Abstract
Cloning by homologous recombination (HR) in Saccharomyces cerevisiae is an extremely efficient and cost-effective alternative to other methods of recombinant DNA technologies. Unfortunately, it is incompatible with all the various specialized plasmids currently used in microbiology and biomedical research laboratories, and is therefore, not widely adopted. In an effort to dramatically improve the versatility of yeast gap-repair cloning and make it compatible with any DNA plasmid, we demonstrate that by simply including a yeast-cloning cassette (YCC) that contains the 2-micron origin of replication (2 μm ori) and the ura3 gene for selection, multiple DNA fragments can be assembled into any DNA vector. We show this has almost unlimited potential by building a variety of plasmid for different uses including: recombinant protein production, epitope tagging, site-directed mutagenesis, and expression of fluorescent fusion proteins. We demonstrate the use in a variety of plasmids for use in microbial systems and even demonstrate it can be used in a vertebrate model. This method is remarkably simple and extremely efficient, plus it provides a significant cost saving over commercially available kits.
Keywords: Ligation-independent DNA cloning, Yeast homologous recombination, Heterologous protein expression, Staphylococcus aureus, Zebrafish
Complex molecular genetic techniques demand efficient and cost-effective DNA cloning methods that can be universally incorporated into any laboratory workflow. This has led to numerous methods for ligase-dependent and independent cloning. Some notable examples include TOPO-TA (Life Technologies) cloning that uses Topoisomerase I to directly insert a PCR-generated DNA fragment into a pre-processed vector. Another popular and versatile commercial method is the Gateway system (Life Technologies) that allows an existing cloned piece of DNA to be transferred from vector to vector. More recently, a number of in vitro cloning methods devised by academic labs have been described. For example, sequence and ligation-independent cloning (SLIC) uses RecA-mediated recombination and was one of the first in vitro recombination methods (Li and Elledge, 2007). The Gibson assembly method combines attributes of a 5′ exonuclease, a thermostable DNA polymerase, and T4 DNA ligase and appears to be a great advancement in constructing large DNA sequences for synthetic biology applications, but is also being widely adopt for smaller cloning experiments (Gibson et al., 2009). SLiCE (Seamless Ligation Cloning Extract) relies on a bacterial extract and is perhaps the best (and least expensive) alternative to routine recombination-mediated cloning, but long oligonucleotides are needed to achieve high efficiencies unless the engineered bacterial PYY strain is used (Zhang et al., 2012). In addition, residual plasmid template in the PCR reaction needs to be removed by DpnI digestion and all fragments have to undergo a gel extraction and purification. CPEC (Circular Polymerase Extension Cloning) is essentially the splicing by overlap extension (SOE) method that is modified to create a plasmid (Horton et al., 1989; Quan and Tian, 2009). Unfortunately, many of these methods have the major drawback of forcing laboratories to purchase expensive kits, maintain a steady supply of reagents, or are complicated to the extent that they require time-consuming optimization and troubleshooting.
A popular and highly efficient cloning alternative utilizes the ease and simplicity of homologous recombination in Saccharomyces cerevisiae. Yeast recombination cloning allows assembly of multiple DNA fragments in a single step (Oldenburg et al., 1997) and many variations on the initial method of yeast recombination/plasmid shuffle/gap repair have been described. It is extremely efficient, requiring only 29 nucleotides of overlapping sequences that can be added to the synthesized oligonucleotides. Despite this, yeast cloning is still limited in its applications because it requires a yeast compatible shuttle vector and many experiments demand specific plasmids with specialized genetic elements for defined applications. Therefore, we set out to make the yeast cloning applicable to any DNA cloning experiment by including a yeast-cloning cassette (YCC). This any-gene-any-plasmid (AGAP) cloning represents a significant improvement that facilitates universal adoption regardless of the plasmid backbone and allows anyone the ability to clone any gene (or combination of DNA fragments) into any conceivable vector in a single step without the need for ligase or molecular cloning kits. We demonstrate the usefulness and versatility of AGAP cloning by generating plasmids for recombinant protein production in Escherichia coli, epitope tagging and site-directed mutagenesis in the opportunistic pathogen Staphylococcus aureus, and we further demonstrate it can be used to create a construct to express a fluorescent fusion protein in the vertebrate model zebrafish (Danio rerio).
