Abstract
The coffee berry borer, Hypothenemus hampei, is the most economically important insect pest of coffee worldwide. We present an analysis of the draft genome of the coffee berry borer, the third genome for a Coleopteran species. The genome size is ca. 163 Mb with 19,222 predicted protein-coding genes. Analysis was focused on genes involved in primary digestion as well as gene families involved in detoxification of plant defense molecules and insecticides, such as carboxylesterases, cytochrome P450, gluthathione S-transferases, ATP-binding cassette transporters, and a gene that confers resistance to the insecticide dieldrin. A broad range of enzymes capable of degrading complex polysaccharides were identified. We also evaluated the pathogen defense system and found homologs to antimicrobial genes reported in the Drosophila genome. Ten cases of horizontal gene transfer were identified with evidence for expression, integration into the H. hampei genome, and phylogenetic evidence that the sequences are more closely related to bacterial rather than eukaryotic genes. The draft genome analysis broadly expands our knowledge on the biology of a devastating tropical insect pest and suggests new pest management strategies.
Worldwide, the genus Hypothenemus (Coleoptera: Curculionidae, Scolytinae) consists of 181 described species, of which only H. hampei (Ferrari), commonly known as the coffee berry borer, is considered an important agricultural pest1. The insect is present in most coffee-producing countries and damage is initiated by a 1.6–1.9 mm long adult colonizing female, which bores a hole in the coffee berry and deposits eggs in galleries within the two seeds inside the berry1. Upon hatching, larvae consume the seeds causing significant losses in yield and quality. Yearly losses in Brazil, the world’s top coffee producer, have been estimated at US$215–358 million2. The insect exhibits sibling mating inside the coffee berry, with females being preponderant, and males never leave the berry1. Even though the insect has been the subject of study for more than 100 years3, it still remains a major threat to coffee production and no consistently reliable pest management strategies are available to manage the insect.
We have sequenced the genome of female coffee berry borers in an attempt to gain a better understanding of the basic biology of the insect. Only two other Coleopteran genomes have been published to date: the red flour beetle, Tribolium castaneum4 (Tenebrionidae), and the mountain pine beetle, Dendroctonus ponderosae5, an insect in the same subfamily (Scolytinae) as the coffee berry borer. Analysis was focused on genes that could reveal important aspects of the insect’s biology, including some that might be useful in developing novel pest management strategies.
Results and Discussion
Gene Content and Draft Assembly
After quality filtering, a total of 62 Gbp of sequence data (36 Gbp from a 350 bp insert library and 26 Gbp from a 550 bp insert library) was used for genome assembly. With an estimated genome size of 200 Mb used for the assembly software (based on the D. ponderosae genome size of 204 Mb5), this corresponds to an average depth of coverage of 180-fold. Sequence reads were assembled using SOAPdenovo2, resulting in 163 Mb in scaffolds (see Table 1 and Table S1 for assembly statistics). The scaffolds N50 was 44.7 Kb and the contigs N50 was 10.5 Kb (Table 1). Nuñez et al.6 reported an estimated coffee berry borer genome size of 170–180 Mb with 20,653 unigenes, while Benavides et al.7 reported a 194 Mb genome, with ~20,500 unigenes. RepeatMasker analysis shows that only 2.7% of the assembled genome contains identifiable repeats and low complexity regions, which is very low for an arthropod genome, suggesting that repeated sequences may be underrepresented in the assembly.
Table 1. Hypothenemus hampei genomic DNA (SOAPdenovo2) and RNA-seq (SOAPdenovo-Trans) assembly statistics.
| DNA | |
| N50 scaffolds, bp | 44,715 |
| Genome size (including N), Mb | 162,950,840 |
| Genome size (without N), Mb | 156,695,323 |
| Scaffold number | 86,848 |
| GC content | 32.46% |
| N50 contigs, bp | 10,499 |
| Longest scaffold, bp | 440,081 |
| Gene models (predicted genes) | 19,222 |
| RNA | |
| N50 | 1,638 |
| Size (including N) | 28,722,952 |
| Size (without N) | 28,327,000 |
| GC content | 35.88% |
Transcriptome
RNA-seq reads were assembled into de novo transcripts using the SOAPdenovo-Trans software8. These predicted transcripts were functionally annotated by BLASTx similarity search against the NCBI non-redundant protein database. An additional set of “genome guided” predicted transcripts was produced from a combination of RNA-seq and genomic data using the TopHat/Cufflinks software package. RNA-seq reads were aligned to the genomic contigs using TopHat29, and Cufflinks210 was used for transcript assembly, yielding a set of 15,546 predicted transcripts. The Cufflinks method also produces gene expression values in units of Fragments per Kilobase per Million reads (FPKM) for each predicted transcript.
Gene Prediction
Genes were predicted on the draft genome assembly with the PASA software system11 using a combination of gene expression, protein homology, and ab inito gene prediction. Ab initio gene predictions on the genomic assemblies were made with GeneMark.hmm-ET12. Potential protein-coding regions on the DNA assemblies were identified by tBLASTn similarity with a set of non-redundant conserved proteins from the UniProt Knowledgebase (UniRef9013). The transcripts built from assembly of RNA-seq reads by SOAPdenovo-Trans were aligned to the genome assembly with BLAT14. The GeneMark models, BLAST similarity, and RNA-seq gene expression information was combined using EVidenceModeler11, which produced a set of 20,301 gene models (predicted genes) and translated predicted proteins.
Predicted proteins were screened for similarity to bacterial proteins in GenBank with blastp, leading to the removal of 1,079 proteins as probable bacterial contaminants. The final set of 19,222 H. hampei predicted proteins are supported by GenMark HMM models, Cufflinks gene models, de novo assembled RNA-seq transcripts, and homology to known GenBank proteins.
One method to estimate the completeness of a genome assembly is to identify orthologs of highly conserved proteins. Using CEGMA (Core Eukaryotic Genes Mapping Approach), a method to identify highly conserved eukaryotic proteins using the NCBI KOGs database15, we aligned the 457 Drosophila CEGMA core proteins to the set of coffee borer proteins predicted from the draft genome. All 457 core proteins had significant BLAST matches, with 455 having e-values lower than 1e−20. This suggests that our gene collection for the coffee berry borer is nearly complete, at least for these ubiquitously expressed conserved genes.
Non-coding RNA (ncRNA)
Functional ncRNA sequences were predicted on the H. hampei genome using the Infernal 1.1 software package16 and the Rfam database (http://rfam.xfam.org)17. A total of 1,085 high quality matches (e-value ≤0.01) were found, with the most abundant classes being 558 microRNAs, 181 snoRNAs, and 64 tRNAs. Sequences similar to ribozymes and CRISPR direct repeat elements were also detected. Predicted ncRNA loci are listed in Table S2.
Biological Function of Predicted Proteins
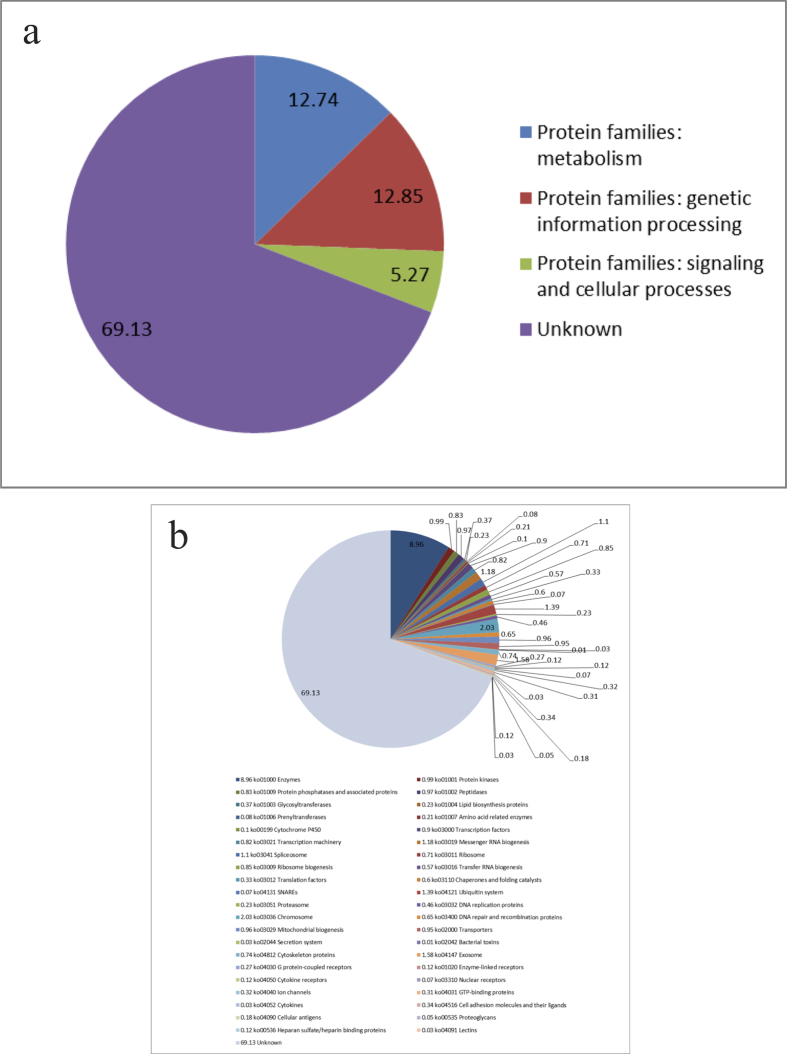
The set of 19,222 predicted H. hampei proteins (https://genome.nyumc.org/CBB.htm) were functionally characterized in the KEGG PATHWAY Database (http://www.genome.jp/kegg/pathway.html) by the BlastKOALA sequence similarity tool (http://www.kegg.jp/blastkoala/) and by alignment to the PANTHER database of protein HMM models (http://pantherdb.org/panther/) with the PANTHER scoring tool (http://pantherdb.org/tools/hmmScoreForm.jsp). The KEGG BlastKOALA tool mapped 27% of the H. hampei predicted proteins (5,149 proteins) to KEGG ortholog groups, while PANTHER assigned 68% of the proteins to PANTHER families. In Fig. 1 we show the distribution of proteins at the top level (Fig. 1a) and second level (Fig. 1b) of the KEGG BRITE hierarchy.
