Abstract
Reinitiation is a strategy used by viruses to express several cistrons from one mRNA. Although extremely weak after translation of long ORFs on cellular mRNAs, reinitiation occurs efficiently on subgenomic bicistronic calicivirus mRNAs, enabling synthesis of minor capsid proteins. The process is governed by a short element upstream of the restart AUG designated “termination upstream ribosomal binding site” (TURBS). It contains the conserved Motif 1 complementary to h26 of 18S rRNA, displayed in the loop of a hairpin formed by species-specific Motifs 2/2*. To determine the advantages conferred on reinitiation by TURBS, we reconstituted this process in vitro on two model bicistronic calicivirus mRNAs. We found that post-termination ribosomal tethering of mRNA by TURBS allows reinitiation by post-termination 80S ribosomes, and diminishes dependence on eIF3 of reinitiation by recycled 40S subunits, which can be mediated either by eIFs 2/1/1A or by Ligatin following ABCE1-dependent or -independent splitting of post-termination complexes.
Keywords: calicivirus, reinitiation, recycling, TURBS, eIF3, ribosome
INTRODUCTION
On most eukaryotic mRNAs, translation initiation occurs by the scanning mechanism (Jackson et al., 2010). The process begins with assembly of a 43S preinitiation complex, comprising a 40S ribosomal subunit, an eIF2-GTP/Met-tRNAiMet ternary complex (eIF2-TC), and eIFs 3, 1 and 1A. 43S complexes attach to the 5′-proximal region of mRNA in a process that involves unwinding of its secondary structure by eIFs 4A, 4B and 4F, and then scan to the initiation codon where they form 48S initiation complexes with established codon-anticodon base-pairing. After start codon recognition, eIF5 and eIF5B promote joining of a 60S subunit, yielding elongation-competent 80S ribosomes.
However, in some instances, protein synthesis begins not with bona fide initiation, but instead results from reinitiation in the vicinity of the stop codon following incomplete recycling of post-termination complexes (post-TCs) (Jackson et al., 2012). Reinitiation enables viral mRNAs to maximize utilization of the coding capacity of their genomes and forms the basis of important mechanisms of translational control (e.g. Barbosa et al., 2013).
During recycling, ABCE1, in concert with eRF1, splits post-TCs into free 60S and tRNA/mRNA-associated 40S subunits (Pisarev et al., 2010). Subsequent release of tRNA can be promoted by eIF1, Ligatin, or MCT1 and DENR (interacting proteins that are homologous to Ligatin’s N- and C-terminal regions), and is followed by dissociation of mRNA (Pisarev et al., 2007, 2010; Skabkin et al., 2010). In addition to releasing tRNA from recycled 40S subunits, Ligatin and MCT1-DENR can promote attachment of Met-tRNAiMet to 40S/mRNA complexes, if the initiation codon is placed directly in the P site (e.g. Skabkin et al., 2010). At low [Mg2], the entire recycling process can also be mediated by eIFs 3, 1 and 1A, with eIF3 being primarily responsible for splitting of post-TCs (Pisarev et al., 2007). If 40S subunits remain on mRNA, termination is followed by reinitiation, usually downstream of the stop codon. Efficient reinitiation commonly occurs only after translation of short ORFs, and depends on the duration of elongation (Jackson et al., 2012). These observations are consistent with the early hypothesis that some eIFs remain associated with ribosomes over several elongation cycles, and those 40S subunits that retain them will be able to reinitiate after dissociation of 60S subunits (Kozak, 1987). Recently, it was suggested that these factors are ribosome-bound eIF3 in association with eIF4F (Pöyry et al., 2004).