A detailed step-by-step protocol of the yeast transformation and other methods needed for AGAP cloning, including the strains and oligonucleotides, can be found in the supplemental material. All the strains used in this report are listed in Supplemental Table 1. S. cerevisiae strain FY2 was used for construct assembly and grown at 30 °C in YPD medium and plated on YMD medium for transformations. S. aureus and E. coli strains were routinely grown at 37 °C on solid (1.5% agar) or liquid TSB or Luria–Bertani media, respectively. Anaerobic growth was conducted on solid media in an anaerobic jar. Oxygen was replaced by a 95% N2 5% H2 headspace using a COY anaerobic chamber. When included in the growth medium, antibiotics were present in the following concentrations; ampicillin 150 μg/mL; chloramphenicol, 30 μg/mL; tetracycline, 3 μg/mL; or spectinomycin, 100 μg/mL.
The plasmids generated in this report were constructed in the following manner and all of the oligonucleotides are listed in Supplemental Table 2. To create the pET24a-Nono, the Nono CDS was first obtained by generating a cDNA library from total Trizol extracted RNA isolated from Zebrafish (Danio rerio) using Superscript III (Invitrogen) with an anchored oligo dT. The Nono CDS was then PCR amplified using oligonucleotides pet24a-NONO-F and pet24aNONO-R. The pET24a vector (EMD Millipore) was digested with Xhol, EcoRI and SphI and then heat-inactivated. The YCC was PCR-amplified from pRS426 using oligonucleotides pET-URA-F and pET-URA-R. The resulting 4 fragments were then transformed into yeast using the detail protocol listed above and plated on YMD plates. Amplification of the YCC with flanking meganuclease sites was also performed as a proof a concept. The resulting vector was ultimately transformed into BL21(DE3)RiL and induced with 0.4 M IPTG for 4 h. The cells were lysed using a French pressure cell and the protein was purified by Ni-NTA chromatography (Qiagen).
A construct containing a N-terminus flag tagged version of the srrA gene was created as follows. The pLL39 vector was linearized with BamHI and Sall. The start codon for the srrA ORF was removed and replaced by a 3X flag sequence. S. aureus strain LAC chromosomal DNA was used as template for PCR amplification of the srrAB promoter and srrAB genes. Following yeast transformation and plasmid isolation from E. coli the pLL39_FLAG_srrAB construct was confirmed by sequencing and positive clones were retained. The pLL39 vector is a single-copy integration vector system for S. aureus that contains an origin of replication for E. coli but, it can only be maintained in S. aureus by chromosomal integration. The pLL39_FLAG_srrAB construct was transformed into RN4220 containing pLL2787 and integrated onto the chromosome of S. aureus at the Φ11 attB site. Correct vector integration was verified using PCR with the Scv8 and Scv9 primers and chromosomal DNA for template. The integrated pLL39 and pLL39_FLAG_srrAB constructs were transduced from RN4220 into an srrAB double deletion mutant created in the S. aureus LAC background using transducing phage α80 creating strains JMB2579 and JMB2592, respectively. The srrAB deletion was verified using PCR with the srrABup5EcoRI and srrABdwn3SalI oligonucleotides and genomic DNA as template.
The FLAG_srrAD53A site-directed mutant was made as follows. A forward and a reverse oligonucleotide were designed that contained an internal mismatch to direct the D53 → A53 mutation (Fig. 2). These primers were used to PCR amplify the srr promoter, FLAG_srrA, and srrB DNA sequences using chromosomal DNA from strain JMB2592 as template. The alleles were integrated onto the chromosome of RN4220 and transduced into an srrABΔ mutant as described above. The directed mutation was confirmed by DNA sequencing.