Figure 1. KEGG families of Hypothenemus hampei predicted proteins assigned by the BlastKOALA tool and grouped at the top level (a) and at the second level (b) of the KEGG BRITE hierarchy.
Numbers indicate the percentage of all proteins in the dataset that received that classification.
Gene Families Analyzed
The coffee berry borer spends most of its life cycle inside the coffee berry, where it consumes the seed. We therefore examined digestive enzyme composition, focusing on two digestive enzymes (amylases and mannanases), essential for catabolism of two representative coffee seed carbohydrates. Mannanases are discussed under horizontal gene transfer (below). Seeds are typically well defended against insect predators, so we also examined different detoxifying systems. While recent research indicates that coffee berry borer relies on bacteria in the midgut to detoxify caffeine (Ceja-Navarro et al., submitted), we focused on genes potentially involved in caffeine detoxification as well as the detoxification of other plant allelochemicals and insecticides, such as the carboxylesterase gene family (CE), the cytochrome P450 gene family (CYP), the gluthathione S-transferase gene family (GST), the ATP-binding cassette transporter gene superfamily (ABC), and a gene (Rdl) that confers resistance to the insecticide dieldrin. In addition, we identified genes involved in pathogen defense, including homologs to antimicrobial genes reported in the Drosophila genome.
Digestive Enzymes
The α-amylases are a family of simple carbohydrate (starch)-metabolizing enzymes common in animals, microorganisms, and plants18. Using the T. castaneum amylase gene (XP_969234) as query, we found exactly one orthologous match to assembled transcript C129694, with a BLAST e-value of 0.0 and 72% conserved amino acid matches. The T. castaneum amylase also matches by tblastn to exactly one genomic location on scaffold 1599, where the gene is predicted by GeneMark as evmscaffold1599.1. The gene is highly expressed, with an FPKM in our RNA-seq data of 52997. This transcript also shows homologous matches to α-amylase proteins from other Coleoptera, including the spruce bark beetle (Ips typographus) amylase B with a tBLASTn e-value of 6e−142 and 70% (predicted) amino acid identity; boll weevil (Anthonomus grandis) α-amylase (AAN77139) at e = 0.0 (55% amino acid identity); rice weevil (Sitophilus oryzae) at e = 0.0 (69% amino acid identity); corn rootworm (Diabrotica virgifera virgifera) α-amylase (AAF20998) at e = 0.0 (56% amino acid identity); azuki bean weevil (Callosobruchus chinensis) (BAD83655) at e = 0.0 (58% amino acid identity); Mexican bean weevil (Zabrotes subfasciatus) (AAF73435) at e = 0.0 (57% amino acid identity); and mustard beetle (Phaedon cochleariae) (CAA76926) at e = 0.0 (54% amino acid identity).
As expected, several additional digestive enzyme genes were present, however some unexpected ones were found that are likely important in the insect’s ability to use the seeds as a food source. Seeds typically have high levels of protease and amylase inhibitors, as well as lectins, designed to interfere with insect digestion and absorption. A wide variety of digestive proteinases were found, and included serine proteinases (e.g., chymotrypsin and trypsin), thiol proteinases (e.g., cathepsins), and metalloproteinases. While we cannot easily determine which of these are involved in digestion and which may have other roles until a gut transcriptome becomes available, it is clear that the insect has a potent suite of proteinases likely to be capable of dealing with plant defensive proteins. Most beetles have mainly thiol proteinases19, but the coffee berry borer appears to have a more extensive composition. Thus, it may be challenging to develop plant-produced protein based controls. Several articles have been published on the presence of amylases and proteases in the midgut of the coffee berry borer with the goal of expressing relevant inhibitors in coffee plants as a possible pest management strategy1. Nevertheless, the development of such a strategy remains a distant goal, complicated by the diversity of proteases present, as indicated by the present study. As would be expected, lipases are also present in the H. hampei genome.
The green coffee seed has high levels of hemicellulose (complex polysaccharides), pectins, and celluloses, which could serve as an important nutrient source if the complex matrix can be degraded1. Relevant digestive enzymes were identified, which are more typically associated with wood-feeding insects. The genome of the phloem feeding D. ponderosae contained polysaccharide endo-β-1,4 glucanases, endopolygalacturonases, glycoside hydrolases in family 48 (microcrystalline cellulases), rhamnogalacturonan lyases, and pectin methylesterases5. We found several of the same genes in the H. hampei genome, namely four glycosyl hydrolases in family 48, seven polygalacturonases, one pectin esterase, five rhamnogalacturonan lyases, and two beta glucanases. Additionally we found two mannanase and two xylanase genes that appear to be the result of horizontal gene transfer (see below). Although microbial associates of insects are reported to produce microcrystalline cellulases to the benefit of the associated insect20 their presence in the beetle host genome is not common, and has otherwise only been reported from the genome of a limited number of species20, including Tribolium4, which does not contain the other polysaccharide degrading enzymes21. The ability to degrade the hemicellulose/cellulose/pectin matrix could give the coffee berry borer a much more efficient use of its limited seed resource, and it has the greatest breadth of such genes so far reported for any beetle species. Developing strategies to interfere with these potentially important digestive enzymes may lead to as specific control mechanism for this insect. Cell walls are also typically lignified, which reduces their ability to be degraded. However, we did not find specific lignin degrading enzymes, such as laccases, although some of the peroxidases found in the genome may be capable of this function.
Genes for gustatory receptors were also found in the H. hampei genome, as were those for olfactory and pheromone receptors. Determination of these sequences allows for the potential development of feeding inhibitors, as well as attractants useful for monitoring, or attraction to insecticides of the free flying females.
Detoxification of Plant Chemical Defenses
Seeds are typically well defended by plant secondary metabolites. The coffee seed is no exception, and contains high concentrations of a variety of chlorogenic acid derivatives22 as well as its principal chemical of human interest, caffeine. The chlorogenic acid derivatives can be toxic and in many cases contain appropriate chemical structures (such as aromatics with ortho- or para-hydroxyl or methoxy groups) that can act as substrates for phenoloxidases or peroxidases, which generate reactive oxygen species and toxic quinones23,24. Although tissue expression location has not been determined, genes to deal with reactive oxygen species, such as glutathione peroxidase, are present in the genome. Both cytochrome P450s and glutathione transferases (see below) would be capable of detoxifying these chlorogenates. Caffeine is a potent anti-insect compound found in plants25,26, and insects that feed on caffeine containing plants may have specialized detoxifying mechanisms.
Caffeine detoxification
In another related study (Ceja-Navarro et al., submitted), we have detected expression of methylxanthine N-demethylase genes (ndm) responsible for caffeine demethylation in H. hampei. However, we observed no homologs of this gene in the draft genome, suggesting that unlike mannanase (which has been reported to be present in the insect genome as a result of horizontal gene transfer from bacteria; discussed below), caffeine demethylation has not been laterally transferred to the genome and that this function resides in the bacterial community. This supports observations of elimination of this function through antibiotic treatment of coffee berry borers (Ceja-Navarro et al., submitted).
ATP-binding cassette transporter gene superfamily (ABC)
ABC transporters are membrane bound proteins that use adenosine triphosphate (ATP) binding and hydrolysis for the translocation of amino acids, lipids, sterols, sugars, peptides, xenobiotics, etc., across membranes, with the efflux of xenobiotics having been implicated in resistance to toxins (including insecticides)27,28.
