that, on average, each reprogrammed neuron was contacted by three or four host neurons (Figure 4N).
Discussion
Recent studies have shown that it is possible to convert endogenous glia into functional neurons in vivo, which offers new possibilities for brain repair (Amamoto and Arlotta, 2014). With few exceptions (Heinrich et al., 2014; Niu et al., 2013), the neuronal properties of the in-vivo-reprogrammed neurons have mostly been assessed based on marker expression (Guo et al., 2014; Magnusson et al., 2014; Torper et al., 2013) or in vitro (Guo et al., 2014).
In this study, we have developed an AAV-based vector system for in vivo neural conversion that results in efficient targeting and conversion of glia in situ. Importantly, the reporters used in this system are designed to be exclusive to the newly reprogrammed neurons and remain stable over time, allowing for long-term analysis of phenotype and function of the new neurons. The reporter can be combined with virus-based tracing methodology, which permits mapping of circuitry integration and allows for the precise identification of the source of host afferent inputs to the reprogrammed neurons.
The vector combination used in this study (ALN) has previously been used to reprogram mouse fibroblasts and glia into dopamine neurons in vitro (Addis et al., 2011; Caiazzo et al., 2011; Torper et al., 2013), yet no TH-expressing neurons could be detected when astrocytes or NG2 glia were converted in vivo. This mimics previous findings on astrocyte conversion, where several factors have been shown to convert astrocytes into neurons in vitro (Berninger et al., 2007; Heinrich et al., 2010) yet fail to do so in vivo (Su et al., 2014).
Nevertheless, the use of ALN results in functionally mature neurons in larger proportions than previously reported for conversion of resident glia using Sox2 alone or in combination with Mash1 (Heinrich et al., 2014), without the need for treatment with neurotrophic factors or histone deacetylase inhibitor as previously reported as a requirement for neuronal maturation (Niu et al., 2013). We made use of a monosynaptic tracing methodology based on modified rabies virus to analyze if and how the new reprogrammed neurons integrate with existing host neurons. Rabies-based tracing has previously been used to trace connectivity of endogenous neural circuitry (Miyamichi et al., 2011; Watabe-Uchida et al., 2012), as well as newly born neurons in the olfactory bulb and hippocampus (Deshpande et al., 2013; Vivar et al., 2012). Here, we used it to map efferent connectivity to the newly reprogrammed neurons and revealed that they are extensively innervated by host striatal projection neurons.
Experimental Procedures
Animals and Surgery
All experimental procedures were carried under the European Union Directive (2010/63/EU) and approved by the ethical committee for the use of laboratory animals at Lund University and the Swedish Department of Agriculture (Jordbruksverket). Surgery was performed under general anesthesia using 2% isoflurane in a mix of air an N2O at a 4:1 ratio. For conversion, 1 μl vector was injected in the striatum of each animal at a rate of 0.2 μl/min and a diffusion time of 2 min at the coordinates A/p = +0.5, M/L = −2.0, and D/V = −2.7, toothbar = flat. For tracing experiments, tracing vectors plus ALN were injected at the same dilutions and volume as above at the same coordinates, and tracing vector alone was injected contra-laterally (M/L 2.0). Animals were kept for 11 weeks, and 1 week prior to perfusion, animals were injected with ΔG-rabies diluted to 5% of stock (20–30 × 106 TU/ml) in three consecutive injections of two deposits each (0.5 μl/deposit) flanking the original injection at coordinates (1) A/P +1.4; M/L −2.0; (2) A/P +0.4; M/L −2.0; or (3) A/P 0.9; M/L −1.5. A depth of D/V −3 and −2 was used for all sites.
Patch-clamp electrophysiology was performed on coronal brain slices at 12 weeks post-conversion. Mice were killed by decapitation, and brains were rapidly taken out and cut on a vibratome at 300 μm. Slices were transferred to a recording chamber and submerged in a continuously flowing Krebs solution gassed with 95% O2 and 5% CO2 at 28°C. The composition of the standard solution was 126 mM NaCl, 2.5 mM KCl, 1.2 mM NaH2PO4-H2O, 1.3 mM MgCl2-6H2O, and 2.4 mM CaCl2-6H2O. Voltage-gated sodium channels were blocked with 1 μM TTX (Tocris). Recordings were made using Multiclamp 700B (Molecular Devices), and signals were acquired at 10 kHz using pClamp10 software and a data acquisition unit (Digidata 1440A, Molecular Devices). Input resistances and injected currents were monitored throughout the experiments.
Converted cells were identified with their GFP fluorescence and patched with borosilicate glass pipettes (3–7 MOhm) filled with the following intracellular solution: 122.5 mM potassium gluconate, 12.5 mM KCl, 0.2 mM EGTA, 10 mM HEPES, 2 mM MgATP, 0.3 mM Na3GTP, and 8 mM NaCl adjusted to pH 7.3 with KOH. Resting membrane potentials were monitored immediately after breaking-in in current-clamp mode. Thereafter, currents were injected from −20 pA to +90 pA with 10-pA increments to induce action potentials. For sodium and potassium current measurements, cells were clamped at −70 mV and voltage-depolarizing steps were delivered for 100 ms at 10-mV increments. Spontaneous activity was measured in voltage-clamp mode using the same internal solution at resting membrane potentials. Group mean values were calculated using GraphPad Prism.