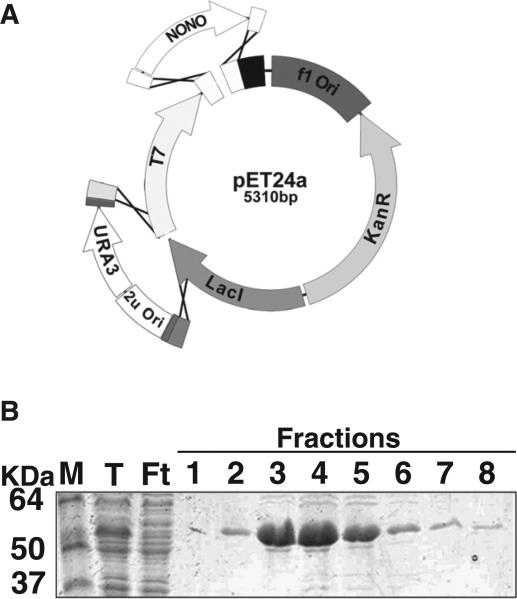
Fig. 2.
Assembly of an E. coli expression construct by AGAP. (A) The bacterial expression vector, pET24a was digested using Sphl and Xhol. The yeast-cloning cassette was inserted into the Sphl site and the Nono cDNA recombined in frame with the methionine start codon and the C-terminal 6x-his tag at the Xhol site. (B) Assembled vector was transformed into the bacterial expression cell line, BL21-RIL (DE3) for ectopic expression. Cells were grown in TSP media and induced with .4 mM IPTG for 4 h. Expressed protein was isolated by Ni-NTA affinity chromatography. Fractions off the column were resolved on a 6.5% SDS-PAGE and stained with Coomassie brilliant blue (M, marker; T, total; Ft, flow through; 1–8, collected fractions).
Detection of FLAG-tagged SrrA was performed by Western blot analysis from overnight cultures of strains JMB2579, JMB2592, and JMB3946 that were diluted 1:100 into 5.0 mL of liquid TSB. Culture aliquots were harvested at specified time points and 500 μL aliquots of the cultures were removed to assess FLAG-SrrA protein abundance. Cells were isolated by centrifugation and suspended in 200 mM Tris–HCl, pH 8.0. Cells were lysed by the addition of 20 μg of lysostaphin and DNasel and incubation at 37 °C for approximately 30 min. Lysates were clarified by centrifugation and protein concentrations of the cell free lysates were determined using a micro bca assay. Proteins were separated using SDS-PAGE chromatography (40 μg protein/lane). Proteins were transferred to a PVDF membrane and incubated with primary anti-FLAG antibody (1:4000 dilution) and subsequently secondary HRP-conjugated anti-mouse antibody (1:12,000 dilution). The blots were developed and visualized using chemiluminescent detection.
For genetic complementation analysis, strains were grown overnight in TSB (~18–20 h) to stationary phase. 2.0 μL of culture was spotted on a TSB agar plate and streaked for colony isolation. Control plates contained nitrate as an electron acceptor. Plates were incubated anaerobically, at 37C for ~18 h. Growth was analyzed for each strain with respect to the small colony variant phenotype for an srrAB deletion mutant.
The pCMV-Nono-eGFP plasmid was constructed in a similar manner. The Nono CDS was PCR-amplified from the same cDNA library used above with oligonucleotides CMVeGFP-NONO-F and CMVeGFP-NONO-R. The YCC was amplified from pRS426 using CMVeGFP-URA-F and CMVeGFP-URA-R. The pEGFP-C1 plasmid (Clonetech) was digested with Asel and EcoRI and then heat inactivated. The 4 fragments were transformed into yeast FY2 using YMD plates as selection. The vector was recovered in E. coli, linearized, then injected into zebrafish embryos and examined by fluorescence microscopy.
All zebrafish husbandry and experimental procedures complied with policies set forth by the NIH Office of Laboratory Animal Welfare and approved by Rutgers University Institutional Animal Care and Use Committees (IACUC).