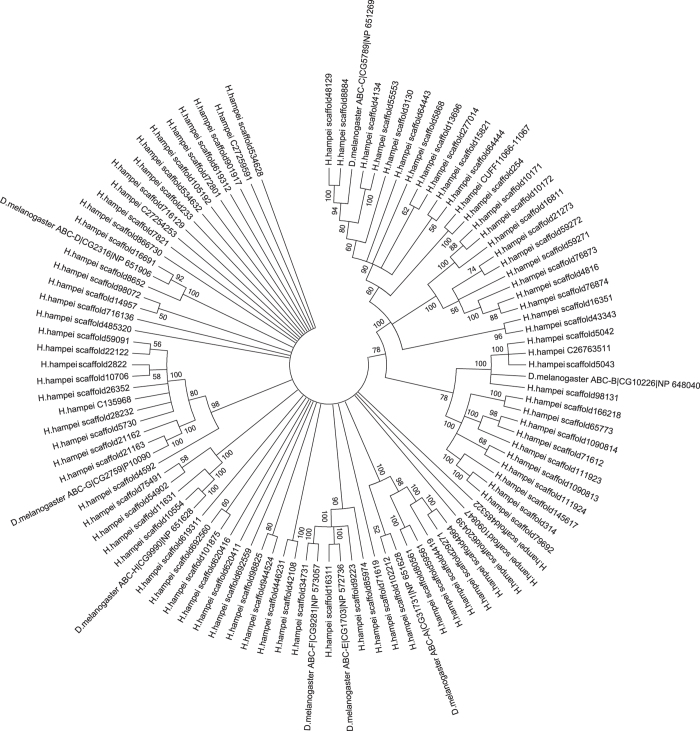
We searched for ABC transporters in the translated set of H. hampei predicted proteins using the hmmsearch program from the HMMER3 software package (http://hmmer.janelia.org)29 with the ABC_tran HMM model (PF00005) from the PFAM website (http://pfam.xfam.org)30. We found a total of 95 putative ABC transporters in the H. hampei genome (Fig. 2). We combined the protein sequences for these genes with annotated protein sequences from D. melanogaster for each of the eight established ABC transporter subfamilies (ABCA-ABCH). The sequences were multiple aligned with CLUSTALW and a maximum likelihood tree (with 100 bootstraps) was built using MEGA6 (http://www.megasoftware.net)31 (Fig. 2). We find the following distribution of subfamilies for the 95 predicted H. hampei ABC proteins ABC-A: six; ABC-B: 14; ABC-C: 24, ABC-D: two; ABC-E/F: four; ABC-G: 11, ABC-H: four; unidentified: 30. A total of 73 ABC genes were reported in T. castaneum27. The findings demonstrate a wide array of ABC transporter subfamilies is present in H. hampei; their specific functions await further study to determine their role in transporting toxic molecules out of cells.
Figure 2. Maximum Likelihood tree (with 100 bootstraps, condensed at 50% bootstrap identity) of translated protein sequences of 95 putative ABC transporter genes identified in the draft genome of Hypothenemus hampei and proteins from Drosophila melanogaster representing the eight established ABC subfamilies (ABCA-ABCH).
Tree built with MEGA6.
Carboxylesterase gene family (CE)
Carboxylesterase genes are involved in insecticide resistance and metabolism32,33, including several pyrethroid insecticides and the organophosphate insecticide malathion. Apis mellifera has 24 genes that are members of this gene family while D. melanogaster has 3534, Aedes aegypti has 4935, Anopheles gambiae has 5132, and Tetranychus urticae has 7636. The CE HMM model from the PFAM database (PF00135.23) was used to search the H. hampei predicted proteins using the hmmsearch program from the HMMER3 software package29. A total of 54 predicted proteins had significant matches to this conserved model (Table S3). In the T. castaneum genome assembly (NCBI version 4.0), there are 10 esterase genes that are highly similar to the H. hampei proteins (blastp e-values range from 0.0 to 1e–144), six of which form a tandem cluster on chromosome ChLG7, indicating a probable ancestral amplification of this gene. This means that the coffee berry borer has the appropriate gene resources to develop resistance to several insecticides.
Cytochrome P450s gene family (CYP)
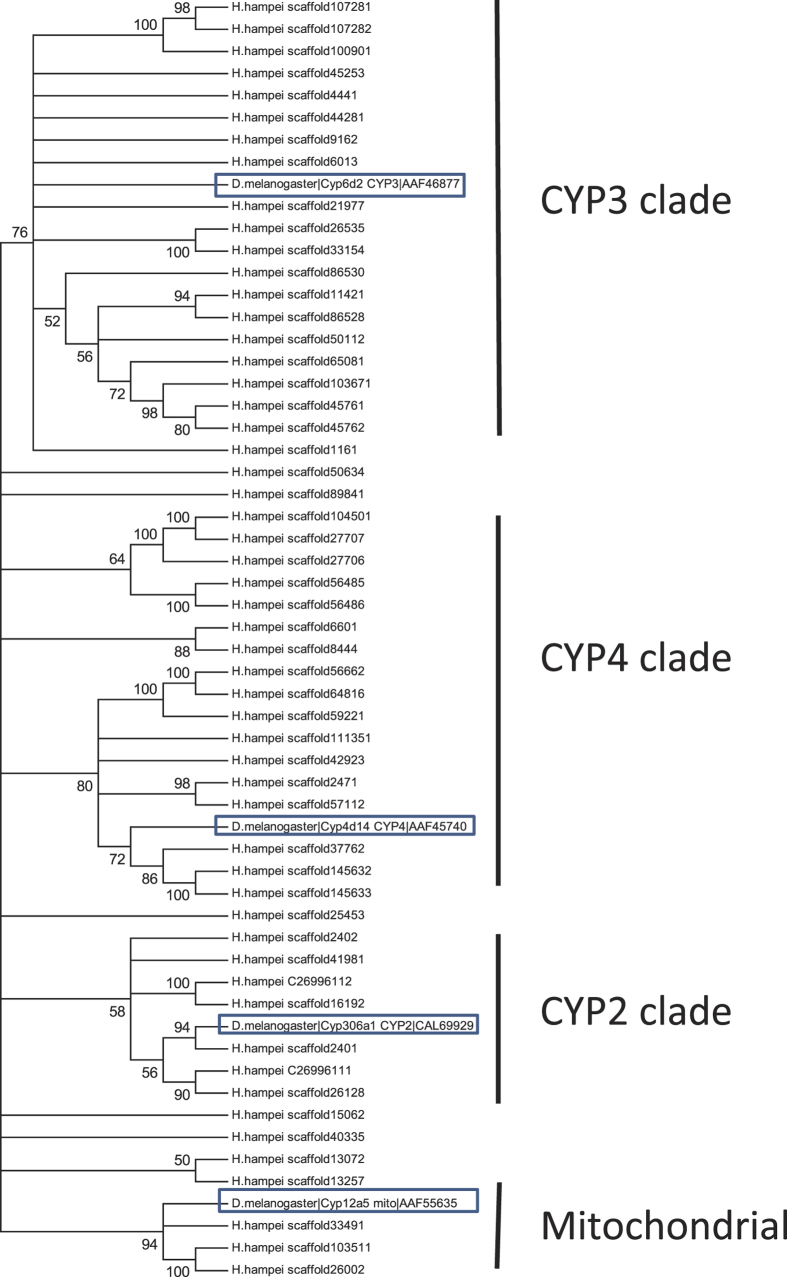
Cytochrome P450s, present in all living systems37, are a superfamily of enzymes (monooxygenases) that, in insects, play a role in the synthesis and metabolism of hormones and pheromones and as well as in the breakdown (detoxification) of exogenous substrates such as plant allelochemicals and insecticides37. CYP families share >40% amino acid similarity, while subfamilies share >55%. Insect CYPs can be divided into four clades: mitochondrial, CYP2, CYP3, and CYP438. The CYP HMM model from the PFAM database (PF00135.23) was used to search H. hampei predicted proteins using the hmmsearch program from the HMMER3 software package29. We identified 54 full-length CYP genes in the H. hampei draft genome (Table 2), with an additional 40 partial sequences. The 54 full-length H. hampei CYP genes were expressed at a wide range of different levels from FPKM 1e6 (the 148th most highly expressed gene) down to FPKM 10. Expression level was not correlated with similarity to the CYP consensus HMM model. Of the 54 full-length CYP genes, three were mitochondrial, seven were CYP2, 20 CYP3, 17 CYP4, and seven undetermined, in contrast to D. ponderosae, whose 85 CYPs showed a different distribution (nine mitochondrial, seven CYP2, 47 CYP3, and 22 CYP49). A phylogenetic reconstruction of Cyp450 genes from H. hampei with Drosophila as a reference shows families with gene loss and expansion (Fig. 3).