Recapitulation of reinitiation in vitro on purified, factor-free pre-termination complexes (pre-TCs) assembled on a β-globin mRNA derivative showed that if splitting of post-TCs proceeds in the presence of eIFs 3/1/1A and eIF2-TC, 40S subunits remain on mRNA and reinitiate at nearby upstream and downstream AUGs (Skabkin et al., 2013). Imposing of 3′-directionality additionally requires eIF4F. eIF3 is essential for the process, likely ensuring ribosomal retention of mRNA (Kolupaeva et al., 2005). Inefficient reinitiation in vivo after translation of long ORFs could therefore result from potentially low relative concentrations of free eIF3, in which case tRNA release from eIF3-unbound 40S subunits would be followed by prompt dissociation of mRNA. Efficient reinitiation after short ORFs, on the other hand, would be consistent with the transient association of eIF3 with ribosomes through several elongation cycles. It was also found that post-termination ribosomes are not stably anchored on mRNA, and can slide to nearby codons that are cognate to the P site tRNA (Skabkin et al., 2013). The mobility is caused by destabilization of P site codon-anticodon base-pairing due to adoption by deacylated tRNA of the P/E hybrid state (McGarry et al., 2005). Association with eRF1, elevated [Mg2+], and the presence of the E site tRNA increase the stability of post-TCs. eEF2, on the other hand, stimulates ribosome migration by destabilizing ribosomal association of eRF1 and promoting the P/E hybrid state. Deacylated tRNA could also spontaneously exchange with tRNAiMet, arresting post-termination ribosomes at nearby AUGs (Skabkin et al., 2013). Thus, some reinitiation events could involve post-termination ribosomes rather than recycled 40S subunits.
Whereas reinitiation after long ORFs on cellular mRNAs is extremely weak, it occurs efficiently on mRNAs from several virus families. The best-characterized example is reinitiation on positive strand RNA viruses of the family Caliciviridae, such as rabbit hemorrhagic disease virus (RHDV) (Meyers 2003, 2007), feline calicivirus (FCV) (Lütterman and Meyers 2007, 2009; Pöyry et al., 2007) and bovine, human and murine noroviruses (NV) (Lütterman and Meyers, 2014; McCormick et al., 2008; Naphthine et al., 2009). In bicistronic subgenomic (sg) calicivirus mRNAs encoding major and minor capsid proteins, translation of the second cistron occurs by reinitiation. Stop and restart codons usually overlap, but can be separated by several nucleotides. The process depends on a ~40–80nt-long element upstream of the restart AUG called TURBS (termination upstream ribosome binding site). It includes two essential elements: the conserved Motif 1, whose UGGGA core is complementary to the apical loop of h26 of 18S rRNA, and species-specific Motif 2 (12–23nt upstream of the restart AUG), which is complementary to Motif 2* immediately preceding Motif 1 (Lütterman and Meyers, 2009; McCormick et al., 2008; Meyers 2003, 2007; Naphthine et al., 2009). The importance of complementarity between Motif 1 and h26 was demonstrated in in vivo experiments involving mutagenesis of 18S rRNA (Lütterman and Meyers, 2009). In the current model, Motif 1 is displayed in the loop of a hairpin formed by Motifs 2/2* at the mRNA exit area of the 40S subunit in such a way that it engages with the apex of h26, tethering mRNA to the ribosome. TURBS was also shown to interact with eIF3 (Pöyry et al., 2007), but the role of this interaction is unknown.
Whereas cis-acting RNA elements required for calicivirus reinitiation have been mapped in considerable detail, the mechanism of the process and its factor requirements remain obscure. To determine the advantages TURBS confers on reinitiation, we recapitulated this process in vitro on two model mRNAs containing RHDV and human NV TURBS elements.
RESULTS
Assembly of pre-termination complexes on model RHDV and NV mRNAs
To investigate the mechanism of calicivirus reinitiation, we employed an in vitro reconstitution approach, in which pre-TCs assembled on the ORF1 stop codon are treated with eRFs in the presence of tRNAiMet and different combinations of eIFs and ABCE1. Formation of ribosomal complexes at the ORF2 restart codon is then assayed by toe-printing. For this we constructed two model mRNAs containing RHDV or NV TURBS elements, in which restart AUGs are located upstream or downstream of the stop codon, respectively. The mRNAs comprised the β-globin 5′UTR with the adjacent coding region (24 nts), followed by minimal elements required for reinitiation (Meyers, 2007; Lüttermann and Meyers, 2009) and the first ~200 nts of ORF2 (Figs. 1A–B).