Conceptually, the AGAP cloning method is remarkably straightforward and is based on yeast gap-repair/plasmid shuffle (Ma et al., 1987). Multiple DNA fragments containing at least 29 nucleotides of overlapping homologous sequence are mixed with a linearized vector backbone along with the YCC. The YCC contains the yeast 2 μm origin of replication (2 μm ori) and the ura3 gene amplified from pRS426. DNA fragments are obtained by PCR amplification using oligonucleotides containing at least 29 nucleotides of sequence identity between each fragment. The amplified YCC, DNA fragments, and linearized plasmid are assembled by transformation into a yeast strain lacking a functional ura3 gene, which provides the selective pressure to facilitate appropriate construct assembly. Plasmids are then recovered from yeast DNA by transforming into E. coli. The YCC and DNA fragments can be assembled in tandem or at separate plasmid locations. Homing meganuclease sites or unique restriction endonuclease sites flanking the YCC can also be included for subsequent removal if desired, but in routine applications, this is unnecessary.
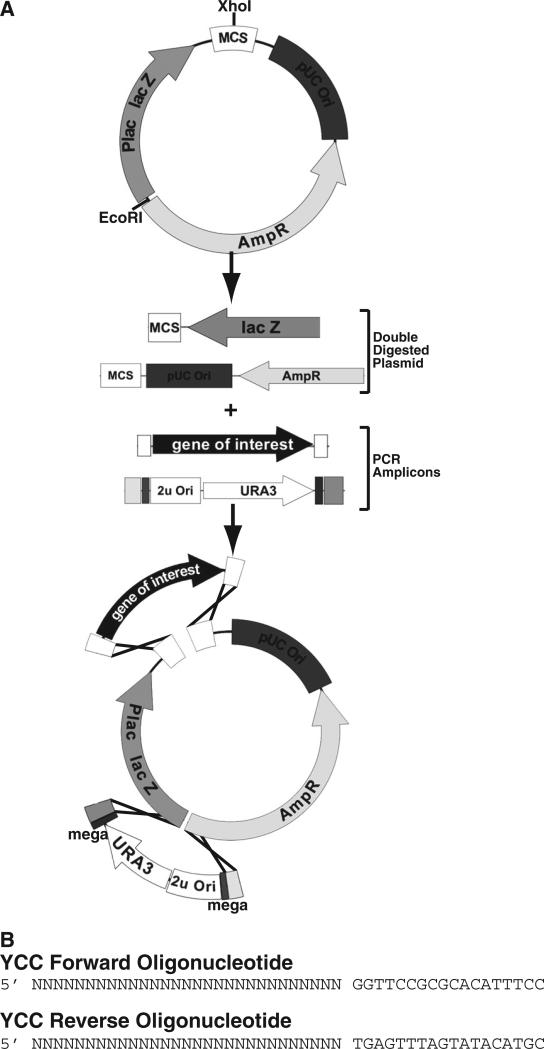
A general outline of the procedure is shown in Fig. 1. Prior to beginning AGAP cloning, one must find a minimum of two unique restriction endonuclease sites within the vector and then double digest the plasmid with both enzymes (This example uses Xhol and EcoRI). One enzyme is also sufficient, but may cause elevated background. The restriction endonuclease recognition sequences can both be located in the multiple cloning site (MCS) and the YCC can be inserted in tandem with the fragment(s). Alternatively, the YCC can be inserted at a separate location (as shown in the figure). If the YCC is inserted in a location separate from the MCS, then care must be taken to ensure no important elements within the vector (regulatory regions, replication origins, selectable markers, etc.) are disrupted by the insertion. The 29 nucleotides of sequence identity (adjacent to the enzyme cut site) needed for homologous recombination is added to the 5′ end of the synthesized oligonucleotide. Once the vector is digested and the fragments amplified, the mixture of linearized DNA fragments is transformed into yeast using the protocol contained in the Supplemental material.
Fig. 1.
Schematic representation of the AGAP method. (A) The desired starting vector is digested with two separate restriction endonuclease and then heat inactivated. Digested vector is mixed with a PCR amplified fragment(s) and the YCC. The fragments are then transformed into yeast for HR-mediated assembly. The YCC can be assembled in tandem with your fragment(s) or located at second unique site within vector. (B) The oligonucleotide sequence to amplify the YCC from pRS426. The “Ns” indicate the homologous nucleotide sequence to the vector integration site.