Table 2. A total of 54 predicted cytochrome P450 genes were identified in the Hypothenemus hampei draft genome.
| H. hampei protein | FPKM | HMMERe-value | CYPclade | CUFFLINKStranscript |
|---|---|---|---|---|
| evmmodelscaffold6481.6 | 1004190 | 1.70E-98 | CYP4 | CUFF.12736.1 |
| evmmodelscaffold2770.7 | 375522 | 5.90E-80 | CYP4 | C125194 |
| evmmodelscaffold865.28 | 240521 | 1.20E-69 | CYP3 | CUFF.14635.1 |
| evmmodelscaffold2197.7 | 183475 | 2.70E-84 | CUFF.4464.1 | |
| evmmodelscaffold1456.32 | 174361 | 1.50E-172 | CYP4 | CUFF.3616 |
| evmmodelscaffold4033.5 | 147147 | 4.10E-54 | Mito | CUFF.9032.1 |
| evmmodelscaffold1325.7 | 101273 | 2.10E-33 | Mito | CUFF.2068.1 |
| evmmodelscaffold2653.5 | 99649 | 2.40E-71 | CYP3 | CUFF.5808.1 |
| evmmodelscaffold1456.33 | 65672 | 4.00E-108 | CYP4 | CUFF.3617 |
| evmmodelscaffold4525.3 | 65455 | 4.10E-83 | CYP3 | CUFF.9829 |
| evmmodelscaffold10450.1 | 60925 | 1.20E-64 | CYP4 | CUTT.985.2 |
| evmmodelscaffold571.12 | 53606 | 1.70E-104 | CYP4 | CUFF.11699.1 |
| evmmodelscaffold1307.2 | 49167 | 5.70E-45 | Mito | — |
| evmmodelscaffold844.4 | 46252 | 6.90E-60 | CUFF.14420.1 | |
| evmmodelscaffold3349.1 | 45619 | 2.50E-64 | Mito | CUFF.7694.1 |
| evmmodelscaffold4576.1 | 40606 | 5.20E-65 | CYP3 | CUFF.9942.1 |
| evmmodelscaffold1506.2 | 40106 | 3.50E-79 | Mito | CUFF.2636.1 |
| evmmodelscaffold4576.2 | 37291 | 1.30E-66 | CYP3 | CUFF.9937.1 |
| evmmodelscaffold247.1 | 36092 | 1.50E-88 | CYP4 | CUFF.5327.1 |
| evmmodelscaffold10728.2 | 35742 | 6.00E-102 | CYP3 | CUFF.1158.1 |
| evmmodelscaffold916.2 | 27657 | 1.50E-100 | CYP3 | CUFF15114.1 |
| evmmodelscaffold4428.1 | 20286 | 2.00E-98 | CYP3 | CUFF.9689.1 |
| evmmodelscaffold8984.1 | 20186 | 5.50E-46 | CYP4 | CUFF.14931.1 |
| evmmodelscaffold4198.1 | 19820 | 4.30E-65 | CYP2 | CUFF.9256.1 |
| evmmodelscaffold4292.3 | 18337 | 2.70E-107 | CYP4 | CUFF.10885 |
| evmmodelscaffold1142.1 | 16123 | 2.30E-66 | C132918 | |
| evmmodelscaffold240.1 | 14007 | 5.20E-92 | CYP2 | CUFF.5111.1 |
| evmmodelscaffold3776.2 | 12574 | 9.00E-101 | CYP4 | CUFF.8596 |
| evmmodelscaffold5922.1 | 11959 | 4.80E-98 | CYP4 | CUFF.11953.1 |
| evmmodelscaffold2612.8 | 11249 | 3.60E-68 | CYP2 | — |
| evmmodelscaffold5063.4 | 10986 | 3.30E-39 | CUFF.10760.1 | |
| evmmodelscaffold10090.1 | 10659 | 1.40E-82 | CYP3 | CUFF.708.1 |
| evmmodelscaffold240.2 | 10581 | 1.00E-110 | CYP2 | CUFF.5112.1 |
| evmmodelscaffold559.1 | 10526 | 2.10E-10 | CUFF.11505 | |
| evmmodelscaffold865.30 | 10475 | 5.30E-85 | CYP3 | CUFF.14635.2 |
| evmmodelscaffold10367.1 | 10240 | 3.40E-61 | CYP3 | CUFF.952.1 |
| evmmodelscaffold2770.6 | 7772 | 8.10E-87 | CYP4 | CUFF.6104.2 |
| evmmodelscaffold9450.1 | 6295 | 4.00E-50 | CUFF.15342.1 | |
| evmmodelscaffold3315.4 | 5399 | 6.40E-76 | CYP3 | CUFF.7475.1 |
| evmmodelscaffold5648.6 | 4430 | 5.50E-79 | CYP4 | CUFF.1157.1 |
| evmmodelscaffold1619.2 | 3247 | 2.40E-64 | CYP2 | CUFF.2900.1 |
| evmmodelscaffold6508.1 | 3198 | 4.40E-87 | CUFF.12657 | |
| evmmodelscaffold2545.3 | 3128 | 6.50E-32 | CUFF.5506.1 | |
| evmmodelscaffold116.1 | 2531 | 4.30E-82 | CYP3 | CUFF.1594.1 |
| evmmodelC2699611.1 | 2099 | 4.90E-14 | CYP3 | CUFF.639.1 |
| evmmodelC2699611.2 | 2099 | 4.40E-68 | CYP2 | — |
| evmmodelscaffold5011.2 | 2098 | 1.20E-54 | CUFF10656.1 | |
| evmmodelscaffold444.1 | 1850 | 1.90E-77 | CYP3 | CUFF.9710.1 |
| evmmodelscaffold10728.1 | 1532 | 3.30E-106 | CYP3 | CUFF.1152.1 |
| evmmodelscaffold2600.2 | 1327 | 4.00E-150 | Mito | CUFF.5658.1 |
| evmmodelscaffold5648.5 | 100 | 2.20E-99 | CYP4 | — |
| evmmodel10351.1 | 20 | 4.10E-13 | Mito | — |
| evmmodelscaffold5666.2 | 10 | 4.70E-95 | CYP4 | — |
| evmmodelscaffold601.3 | 2 | 5.40E-80 | CYP3 | — |
Gene expression is shown as FPKM values generated by Cufflinks. Cytochrome protein function is inferred by motif analysis of translated PASA/EVM predicted gene coding regions with HMMER hmmsearch (v.3.0) using the consensus cytochrome HMM model from PFAM (PF00067.17). Cytochrome clade was inferred by phylogenetic clustering with reference genes from D. melanogaster by minimum evolution with MEGA6.
Figure 3. The 54 full-length cytochrome P450 genes predicted in the Hypothenemus hampei genome clustered with Drosophila melanogaster genes representing the four insect clades: CYP2, CYP3, CYP4, and mitochondrial.
Predicted proteins in the cytochrome P450 family were identified by HMMER3 search with the Pfam CYP consensus model (PF00135.23). Evolutionary analyses were conducted using the Maximum Likelihood method in MEGA6. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. Branches with less than 50% bootstrap support are condensed.
Cyp301a1 is a protein involved in cuticle formation and has been detected in all insect genomes39. An ortholog of the D. melanogaster Cyp301a1 was detected in scaffold2600.2 of the draft genome (e = 0.0, amino acid identities = 331/480 (69%), positives = 397/480 (83%), gaps = 3/480 (1%)).
The proteins coded by some of these CYP genes are likely responsible for detoxifying the chlorogenic acids present in the seed. Again, some of these could potentially contribute to insecticide resistance. Insect P450s involved in the detoxification of a wide range of xenobiotics have nonpolar residues in the hydrophilic cage surrounding the aromatic network responsible for substrate positioning, and nonaromatic residues in regions SRS2, SRS5 and SRS640. An epoxide hydrolase gene was also found in the genome. Epoxides can be generated by P450s, and if a molecule such as aflatoxin (which has been reported in coffee seeds41) is present, will produce a compound that will intercalate into DNA and result in toxic DNA effects42. Epoxide hydrolases will break this epoxide and prevent such adverse reactions on the DNA42.
Determining the substrate capabilities of the P450s may help guide choices for insecticides for control and selecting those that are less likely to develop resistance due to an absence of capable P450s. For example, based on studies reported above, it does not appear that the H. hampei has N-demethylase capability of its own. Thus, using contact insecticides that contain N-methyl groups attached to aromatic molecules, compared to other structural moieties, may be preferable for longer lifespan controls.
Glutathione S-transferase gene family (GST)
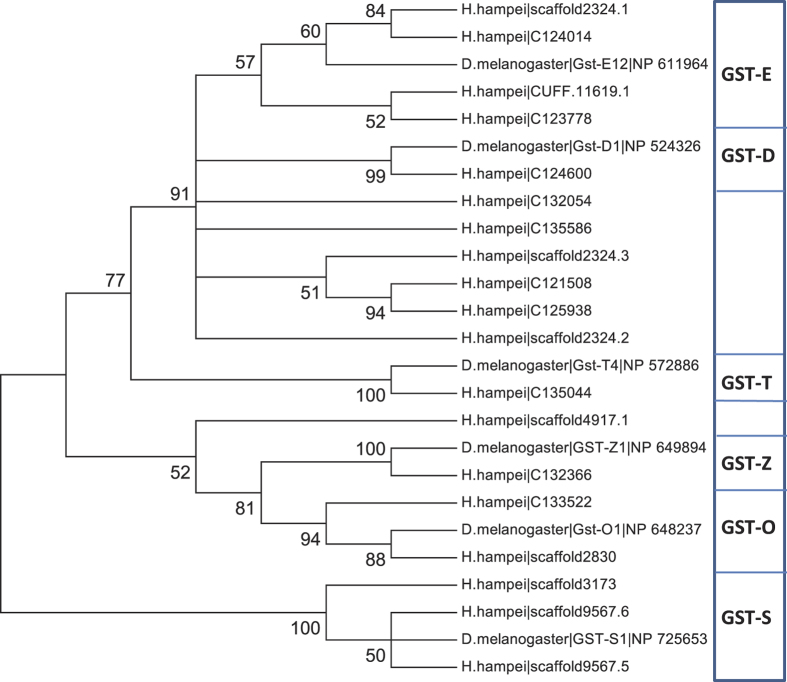
The GST gene family is present in all aerobic organisms and is involved in the mediation of oxidative stress responses, hormone biosynthesis, and in the detoxification of plant allelochemicals and insecticides43. Six GST classes have been identified in insects, with members within a class sharing >40% amino acid identity43,44. Using a combination of BLAST searches with annotated GST genes from Drosophila and HMM searches with PFAM GST families, 19 predicted GST genes were identified in the H. hampei genome and transcriptome, distributed throughout all six classes as follows: one Delta, four Epsilon, two Omega, three Sigma, one Theta, and one Zeta (Fig. 4). Five of the H. hampei genes are similar to both Epsilon and Delta classes (44% amino acid identity to both Drosophila GST D and E genes) but cannot be clearly distinguished between them. Homologs of these unclassified genes (63% amino acid identity) in D. ponderosae are annotated as hypothetical proteins. At least some of these GSTs are likely involved in detoxifying the chlorogenates in the seed and could contribute to detoxification of insecticides as well. Delta class GSTs are associated with resistance to DDT and are involved in resistance to some organophosphate insecticides, while omega forms can readily act on aromatic substrates45, so both classes are likely to be involved in detoxification. The Zeta class GSTs have a very small active site; the Sigma class are involved in prostaglandin synthesis, and the Theta class are inactive toward chlorinated aromatics45, so are less likely to be involved in detoxification. Thus, H. hampei has GSTs likely to be involved in plant defensive compound and insecticide detoxification.
