Abstract
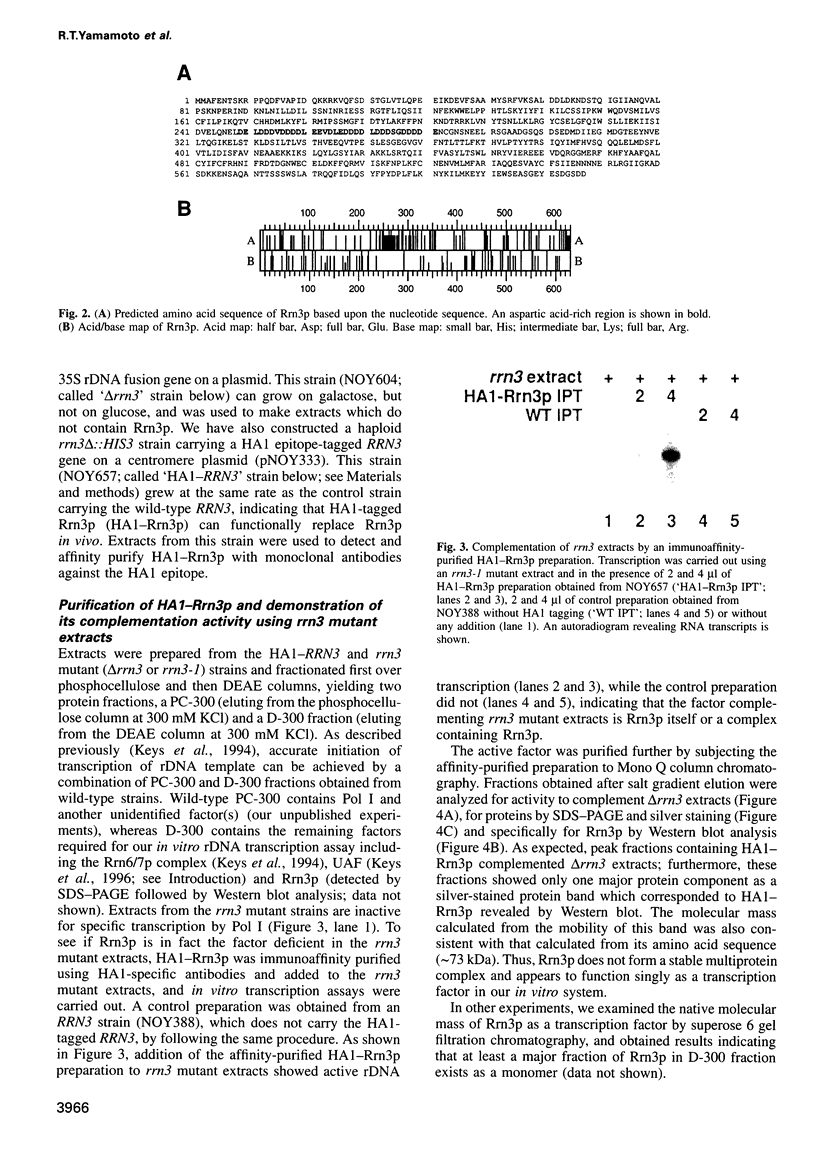
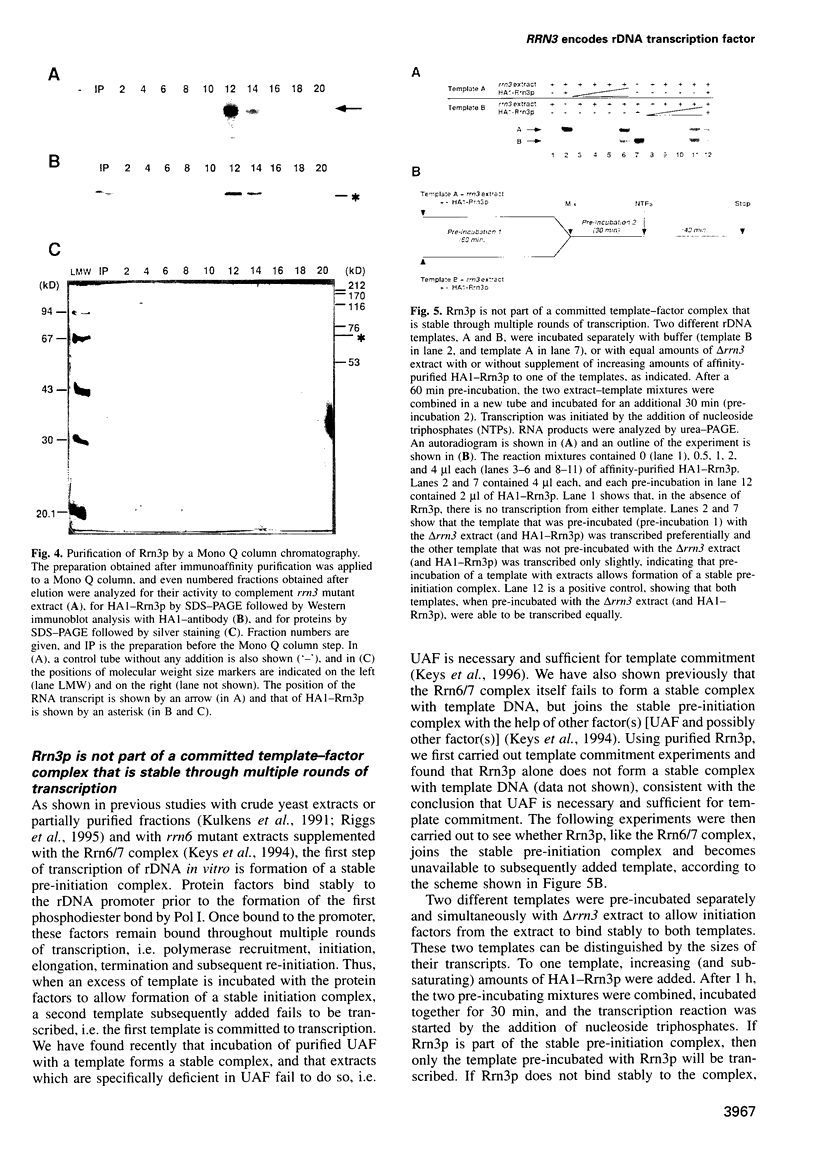
RRN3 is one of the RRN genes specifically required for the transcription of rDNA by RNA polymerase I (Pol I) in Saccharomyces cerevisiae. We have cloned the gene, determined the nucleotide sequence, and found that it is an essential gene which encodes a protein of calculated molecular weight of 72 369. Extracts prepared from rrn3 mutants were defective in in vitro transcription of rDNA templates. We used extracts from a strain containing an epitope-tagged Rrn3 protein to purify a factor that could complement the mutant extracts. Using immunoaffinity purification combined with Mono Q chromatography, we obtained an essentially pure preparation of Rrn3p which complements the mutant extracts. By carrying out template commitment experiments, we found that Rrn3p is not part of the pre-initiation complex that is stable through multiple rounds of transcription. We also found that pre-incubation of Rrn3p with purified Pol I leads to stimulation of transcription upon subsequent mixing with DNA template and other transcription reaction components. Single-round transcription experiments using the detergent Sarkosyl showed that this stimulation is due to increased efficiency of formation of a Sarkosyl-resistant pre-initiation complex. Thus, Rrn3p appears to interact directly with Pol I, apparently stimulating Pol I recruitment to the promoter, and is distinct from two other Pol I-specific transcription factors, the Rrn6/7 complex and the Rrn5/9/10 complex (UAF), characterized previously.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aso T., Vasavada H. A., Kawaguchi T., Germino F. J., Ganguly S., Kitajima S., Weissman S. M., Yasukochi Y. Characterization of cDNA for the large subunit of the transcription initiation factor TFIIF. Nature. 1992 Jan 30;355(6359):461–464. doi: 10.1038/355461a0. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D. P., Leblanc B., Herfort M., Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994 May 20;264(5162):1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- Brun R. P., Ryan K., Sollner-Webb B. Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol Cell Biol. 1994 Jul;14(7):5010–5021. doi: 10.1128/mcb.14.7.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994 Apr 8;77(1):1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Chen P., Lee K. S., Levin D. E. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol Gen Genet. 1993 Jan;236(2-3):443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Comai L., Zomerdijk J. C., Beckmann H., Zhou S., Admon A., Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994 Dec 23;266(5193):1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- Copenhaver G. P., Putnam C. D., Denton M. L., Pikaard C. S. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 1994 Jul 11;22(13):2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., Alexandraki D., André B., Ansorge W., Baladron V., Ballesta J. P., Banrevi A., Bolle P. A., Bolotin-Fukuhara M., Bossier P. Complete DNA sequence of yeast chromosome XI. Nature. 1994 Jun 2;369(6479):371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- Eberhard D., Tora L., Egly J. M., Grummt I. A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res. 1993 Sep 11;21(18):4180–4186. doi: 10.1093/nar/21.18.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A., Kostrub C. F., Li J., Chavez D. P., Wang B. Q., Fang S. M., Greenblatt J., Burton Z. F. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature. 1992 Jan 30;355(6359):464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987 Mar 15;262(8):3452–3461. [PubMed] [Google Scholar]
- Henry N. L., Campbell A. M., Feaver W. J., Poon D., Weil P. A., Kornberg R. D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994 Dec 1;8(23):2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Fujita H., Wang J., Takada R., Roeder R. G. Nucleotide and amino acid sequence of RAP30. Nucleic Acids Res. 1991 Oct 11;19(19):5436–5436. doi: 10.1093/nar/19.19.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. H., McStay B., Jeong S. W., Reeder R. H. xUBF, an RNA polymerase I transcription factor, binds crossover DNA with low sequence specificity. Mol Cell Biol. 1994 May;14(5):2871–2882. doi: 10.1128/mcb.14.5.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D. A., Lee B. S., Dodd J. A., Nguyen T. T., Vu L., Fantino E., Burson L. M., Nogi Y., Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 1996 Apr 1;10(7):887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Keys D. A., Vu L., Steffan J. S., Dodd J. A., Yamamoto R. T., Nogi Y., Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes Dev. 1994 Oct 1;8(19):2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Killeen M. T., Greenblatt J. F. The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol Cell Biol. 1992 Jan;12(1):30–37. doi: 10.1128/mcb.12.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis K. J., Goldin A. L. Site-directed mutagenesis of the putative pore region of the rat IIA sodium channel. Mol Pharmacol. 1993 Apr;43(4):635–644. [PubMed] [Google Scholar]
- Kulkens T., Riggs D. L., Heck J. D., Planta R. J., Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991 Oct 11;19(19):5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Greenblatt J. Related RNA polymerase-binding regions in human RAP30/74 and Escherichia coli sigma 70. Science. 1991 Aug 23;253(5022):900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- Moss T., Stefanovsky V. Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A. F., van Heerikhuizen H., Planta R. J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989 Dec 11;17(23):9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Miyajima A., Arai K. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 1985 Oct;4(10):2643–2648. doi: 10.1002/j.1460-2075.1985.tb03982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Vu L., Nomura M. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol Cell Biol. 1993 Jan;13(1):114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam C. D., Copenhaver G. P., Denton M. L., Pikaard C. S. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol. 1994 Oct;14(10):6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs D. L., Peterson C. L., Wickham J. Q., Miller L. M., Clarke E. M., Crowell J. A., Sergere J. C. Characterization of the components of reconstituted Saccharomyces cerevisiae RNA polymerase I transcription complexes. J Biol Chem. 1995 Mar 17;270(11):6205–6210. doi: 10.1074/jbc.270.11.6205. [DOI] [PubMed] [Google Scholar]
- Riva M., Buhler J. M., Sentenac A., Fromageot P., Hawthorne D. C. Natural variation in yeast RNA polymerase A. Formation of a mosaic RNA polymerase A in a meiotic segregant from an interspecific hybrid. J Biol Chem. 1982 Apr 25;257(8):4570–4577. [PubMed] [Google Scholar]
- Schnapp A., Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991 Dec 25;266(36):24588–24595. [PubMed] [Google Scholar]
- Schnapp A., Schnapp G., Erny B., Grummt I. Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol Cell Biol. 1993 Nov;13(11):6723–6732. doi: 10.1128/mcb.13.11.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp G., Schnapp A., Rosenbauer H., Grummt I. TIF-IC, a factor involved in both transcription initiation and elongation of RNA polymerase I. EMBO J. 1994 Sep 1;13(17):4028–4035. doi: 10.1002/j.1460-2075.1994.tb06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid A., Riva M., Bouet F., Sentenac A., Carles C. The association of three subunits with yeast RNA polymerase is stabilized by A14. J Biol Chem. 1995 Jun 2;270(22):13534–13540. doi: 10.1074/jbc.270.22.13534. [DOI] [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989 Oct 5;341(6241):410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Tan S., Garrett K. P., Conaway R. C., Conaway J. W. Cryptic DNA-binding domain in the C terminus of RNA polymerase II general transcription factor RAP30. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9808–9812. doi: 10.1073/pnas.91.21.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Mariotte S., Buhler J. M., Sentenac A., Vu L., Lee B. S., Nomura M. Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J Biol Chem. 1995 Oct 13;270(41):24252–24257. doi: 10.1074/jbc.270.41.24252. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Zawel L., Kumar K. P., Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995 Jun 15;9(12):1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- Zomerdijk J. C., Beckmann H., Comai L., Tjian R. Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science. 1994 Dec 23;266(5193):2015–2018. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]