The elegance of this system is that multiple PCR-amplified DNA fragments can be assembled into any E. coli compatible shuttle vector by simply adding the YCC in a transformation reaction. Thus AGAP can assemble complex constructs with ease, simplicity, and high efficiency in any desired plasmid. This method eliminates the need for expensive reagents or kits and can easily be adopted by any laboratory.
A common use for cloning is the heterologous expression of protein in E. coli. There are a plethora of expression vectors to choose from, many with different affinity purification epitopes. Unfortunately, none of these can be used with yeast cloning because the plasmids lack the appropriate genetic material for selection and amplification in yeast. Therefore, to demonstrate the ease and applicability of AGAP cloning, we constructed a bacterial expression construct for recombinant protein expression and purification (Fig. 2A). For this application, we inserted a fragment containing the nono coding sequence (CDS) from zebrafish (Danio rerio) (GenBank #NM_201579) into pET24a (EmD Millipore). The nono CDS was amplified from a cDNA library generated with an anchored oligo dT. The pET24a vector was linearized by enzymatic digestion with three restriction enzymes SphI, EcoRI and Xhol. The digested plasmid (2 fragments) was combined with PCR amplified YCC and the nono CDS amplicon (four fragments total) and transformed into yeast strain FY2 (Winston et al., 1995) (Supplemental Table 1). The oligonucleotides used to amplify the nono CDS were designed to ensure insertion occurred in-frame with the poly-histidine affinity tag. The assembled plasmid was recovered from a yeast genomic DNA preparation and subsequently transformation into E. coli strain BL21 (DE3)-RIL. Protein production was achieved by induction with IPTG. Recombinant Nono protein was purified using standard Ni-NTA chromatography and visualized by SDS-PAGE (Fig. 2B). It is clear from the results that this method allows for easy and efficient generation of protein expression constructs for recombinant protein production. This may have useful applications in the structural biology field because yeast recombination cloning has been successfully used on robotic platforms for high throughput construct assembly (Colot et al., 2006).
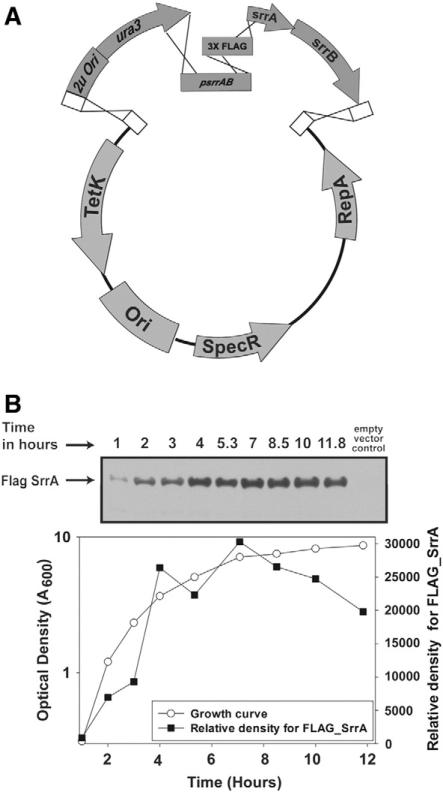
In addition to commercially available or commonly used plasmids, there are many vector backbones that are designed for specific organisms. This poses a significant obstacle for many laboratories and makes them reliant on conventional ligase-dependent cloning with few alternatives. An example of this is the S. aureus vector pLL39 (Luong and Lee, 2007). To highlight some of the broader applications of AGAP cloning, we assembled numerous DNA fragments in a single cloning step for monitoring endogenous protein abundance in the pathogenic bacterium S. aureus. The S. aureus srrAB genes encode a two component regulatory system that is arranged in an operon (Throup et al., 2001; Yarwood et al., 2001). SrrA is the DNA binding protein and SrrB is the cognate sensor histidine kinase. In order to monitor SrrA protein abundance in cells, we generated a construct that attached a FLAG epitope at the amino terminus. This was performed by first digesting the plasmid pLL39 (Luong and Lee, 2007) with BamHI and SalI. Digested pLL39 was then combined with PCR amplicons consisting of the YCC, srrAB promoter, a codon optimized 3X FLAG sequence, and srrAB CDS. The plasmid was generated by transforming all the fragments into yeast FY2 and recovered in E. coli (Fig. 3A). The epitope tagged FLAG-SrrA construct was integrated onto the chromosome of S. aureus and transduced into an srrABΔ strain. The srrAFLAGsrrB alleles genetically complemented the anaerobic growth defect of an srrAB mutant confirming the functionality of FLAG-SrrA (data not shown). Cytosolic, soluble FLAG-SrrA was confirmed using Western blots and protein abundance was found to increase over the growth period (Fig. 3B). This documents a simple and efficient method to attach amino-terminal epitope to a protein in a pathogenic bacterium where no commercially available vector exists.