Figure 4. Maximum likelihood tree of 19 predicted Hypothenemus hampei glutathione S-transferase proteins with selected Drosophila melanogaster proteins representative of the six characterized GST classes.
Hypothenemus hampei proteins were identified by significant hmmsearch similarity scores using the Pfam GST-C (PF00043) and GST-N (PF02798) HMM models. The tree was constructed with MEGA 6 using the JTT matrix-based model, with 100 bootstraps. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Branches with less than 50% bootstrap support are condensed. Evolutionary analyses were conducted in MEGA6.
Resistance to dieldrin gene (Rdl)
The broad-spectrum chlorinated cyclodiene insecticide endosulfan (C9H6Cl6O3S) has been used in many countries in an attempt to control H. hampei, but due to its human and environmental toxicity, the insecticide has been banned in at least 70 countries1. Mutations (Ala-302 → Ser) in the Rdl gene (resistance to dieldrin), which encodes the γ-aminobutyric acid subtype A (GABAA) receptor, are responsible for the resistance to the cyclodiene group insecticides in many insects, including A. aegypti, D. melanogaster, Musca domestica, Periplaneta americana, and T. castaneum46. A genomic sequence of the H. hampei Rdl gene from an endosulfan resistant strain was deposited in GenBank (AF037324)47 and contains the Ala → Ser resistance mutation at an equivalent position. Hypothenemus hampei resistance to endosulfan has been reported in New Caledonia48 and molecular detection of the cyclodiene resistance gene Rdl has been demonstrated in insects from New Caledonia47,49 and Colombia50.
The H. hampei Rdl gene is located on scaffold 355 in the draft genome. The gene predicted by GenMark/EVM shows 98% nucleic acid identity to GenBank accession AF037324 over its entire length. However, very few RNA-seq reads align to this locus and the transcript is not fully reconstructed using SOAPdenovo-Trans or Cufflinks. The translation of the predicted gene aligns with the protein sequence of the T. castaneum GABA receptor isoform X1 (BLAST e-value 0.0, 77% amino acid identity) and shows a serine at an equivalent site to the resistance mutation that has been reported in endosulfan resistant flies, beetles and other insects. The insects used to establish the laboratory colony used in the present study originated in Colombia and have been under continuous rearing in the laboratory for ca. 10 years and hundreds of generations, indicating that even though there is no longer selective pressure, the resistant form of the gene has been maintained.
Pathogen Defense Genes
Insects have a complex pathogen defense system that consists of pathogen recognition, humoral responses (antimicrobial peptides), cellular responses, melanization, and intracellular defenses (RNAi)51. A variety of lectin genes potentially involved in pathogen recognition were detected in the H. hampei draft genome. These include lectin C, mannose and galactose binding lectins, as well as peptidoglycan receptor proteins (see below). The regulatory protein gene Spätzle was also present. A prophenoloxidase gene with 77% homology to two Tribolium prophenoloxidases (XM_008196766.1 and NM_001039433) was present in the draft genome, although it was annotated during our analysis as Hemocyanin_C , 6586. Several serpin genes, which can regulate the phenoloxidase activity in insects and shrimps52, do occur in the draft genome. Two Dicer genes, which are involved in the RNA interference (RNAi) process, were also present in the draft genome.
Homologs to Drosophila antimicrobial peptides and related genes
We identified additional putative antimicrobial-related genes by BLAST using the set of all D. melanogaster genes annotated with the following Gene Ontology (GO) database terms (http://amigo.geneontology.org/amigo/landing): antimicrobial humoral response (GO:0019730), antibacterial humoral response (GO:0019731), or defense response to bacterium (GO:0042742). A total of 175 genes were identified including 18 w, akirin, andropin, attacin, cecropin, defensin, diptericin, drosomycin, listericin, lysozyme, etc. Of these 175 Drosophila antimicrobial genes, we find that 120 of them match predicted H. hampei proteins with BLAST significance scores of 5e−10 or lower. For the reciprocal search, a total of 749 predicted H. hampei proteins match the set of Drosophila anti-microbial genes with BLAST significance scores of 5e−10 or lower, showing that these gene families are conserved and have abundant members in the H. hampei genome. A search of the T. castaneum proteome (21,468 GenBank proteins) with the same set of 175 D. melanogaster antimicrobial proteins finds 1,075 matches with BLAST e-values of 5e−10 or lower.
Drosophila melanogaster has five attacin proteins (attacin, attacin-A, -B, -C, and -D). Four uncharacterized homologs are found in the genome of T. castaneum and three in the genome of D. ponderosae. Three significantly similar H. hampei predicted proteins (evm.model.scaffold8269.1, evm.model.scaffold8096.1, evm.model.scaffold3795.1; Fig. 5) were found by BLAST searches with this set of query sequences. All three H. hampei proteins were highly expressed with FKPM values of 12524, 12025, and 2098 respectively. The same three proteins were found by hmmsearch with PFAM HMM model PF03769 (attacin-C). Phylogenetic analysis does not show a clear orthology relationship among the attacin proteins from these species, rather the sequences from each species cluster together.
Figure 5. Maximum likelihood tree of attacin protein sequences from Dendroctonus ponderosae, Drosophila melanogaster, and Tribolium castaneum, and three predicted proteins from Hypothenemus hampei.

Tree was constructed with MEGA 6 using the JTT matrix-based model, with 100 bootstraps. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Branches with less than 50% bootstrap support are condensed. Evolutionary analyses were conducted in MEGA6.
Tribolium castaneum has an annotated cecropin gene (NP_001164146), but no significant similarity to this gene was found by blastp to H. hampei predicted proteins or by tblastn to the H. hampei transcript assemblies or to the H. hampei draft genome sequence. No PFAM matches to the cecropin HMM model (PF00272) were found for H. hampei predicted proteins.
Coleoptericin is a glycine rich antimicrobial protein found in many Coleopteran species. Homologs have been identified in the draft genomes of T. castaneum (XP_008199794) and D. ponderosae (ERL87200). All of these proteins have significant blastp matches to H. hampei predicted protein evm.model.scaffold348.1 (7e−37, 70% amino acid identity). No PFAM matches to the coleoptericin HMM model (PF06286) were found for H. hampei predicted proteins.
A blastp search of H. hampei predicted proteins with the defensin peptide sequence from the Japanese rhinoceros beetle Allomyrina dichotoma (gb|AAB36306) produced one significant match (evm.scaffold2042.7), which is highly expressed (FPKM 10520). A hmmsearch with PFAM HMM model PF01097 (Defensin_2) also matches only to evm.model.scaffold2042.7, whose predicted protein has significant blastp similarity to defensin from several insect species with 50–60% amino acid identity, including holotricin (T. castaneum), coprisin (Copris tripartitus), tenecin (Tenebrio molitor), and phormin (Protophormia terraenovae).
Drosomycin is found primarily in flies. The Drosophila drosomycin protein (AAF47767) was used as a query, but no significant similarity was found by blastp to H. hampei predicted proteins or by tblastn to the H. hampei transcript assemblies or to the H. hampei draft genome sequence. Similarly, moricins and gloverins have been found only in Lepidoptera (e.g., BAB13508 and CAL25129). BLAST searches of H. hampei predicted proteins, assembled transcripts, and draft genome sequence did not yield any significant similarities to these proteins. Proline rich antimicrobial proteins such as lebocin (NP_001037468) and drosocin (CAA79936) are found in Diptera, Hymenoptera, Hemiptera, and Lepidoptera. No homologs of these genes have been reported in Coleoptera. BLAST searches with sequences of drosocin and lebocin did not find any significant matches in the H. hampei predicted proteins, assembled transcripts, or draft genome sequence.
The detection of antimicrobial peptides genes reveals that the coffee berry borer possesses a wide array of mechanisms that could be used for antimicrobial defence. These peptides have been reported to have activity against both fungi and bacteria53.