For details on cloning, virus production, immunohistochemistry, and quantifications, please see the Supplemental Experimental Procedures.
Author Contributions
O.T., D.R.O, M. Pereira, S.L., and T.C. performed and designed experiments. O.T., D.R.O., S.G., and M. Parmar designed experiments, guided preparation of the data and figures, and led data interpretation and analysis. O.T., S.G., and M. Parmar wrote the manuscript. All authors gave input to manuscript.
Acknowledgments
We would like to thank Ed Callaway for valuable help establishing monosynaptic tracing and producing modified rabies virus; Johan Jakobsson and Anders Björklund for scientific discussions and proof reading of manuscript; and Ulla Jarl, Bengt Mattsson, Michael Sparrenius, Jenny Johansson, and Ingar Nilsson for excellent technical assistance. This study was supported by grants from the European Community’s 7th Framework Programme through NeuroStemcellRepair (nr. 602278), The Strategic Research Area at Lund University Multipark (multidisciplinary research in Parkinson’s disease), the Swedish Research Council (70862601/Bagadilico and K2014-61X-20391-08-4), and the Swedish Parkinson Foundation (Parkinsonfonden). S.G. was supported by a postdoctoral stipend from the Swedish Brain Foundation (Hjärnfonden). The research leading to these results has received funding from the European Research Council under the European Union’s 7th Framework Programme (FP/2007-2013)/ERC grant agreement 309712.
Published: July 9, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.06.040.
Supplemental Information
References
- Addis R.C., Hsu F.C., Wright R.L., Dichter M.A., Coulter D.A., Gearhart J.D. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS ONE. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amamoto R., Arlotta P. Development-inspired reprogramming of the mammalian central nervous system. Science. 2014;343:1239882. doi: 10.1126/science.1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D., Aponte Y., Su H.H., Sternson S.M. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard R.J., Elleder D., Young J.A. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology. 2006;344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Götz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Giannelli S., Valente P., Lignani G., Carissimo A., Sessa A., Colasante G., Bartolomeo R., Massimino L., Ferroni S. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports. 2015;4:25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S., Ang C.E., Davila J., Pak C., Mall M., Lee Q.Y., Ahlenius H., Jung S.W., Südhof T.C., Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A., Bergami M., Ghanem A., Conzelmann K.K., Lepier A., Götz M., Berninger B. Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proc. Natl. Acad. Sci. USA. 2013;110:E1152–E1161. doi: 10.1073/pnas.1218991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., Sánchez R., Tiedt S., Schroeder T., Götz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Bergami M., Gascón S., Lepier A., Viganò F., Dimou L., Sutor B., Berninger B., Götz M. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Reports. 2014;3:1000–1014. doi: 10.1016/j.stemcr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson J.P., Göritz C., Tatarishvili J., Dias D.O., Smith E.M., Lindvall O., Kokaia Z., Frisén J. A latent neurogenic program in astrocytes regulated by Notch signaling in the mouse. Science. 2014;346:237–241. doi: 10.1126/science.346.6206.237. [DOI] [PubMed] [Google Scholar]
- Miyamichi K., Amat F., Moussavi F., Wang C., Wickersham I., Wall N.R., Taniguchi H., Tasic B., Huang Z.J., He Z. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Zang T., Zou Y., Fang S., Smith D.K., Bachoo R., Zhang C.L. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Björklund A., Lindvall O., Jakobsson J., Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Niu W., Liu M.L., Zou Y., Zhang C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M., Wörsdörfer P., Lakes Y.B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M.M. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Torper O., Pfisterer U., Wolf D.A., Pereira M., Lau S., Jakobsson J., Björklund A., Grealish S., Parmar M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA. 2013;110:7038–7043. doi: 10.1073/pnas.1303829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Wernig M. Direct lineage conversions: unnatural but useful? Nat. Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivar C., Potter M.C., Choi J., Lee J.Y., Stringer T.P., Callaway E.M., Gage F.H., Suh H., van Praag H. Monosynaptic inputs to new neurons in the dentate gyrus. Nat. Commun. 2012;3:1107. doi: 10.1038/ncomms2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M., Zhu L., Ogawa S.K., Vamanrao A., Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wickersham I.R., Finke S., Conzelmann K.K., Callaway E.M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham I.R., Lyon D.C., Barnard R.J., Mori T., Finke S., Conzelmann K.K., Young J.A., Callaway E.M. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Zuchero J.B., Ahlenius H., Marro S., Ng Y.H., Vierbuchen T., Hawkins J.S., Geissler R., Barres B.A., Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Bergles D.E., Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.