Fig. 3.
Adding a protein epitope for protein detection. (A) AGAP was used to insert an N-termial epitope on SrrA in S. aureus. The pLL39 plasmid was digested with BamHI and Sall and mixed with 3 separate PCR generated fragments. The construct was assembled by transforming all the linearized DNA fragments into yeast, recovered in E. coli, and integrated onto the S. aureus chromosome. The construct contains the native srrAB promoter and codon optimized 3X FLAG sequence fused in frame with N-terminus of SrrA. (B) SrrA protein production increases during growth. Culture density (A600 nm) was monitored over 12 h. Every hour cells were harvested, lysed, and SrrA protein abundance monitored. FLAG-SrrA protein abundance was monitored by Western blot analysis with antibodies raised against the FLAG epitope (SIGMA).
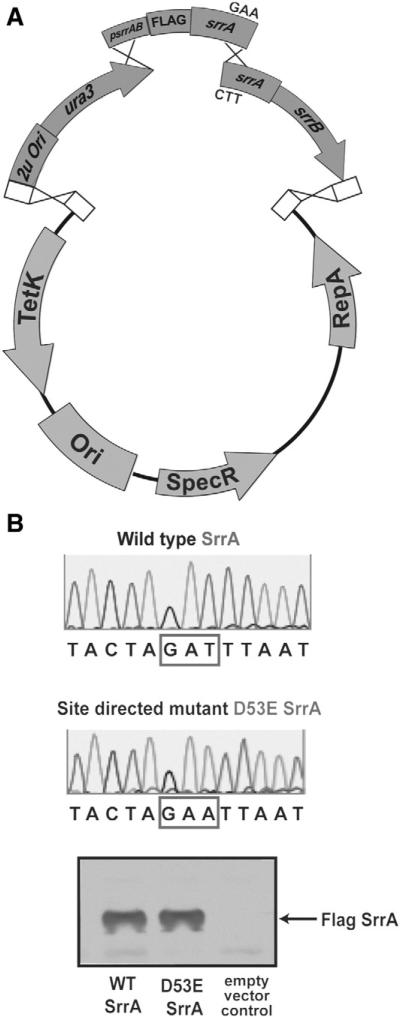
Another routine technique in molecular genetics is the use of site-directed mutagenesis that allows one to identify critical amino-acids contained within a protein. A common method to create site-directed mutants uses the site-directed mutagenesis kit (Statagene) that relies on the ability of DpnI restriction enzyme to preferentially digest adenine methylated DNA. This kit is extremely efficient and is a widely used method. However, it can be difficult when working with large genes or operons. Therefore, as a further demonstration of the versatility of AGAP, we conducted site directed mutagenesis to create a mutant srrA allele. AGAP is ideally suited to incorporate site directed mutants because base-pair changes can be designed into the overlapping oligonucleotide sequences. We designed our oligonucleotide primers to change the coding sequence for aspartate at amino-acid 53 to alanine creating the srrAD53A allele. Amplified products were mixed with linearized pLL39 and the YCC, transformed into yeast, and recovered with E. coli (Fig. 4A). Incorporation of the site-directed mutation was sequence verified (Fig. 4B) and the srrAFLAG-D53EsrrB alleles were mobilized into S. aureus. Western blot analysis confirmed variant protein production and stability in S. aureus (Fig. 4C). These results demonstrate that AGAP cloning is a suitable and less expensive method to create a site-directed mutation.