Horizontal Gene Transfer (HGT)
A mannanase gene (GenBank ID ACU52527) similar to a Bacillus circulans protein was identified from a H. hampei cDNA54 and a xylanase (GenBank ID ADN94682) highly similar to Streptomyces bingchenggensis was isolated from a H. hampei genomic clone55. All predicted H. hampei proteins from the draft genome were screened for HGT candidates by identifying proteins with more significant BLAST e-values to a set of bacterial proteins (UniRef50 bacterial proteins) than to a set of arthropod proteins (UniRef50 arthropod proteins). To remove genes originating from contaminating bacterial DNA in the H. hampei genomic library, only gene models with >10% of exon bases covered by aligned RNA-seq reads were considered as HGT candidates.
A total of 10 HGT candidate genes were identified with evidence for expression, integration into the H. hampei genome, and phylogenetic evidence that the sequences are more closely related to bacterial rather than eukaryotic genes (Table 3). Two copies of endo-1,4-beta-xylanase genes with high similarity to Streptomyces sp. were found on two different draft genome contigs (BLAST e-value ~1e−107, 46–56% amino acid identity). These predicted proteins have 89% and 86% amino acid identity respectively to the xylanase gene reported by Padilla-Hurtado et al.55, but only 75% amino acid identity to each other. These predicted H. hampei genes are highly expressed (FPKM 1.5e6 and 3.1e5, based on Cufflinks analysis of aligned RNA-seq reads), contain introns, and are on contigs that contain other predicted H. hampei genes. A similar endo-1,4-beta-xylanase (BLAST e-value 8e−40) was identified in the gut microbiota of the termite Globitermes brachycerastes, but is incorrectly annotated in GenBank ID 4HU8_A as of arthropod origin56. Xylanases present in the mustard leaf beetle, Phaedon cochleariae (Chrysomelidae) also appear to have been acquired from bacteria57. Polygalacturonase genes apparently acquired from bacterial or fungal sources have been reported for some species of cerambycids, chrysomelids, and curculionids58.
Table 3. Ten candidate horizontally transferred genes detected in Hypothenemus hampei.
| Gene ID | RNA-seq expression (FPKM) | Bacterial BLAST match | e-value | Closest arthropod BLAST match | e-value | Predicted gene function |
|---|---|---|---|---|---|---|
| scaffold769.3 | 1.5e6 | Streptomyces | 1e−107 | Musca domestica (Diptera) | 0.016 | Xylanase |
| scaffold2902.1 | 3.1e5 | Streptomyces | 1e−107 | Stegodyphus (Araneae) | 1.0 | Xylanase |
| scaffold1203.1 | 4.3e6 | Bacillus | 2e−30 | Anopheles darling (Diptera) | 1.1 | Mannanase |
| scaffold1827.1 | 3.7e4 | Bacillus | 1e−57 | Cerapachys biroi (Hymenoptera) | 3e−7 | Mannanase |
| scaffold3436.2 | 1.5e4 | Streptomyces | 0.0 | Nilaparvata lugens (Homoptera) | 2.2 | Hydrolase |
| scaffold7971.1 | 4.9e6 | Protobacteria | 1e−37 | Tribolium castaneum (Coleoptera) | 1e−21 | Lipase/acyl-hydrolase |
| scaffold537.2 | — | Rickettsia | 2e−31 | none | — | Hypothetical |
| scaffold3881.7 | 3.2e4 | Wolbachia | 9e−34 | none | — | Regulatory protein RepA |
| scaffold1344.1 | 1.9e4 | Citrobacter | 2e−23 | none | — | Ig-like protein, Invasin |
| C2660673.1 | 2.3e3 | Lactobacillus | 1e−53 | Ixodes scapularis (Ixodida) | 6e−22 | Permease |
Two copies of mannanase genes of possible bacterial origin were found on two different contigs of the draft genome (Table 3; Fig. 6). The previously reported HhMAN1 gene (GenBank ID GQ37515654) matches scaffold1203.1 with 100% DNA sequence identity over a region of 5 kb, including a predicted transcript, which is highly expressed (FPKM 4.3e6). A second predicted gene on scaffold1827.1 was identified with similarity to mannanase from Bacillus (BLAST e-value 2e−30, ~59% amino acid identity). This gene has ~94% DNA sequence identity to GQ375156 over a much smaller region of 562 bases and 91% amino acid identity to the HhMAN1 protein (GenBank ID ACU52526) over the coding sequence. This second mannanase gene is expressed at FPKM 3.7e4.
Figure 6. Maximum likelihood tree including the protein sequence of the previously reported Hypothenemus hampei mannanase gene (GenBank ID ACU52527), two highly similar predicted proteins from the H. hampei draft genome (scaffold1203.1 and scaffold1827.1) with homologs from eubacteria, fungal, and insect proteins.
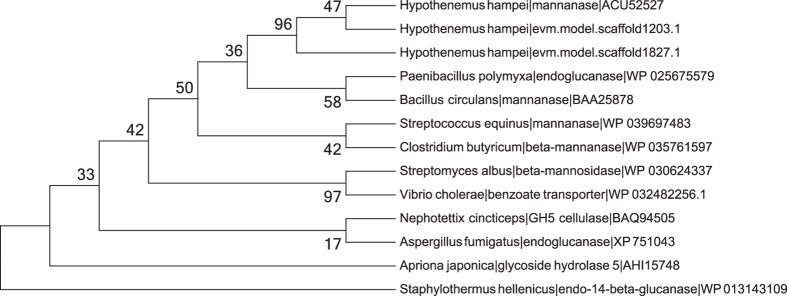
The beta-glucanase protein from the archaebacteria Staphylothermus hellenicus is included as an outgroup. The H. hampei proteins clearly cluster with the eubacterial proteins. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. Evolutionary analyses were conducted using MEGA6.
Predicted gene scaffold3436.2 has strong similarity to hydrolase proteins from Streptomyces (BLAST e-value 0.0, 43% amino acid identity) and no significant similarity to any arthropod genes (best BLAST value >2.0) (Table 3). The gene is highly expressed (FPKM 1.5e4) and is located on a scaffold with other predicted genes that have homology with insect proteins.
A predicted H. hampei gene (scaffold7971.1) has stronger similarity to a Proteobacteria lipase/acylhydrolase (BLAST e-value 1e−37, 37% amino acid identity) than to its closest arthropod match, platelet activating factor acetylhydrolase (BLAST e-value 1e−21, 32% amino acid identity) (Table 3). A maximum likelihood tree including a sample of protein sequences from KEGG orthology group K01137 (EC:3.1.6.14) clearly shows that the H. hampei gene is much more closely related to bacterial than to eukaryotic genes (Fig. 7). Aligned RNA-seq reads show that the gene is highly expressed (FPKM 4.9e6) and has a spliced intron.
Figure 7. Maximum likelihood tree of the translated protein sequence of the predicted Hypothenemus hampei GDSL lipase/acylhydrolase gene (scaffold7971.1) with homologs from eubacteria, animal, and insect proteins.
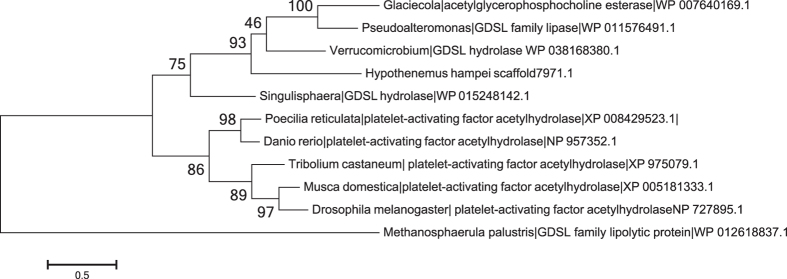
The GDSL family lipolytic protein from the archaebacteria Methanosphaerula palustris is included as an outgroup. The H. hampei protein clearly clusters with the eubacterial proteins. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. Evolutionary analyses were conducted using MEGA6.
A predicted gene on scaffold 537.2 has homology to hypothetical proteins from Rickettsia (BLAST e-value 2e−31) (Table 3), but is supported by only a few RNA-seq reads and is not included among transcript models created by Cufflinks or SOAPdenovo-Trans.
Predicted protein scaffold 3881.7 is similar to Wolbachia regulatory protein RepA (BLAST e-value 9e−34) (Table 3), but has lesser levels of similarity with other bacteria such as Ralstonia, Candidatus, Xanthomonas, as well as gp5 originating from bacteriophage WO. It has no significant similarity to any arthropod genes. It is expressed at a moderate level (FPKM 3.2e4).
Predicted protein scaffold1344.1is similar to Citrobacter Ig-like protein (BLAST e-value 2e−23) invasin (Table 3), with no significant similarity to any arthropod genes. It is expressed at FPKM 1.9e4.
Predicted protein C2660673.1 has strong similarity to permease from Lactobacillus and other Firmicutes (BLAST e-value 2e−53, 66% amino acid identity) (Table 3). The closest similarity to an arthropod gene is a BLAST e-value of 6e−22 with a short 88 amino acid segment of hypothetical protein from the tick Ixodes scapularis. The gene is on a small contig (1.1 kb) with moderate RNA-seq coverage (FPKM 2.3e3), gene model prediction by Cufflinks, SOAPdenovo-Trans, and PASA/EvidenceModeler, but no evidence of splicing.
The 10 HGT candidate genes in the H. hampei genome confirm the close relationship the insect has had with bacterial populations. For example, Ceja-Navarro et al. (submitted) identified several bacteria isolated from the alimentary canal that are able to subsist on caffeine as the sole source of carbon and nitrogen, and demonstrated that caffeine detoxification by the coffee berry borer is meditated by its microbiota.
Conclusions
The draft H. hampei genome is the third Coleoptera genome and the second bark beetle genome to have been analyzed. The most important findings in the coffee berry borer genome include (1) evidence for 10 cases of horizontal gene transfer from diverse bacteria; (2) paralogous expansion of antimicrobial peptides; (3) presence of a broad range of enzymes capable of degrading complex polysaccharides; and (4) the presence of a gene (Rdl) that confers resistance to a cyclodiene insecticide. The genome should be a useful tool for other researchers studying Coleoptera, and in particular, to those studying economically important bark beetles. The genome has also provided further understanding as to how H. hampei is able to successfully exploit its food resource and defend against pathogens. The information provided also can help guide the development of management strategies related to use of insecticides, biological control agents, attractants and plant resistance mechanisms that could be developed by breeding or genetic engineering.
Methods
Nucleic Acid Extraction and Purification
Female coffee berry borers had been continuously reared on a meridic diet in the laboratory59 for ca. 10 years. Ten batches of 100 female insects were placed in microcentrifuge tubes containing 1 ml of RNAlater® (Ambion, Grand Island, NY, USA) and incubated overnight at 4°C. After the initial incubation, the RNAlater® was removed, and the insects’ cuticle was sterilized by treating each batch with four sequential washing steps consisting of 500 μl of DEPC-treated water, commercial bleach, absolute ethanol and DEPC-water. At each step, the sample was vigorously shaken for ca. 30 sec. The washing of the samples was done twice. The insects from each batch were then transferred to a Lysing Matrix E tube (MP Biomedicals, Santa Clara, CA, USA) containing 500 μl of lysis solution (2% CTAB in 0.5 M NaCl). The samples were bead-beaten in a FastPrep Instrument (MP Biomedicals) at 4 m/s for 30 seconds. Lysozyme buffer (180 μl; 20 mM Tris-HCl (pH 8.0), 2mM EDTA, 1.2% Triton X-100) containing 10 mg/ml of lysozyme was added to each tube and the mixture incubated at 37 °C for 30 min. Five μl of proteinase K (Ambion, Grand Island, NY, USA) was added to the tubes, mixed by tapping and incubated at 56°C for 30 minutes. After the proteinase K lysis, 500 μl of phenol:chloroform:isoamyl alcohol (25:24:1) was added to each tube, and the samples bead-beaten in the FastPrep instrument as before. The samples were centrifuged at 10,000 × g for 1 min at 4 °C and the supernatants transferred to a MaXtract high density tube (Qiagen, Germantown, MD, USA) containing 500 μl of chloroform:isoamyl alcohol (24:1). The samples were centrifuged 10,000 × g for 1 min at 4 °C, and the supernatants transferred to a microcentrifuge tube containing 1 ml of isopropanol and 1 μl of linear acrylamide (Ambion, Grand Island, NY, USA). The DNA/RNA mixture was precipitated by incubating for 10 min at room temperature and centrifuged at 10,000 × g for 5 min at 4 °C, and the isopropanol removed. The obtained pellet was washed with 70% ethanol and centrifuged at 10,000 × g for 1 min at 4 °C. The ethanol was completely removed, and the pellet dissolved in DEPC-treated water. The DNA and RNA were purified and separated with the AllPrep DNA/RNA kit (Qiagen) following the manufacturers protocol that was modified to treat the samples with either DNase or RNase (Ambion) for 5 min before the washing steps. The DNA and RNA concentration was determined with the use of a Qubit fluorometer (Qiagen) and the RNA quality assessed by bioanalyzer. The extracted DNA and RNA were finally pooled to a final concentration of 10 μg for genomic and transcriptomic library preparation.
Protocol for Coffee Berry Borer noPCR Libraries
Genomic DNA from whole coffee berry borers was sheared to 350 and 550 bp sized fragments. To remove any amplification bias at the PCR step of the library preparation, noPCR libraries were prepared from coffee berry borer DNA using Illumina TruSeq DNA PCR-Free LT Sample Preparation Kit according to manufacturer’s instructions. Briefly, 1–2 ug of genomic DNA was fragmented using Covaris shearing with prescribed settings for 350 or 550 bp insert sizes. The fragmented DNA was cleaned, end-repaired, and selected for appropriate insert sizes (350 bp and 550 bp) using different ratios of Sample Purification Beads. This was followed by 3′ end adenylation and adapter ligation. Libraries were sequenced in Illumina Hiseq2000 with 100 bp paired-end reads.
Protocol for Coffee Berry Borer ssRNAseq Library
Strand-specific mRNA sequencing libraries were prepared from total RNA of coffee berry borer using TruSeq Stranded mRNA Sample Prep Kit LT (Illumina) according to manufacturer’s instructions. Briefly, polyA + mRNA was purified from total RNA using oligo(dT) Dynabeads (Life Technologies) selection. First strand cDNA was synthesized using randomly primed oligos followed by second strand synthesis where dUTPs were incorporated to achieve strand-specificity. The cDNA was adapter-ligated and the libraries amplified by PCR. Libraries were sequenced in Illumina Hiseq2000 with paired-end 100 bp read chemistry.
Genome Assembly
The genomic reads were cleaned with Trimmomatic60 to trim low quality reads (including leading or trailing sequences with the quality less than 20, any 4-base window with the quality less than 25) and Illumina TruSeq 3 adapter artifacts, filtered for bacterial contamination by BLAST searching.
After quality filtering, a total of 62 Gbp of sequence data (36 Gbp from the 350 bp insert library and 26 Gbp from the 550 bp insert library) was used for genome assembly. With an estimated genome size of 200 Mb, this corresponds to an average depth of coverage of 180-fold. Sequence reads were assembled using SOAPdenovo2 to form contigs and scaffolds resulting in 156 Mb in contigs and 163 Mb in scaffolds (see Table 1 and Table S1 for assembly statistics).
Gene Prediction
Ab initio gene prediction on the assembled genomic contigs and scaffolds was made with GeneMark.hmm-ET12. Potentially protein-coding regions on the DNA assemblies were identified by tBLASTn similarity with a set of non-redundant conserved proteins from the UniProt Knowledgebase (UniRef9013).
RNA-seq data was used to de novo assemble transcripts using the SOAPdenovo-Trans software8, producing an overall count of 54,068 transcript assemblies. The transcript assemblies produced by SOAPdenovo-Trans were mapped to the genome with BLAT v3514 and then analyzed by PASA to optimize intron splicing sites, producing genome aligned spliced transcripts. EVidenceModeler11 was used to combine the PASA transcripts with GeneMark predicted genes, and with the genomic regions showing BLAST similarity to known proteins, which produced a final set of 20,301 gene models and translated predicted proteins.
An additional set of “genome guided” predicted transcripts was produced from a combination of RNA-seq and genomic data using the TopHat/Cufflinks software package. RNA-seq data were aligned to the genomic contigs using TopHat29, then Cufflinks210 was used for transcript assembly, yielding a set of 18,797 predicted transcripts.
The transcripts produced by Cufflinks and the predicted protein sequences produced by GeneMark/EVM were annotated by BLAST comparison with the NCBI non-redundant protein database. Gene functions (Fig. 1) were inferred from Genome Ontology information of these matching proteins using Blast2GO software.
Data Availability
The assembled genomic contigs and predicted proteins for the H. hampei genome are available on a GBrowse server hosted at NYU Medical Center: https://genome.med.nyu.edu/gb2/gbrowse/hham. Tracks are available for the DNA contigs, de novo assembled RNA transcripts, Cufflinks predicted transcripts, and GeneMark/EVidenceModeler predicted gene models as well as regions of homology to UniProt proteins.
Additional Information
How to cite this article: Vega, F. E. et al. Draft genome of the most devastating insect pest of coffee worldwide: the coffee berry borer, Hypothenemus hampei. Sci. Rep. 5, 12525; doi: 10.1038/srep12525 (2015).
Supplementary Material
Acknowledgments
Supported in part by Research Support Agreement 58-1245-3-309 between USDA-ARS and the Center for Health Informatics and Bioinformatics at New York University. Part of this work was performed at Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract No. DE-AC02-05CH11231. The work was also supported by faculty baseline research funding (BRF) by King Abdullah University of Science and Technology (KAUST). J.A.C.-N. was supported in part by the Laboratory Directed Research and Development Program of Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract DE-AC02-05CH1121231. This work has used computing resources at the High Performance Computing Facility of the Center for Health Informatics and Bioinformatics at the NYU Langone Medical Center.