Fig. 4.
AGAP to construct site-specific mutations (A) Site-directed mutagenesis of the srrA allele using AGAV cloning. The pLL39 plasmid was digested with BamHI and Sall. The yeast cloning cassette, the native srrAB promoter, 3X FLAG sequence and part of srrA ORF was amplified as one piece. Oligonucleotides, containing internally directed mismatch mutations that changed aspartate 53 of SrrA were used to amplify the remaining portion of srrA and srrB ORFs. The two pieces were combined with digested vector and the YCC to obtain the srrAFLAG-D53AsrrB construct. (B) DNA sequence analysis was used to verify the D53A mutation in the srrA coding sequence. (C) The mutant FLAG-SrrAD53A variant protein is soluble and could be detected in S. aureus cellular lysates by Western blotting.
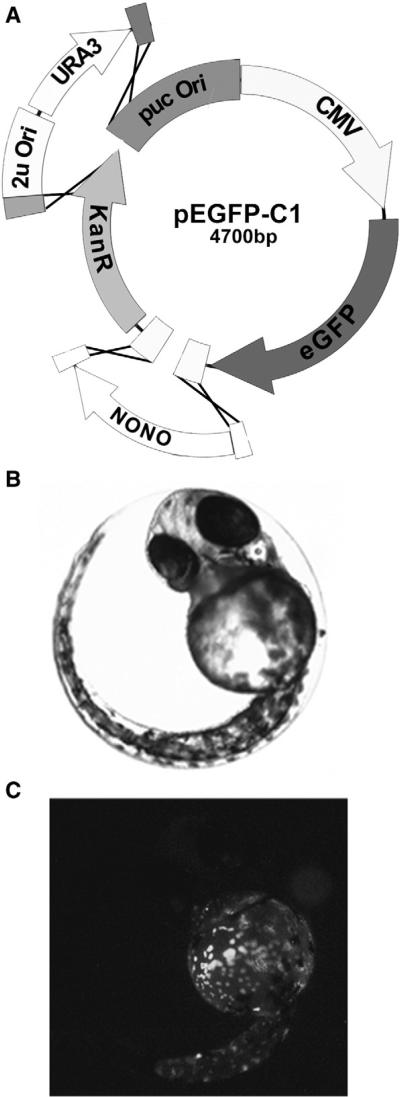
To demonstrate that this method is not limited to just microbial systems, we sought to create a fusion protein that could be used for in vivo visualization in mammalian cell culture or whole organisms. For this example we choose to use the enhanced green fluorescent protein (eGFP) that is used to track proteins in cell culture or visualize proteins in organisms such as zebrafish. Normally, constructing an eGFP fusion protein involves cloning a cDNA fragment in-frame with the egfp gene. There are numerous commercially available vectors that contain egfp and other fluorescent protein genes, but none are currently adapted for ligase independent cloning. Therefore, to demonstrate how AGAP is ideally suited for this application, we constructed a vector that contains Nono CDS fused in frame with eGFP for visualization in zebrafish. The nono cDNA sequence was cloned into the pEGFP-C1 (Clonetech) plasmid, which contains the cytomegalovirus promoter (CMV) and an eGFP. The nono CDS was PCR amplified with primers designed to ensure in-frame insertion with the egfp gene. The nono CDS and YCC were combined with linearized pEGFP-C1 digested with EcoRI and AseI (four fragments total; Fig. 5A). Following passage and recovery in E. coli, purified plasmid was microinjected into zebrafish (fertilized) eggs for visualization (Fig. 5B and C). Nono-GFP accumulation exhibited a punctate pattern that appeared consistent with paraspeckle nuclear body formation (Fox and Lamond, 2010; Passon et al., 2012). Thus we demonstrate that AGAP is ideally suited for construction of fluorescent fusion protein in pre-existing plasmids. Implementation of this cloning technique is likely to prove extremely useful to the vast number of laboratories that routinely use fluorescent microscopy especially for host–pathogen interaction studies.
Fig. 5.
Construction of a fluorescent fusion protein with AGAP. (A) The Nono CDS was cloned into pCMVeGFP for in vivo detection in zebrafish embryos. The vector was digested with AseI, and EcoRI and the YCC were inserted into AseI site and Nono CDS into the EcoRI site by homologous recombination in yeast. (B) The recovered plasmid was injected into fertilized zebrafish eggs and observed by light and fluorescence microscopy 48 h post-fertilization to examine NONO-eGFP expression and localization.
In this report, we demonstrate a modification to yeast cloning that makes it applicable to almost all conceivable recombinant DNA cloning experiments, yet it does not require the need for any specific molecular biology kit or predefined vector. This makes it a truly inexpensive alternative to other techniques that have been described over the past decade. Moreover, we have found that AGAP cloning is more efficient than standard gap-repair cloning and we routinely found that nearly 100% of the recovered plasmids contained the properly assembled inserts when the YCC is included as an independent fragment. This is significantly higher than the 50–80% efficiency observed using traditional gap-repair (Shanks et al., 2006) and far exceeds the efficiency of ligase-mediated cloning. This accentuates how AGAP cloning is a more efficient, reliable, and cheaper than any other cloning technique currently available. Moreover, the AGAP cloning method enables direct assembly of any DNA fragment(s) into any E. coli compatible plasmid in a single step without the need for ligase, a yeast shuttle vector, or costly molecular cloning kits.
AGAP cloning imposes only minor theoretical limitations. Most notable are difficulties with highly repetitive sequences or direct cloning of yeast genes. However, conceptually and experimentally, we have bypassed the need for a preexisting yeast shuttle vector, paving the way for a remarkably simple procedure that facilitates widespread adoption by any molecular biologist regardless of educational background. In addition, the method limits the amount of time needed to prepare the fragments. There is no need to gel purify PCR generated fragments provided the PCR results in a single band on an agarose gel. This further eliminates costs and time-consuming steps. The AGAP method has broad versatility and almost unlimited applications which we demonstrate by cloning basic and complex DNA constructs for recombinant protein production, expression of fusion proteins, epitope tagging, and site-directed mutagenesis in a single step. Importantly, construction of these plasmids required little bench time (~6 h including the time to amplify the fragments and recover the final plasmid) and the reagent costs are less than would be required for conventional cloning. Multiple reactions can be carried through with no major increase in effort. Like all molecular cloning experiments, this method requires careful attention in the design of the oligonucleotides sequences. Typically, the only error that arises occurs in poor oligonucleotides design. Therefore, we highly encourage careful and systematic in silico review of the designed constructs and primer sequences prior to ordering oligonucleotides.
Supplementary Material
Acknowledgments
Work presented in this report was made possible by funds received from the School of Environmental and Biological Sciences at Rutgers University, and New Jersey Agriculture Experiment Station to TMJ, JMB, and WJB. WJB is supported by the NIH (RO1GM101378). WJB is part of NIEHS Center for Environmental Exposure and Disease (ceed) (P30ES005022). Parts of this work were also made possible by funding from the Charles and Joanna Busch Memorial fund (WJB and JMB).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mimet.2013.11.013.
References
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Luong TT, Lee CY. Improved single-copy integration vectors for Staphylococcus aureus. J. Microbiol. Methods. 2007;70:186–190. doi: 10.1016/j.mimet.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl. Environ. Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throup JP, Zappacosta F, Lunsford RD, Annan RS, Carr SA, Lonsdale JT, Bryant AP, McDevitt D, Rosenberg M, Burnham MK. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry. 2001;40:10392–10401. doi: 10.1021/bi0102959. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 2001;183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.