Footnotes
Author Contributions F.E.V. conceived the study. F.E.V. collected samples and J.A.C.-N. and E.L.B. extracted DNA/RNA. A.P. and M.B.N. sequenced the genome. S.M.B., H.C. and E.S. coordinated the assembly and annotation. F.E.V., S.M.B., J.A.C.-N., E.L.B., F.I., P.F.D. and A.P. wrote the paper.
References
- Vega F. E. Infante F. & Johnson A. J. in Bark Beetles: Biology and Ecology of Native and Invasive Species (eds Vega F. E. & Hofstetter R. W.) Ch. 11, 427–494. (Academic Press, 2015). [Google Scholar]
- Oliveira C. M., Auad A. M., Mendes S. M. & Frizzas M. R. Economic impact of exotic insect pests in Brazilian agriculture. J. Appl. Entomol. 137, 1–15 (2013). [Google Scholar]
- Infante F., Pérez J. & Vega F. E. The coffee berry borer: the centenary of a biological invasion in Brazil. Braz. J. Biol. 74, S125–S126 (2014). [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 (2008). [DOI] [PubMed] [Google Scholar]
- Keeling C. I. et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 14, R27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez J. et al. First draft genome sequence of coffee berry borer: the most invasive insect pest of coffee crops. Paper presented at Sixth Annual Arthropod Genomics Symposium and i5k Community Workshop, Kansas City. (2012, May 30-June 2).
- Benavides P. et al. The genome of the coffee berry borer, Hypothenemus hampei, the major insect pest of coffee worldwide. Paper presented at Plant & Animal Genome XXII Conference, San Diego. (2014, January 10–15).
- Xie Y. et al. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 12, 1660–1666 (2014). [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. & Salzberg S. L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pimentel H., Trapnell C. & Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011). [DOI] [PubMed] [Google Scholar]
- Haas B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A., Burns P. D. & Borodovsky M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42, e119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek B. E., Huang H., McGarvey P., Mazumder R. & Wu C. H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 10, 1282–1288 (2007). [DOI] [PubMed] [Google Scholar]
- Kent W. J. BLAT - the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K. & Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- Nawrocki E. P. & Eddy S. R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge S. W. et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi-de-sá M. F. & Chrispeels M. J. Molecular cloning of bruchid (Zabrotes subfasciatus) α-amylase cDNA and interactions of the expressed enzymes with bean amylase inhibitors. Insect Biochem. Mol. Biol. 27, 271–281 (1997). [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K. & Zeng R. Insect response to plant defensive protease inhibitors. Annu. Rev. Entomol. 60, 233–252 (2015). [DOI] [PubMed] [Google Scholar]
- Watanabe H. & Tokuda G. Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632 (2010). [DOI] [PubMed] [Google Scholar]
- Pauchet Y., Wilkinson P., Chauhan R. & ffrench-Constant R. H. Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS ONE 5, e15635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmach A., Clifford M. N., Williamson G. & Crozier A. in Teas, Cocoa and Coffee: Plant Secondary Metabolites and Health (eds Crozier A., Ashihara H. & Tomás-Barbéran F.) Ch. 5, 143–168 (Blackwell Publishing Ltd., 2012). [Google Scholar]
- Gaspar T., Penel C., Thorpe T. & Greppin H. Peroxidases 1970-1980. A survey of their biochemical and physiological roles in higher plants. (Université de Genéve, 1982). [Google Scholar]
- Vaughn K. C. & Duke S. O. Function of polyphenol oxidase in higher plants. Physiol. Plant. 60, 106–112 (1994). [Google Scholar]
- Nathanson J. A. Caffeine and related methylxanthines: possible naturally occurring pesticides. Science 226, 184–187 (1984). [DOI] [PubMed] [Google Scholar]
- Vega F. E., Blackburn M. B., Kurtzman C. P. & Dowd P. F. Identification of a coffee berry borer-associated yeast: does it break down caffeine? Entomol. Exp. Appl. 107, 19–24 (2003). [Google Scholar]
- Broehan G., Kroeger T., Lorenzen M. & Merzendorfer H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics 14, 6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W. & Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110 (2014). [DOI] [PubMed] [Google Scholar]
- Eddy S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7(10), e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H. et al. Evolution of supergene families associated with insecticide resistance. Science 298, 179–181 (2002). [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Horne I., Sutherland T. D. & Russell R. J. The genomics of insecticide resistance. Genome Biol. 4, 202 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudianos C. et al. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 15, 615–636 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strode C. et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 38, 113–123 (2008). [DOI] [PubMed] [Google Scholar]
- Grbić M. et al. The genome of Tetranychus urticae reveals herbivore pest adaptations. Nature 479, 487–492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. G. & Wen Z. Cytochromes P450 of insects: the tip of the iceberg. Pest Manag. Sci. 57, 958–967 (2001). [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Evolution of insect P450. Biochem. Soc. Trans. 34, 1252–1255 (2006). [DOI] [PubMed] [Google Scholar]
- Sztal T., Chung H., Berger S., Currie P. D., Batterham P. & Daborn P. J. A cytochrome P450 conserved in insects is involved in cuticle formation. PLoS ONE 7 (5), e36544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M. A. P450s in plant-insect interactions. Biochim. Biophys. Acta 1814, 36–45 (2010). [DOI] [PubMed] [Google Scholar]
- Soliman K. M. Incidence, level, and behavior of aflatoxins during coffee bean roasting and decaffeination. J. Agric. Food Chem. 50, 7477–7481 (2002). [DOI] [PubMed] [Google Scholar]
- Wild C. P. & Turner P. C. The toxicology of aflatoxins as a basis for public health decisions. Mutagenesis 17, 471–481 (2002). [DOI] [PubMed] [Google Scholar]
- Enayati A. A., Ranson H. & Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8 (2005). [DOI] [PubMed] [Google Scholar]
- Ranson H. & Hemingway J. in Comprehensive Molecular Insect Science, Vol. 5 (eds Gilbert L. I., Iatrou K. & Gill S. S.), 383–402 (Elsevier Ltd., 2005). [Google Scholar]
- Sheehan D., Meade G., Foley V. M. & Dowd C. A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M., Steichen J. C. & ffrench-Constant R. H. Conservation of cyclodiene insecticide resistance-associated mutations in insects. Insect Mol. Biol. 2, 149–154 (1993). [DOI] [PubMed] [Google Scholar]
- Andreev D., Breilid H., Kirkendall L., Brun L. O. & ffrench-Constant R. H. Lack of nucleotide variability in a beetle pest with extreme inbreeding. Insect Mol. Biol. 7, 197–200 (1998). [DOI] [PubMed] [Google Scholar]
- Brun L. O., Marcillaud C., Gaudichon V. & Suckling D. M. Endosulfan resistance in Hypothenemus hampei (Coleoptera: Scolytidae) in New Caledonia. J. Econ. Entomol. 82, 1311–1316 (1989). [Google Scholar]
- ffrench-Constant R. H., Steichen J. C. & Brun L. O. A molecular diagnostic for endosulfan insecticide resistance in the coffee berry borer Hypothenemus hampei (Coleoptera: Scolytidae). Bull. Entomol. Res. 84, 11–16 (1994). [Google Scholar]
- Navarro L., Gongora C. & Benavides P. Single nucleotide polymorphism detection at the Hypothenemus hampei Rdl gene by allele-specific PCR amplification with Tm-shift primers. Pestic. Biochem. Physiol. 97, 204–208 (2010). [Google Scholar]
- Lundgren J. G. & Jurat-Fuentes J. L. in Insect Pathology, 2nd edn, (eds Vega & Kaya) Ch. 13, 461–480 (Academic Press, 2012). [Google Scholar]
- Somnuk S., Tassanakajon A. & Rimphanitchayakit V. Gene expression and characterization of a serine proteinase inhibitor PmSERPIN8 from the black tiger shrimp Penaeus monodon. Fish Shellfish Immunol. 33, 332–341 (2012). [DOI] [PubMed] [Google Scholar]
- Yi H.-Y., Chowdhury M., Huang Y.-D. & Yu X.-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 98, 5807–5822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuña R. et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. USA 109, 4197–4202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Hurtado B. et al. Cloning and expression of an endo-1,4-β-xylanase from the coffee berry borer, Hypothenemus hampei. BMC Res. Notes 5, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. et al. Microbiome of fungus-growing termites: a new reservoir for lignocellulase genes. Appl. Environ. Microbiol. 77, 48–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauchet Y. & Heckel D. G. The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacterial through horizontal gene transfer. Proc. R. Soc. B 280, 20131021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch R. et al. Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: key events in the evolution of herbivory in beetles. Insect Biochem. Mol. Biol. 52, 33–50 (2014). [DOI] [PubMed] [Google Scholar]
- Vega F. E., Kramer M. & Jaramillo J. Increasing coffee berry borer (Coleoptera: Curculionidae: Scolytinae) female density in artificial diet decreases fecundity. J. Econ. Entomol. 104, 87–93 (2011). [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled genomic contigs and predicted proteins for the H. hampei genome are available on a GBrowse server hosted at NYU Medical Center: https://genome.med.nyu.edu/gb2/gbrowse/hham. Tracks are available for the DNA contigs, de novo assembled RNA transcripts, Cufflinks predicted transcripts, and GeneMark/EVidenceModeler predicted gene models as well as regions of homology to UniProt proteins.