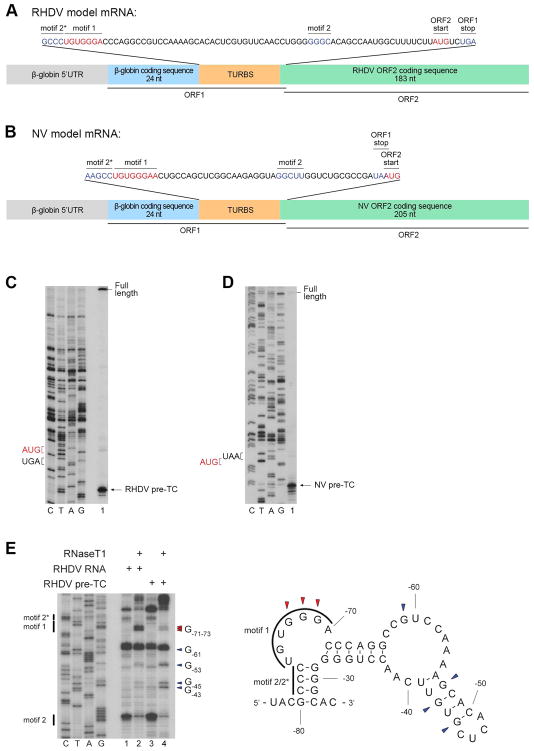
Figure 1. Characterization of pre-TCs assembled on model RHDV and NV mRNAs.
(A, B) Structural diagrams of model RHDV and NV mRNAs with sequences of TURBS and the adjacent stop (blue) and restart (red) codons shown in the inserts. (C, D) Toe-printing analysis of pre-TCs assembled on (C) RHDV and (D) NV mRNAs in RRL in the presence of eRF1AGQ and purified by SDG centrifugation. Black arrows indicate the positions of pre-TCs. (E) Left panel: RNase T1 foot-printing analysis of TURBS in free and pre-TC-engaged RHDV mRNA. Cleavage sites are indicated on the right (red arrows correspond to residues protected in pre-TCs). Right panel: cleavage sites mapped onto the predicted mFold secondary structure of TURBS.
The attempt to assemble pre-TCs on these mRNAs in an in vitro reconstituted system (from ribosomal subunits, eIFs, eEFs and Σaa-tRNAs) was not successful because elongation was strongly arrested at some codons due to low levels of corresponding tRNAs in commercial tRNA preparations. Since mutagenesis of these codons could potentially disrupt the TURBS structure, pre-TCs were instead formed by translation of RHDV and NV mRNAs in rabbit reticulocyte lysate (RRL) in the presence of eRF1AGQ mutant (with a G183A substitution in the GGQ motif), which can bind to pre-TCs but cannot trigger peptide release and therefore arrests ribosomal complexes at the pre-termination stage. Subsequent sucrose density gradient (SDG) centrifugation dissociated eRF1AGQ, yielding factor-free pre-TCs with characteristic toe-prints +16nt downstream of the P-site codon (Figs. 1C–D).
To evaluate the structural functionality of pre-TCs, we used RNase T1 footprinting to verify that Motif 1 is engaged in base-pairing. In free RHDV mRNA, the pattern of TURBS cleavage correlated well with the predicted mFold secondary structure, with strong cuts at GGG-71-73 and G-61 in the single-stranded regions, and moderate cleavage at G-43, G-45 and G-53 in the weak distal stem-loop (Fig. 1E). In pre-TCs, GGG-71-73 in Motif 1 became strongly protected (Fig. 1E), consistent with their base-pairing with 18S rRNA. The moderate enhancement of cleavage at G-43, G-45 and G-53 in the distal stem-loop was likely caused by the slightly elevated concentration of RNase T1 that was used in the case of pre-TCs to compensate for the additional presence of rRNA. Similar protection of Motif 1 was observed in pre-TCs formed on NV mRNA (data not shown).
However, RRL-derived pre-TCs assembled on RHDV and NV mRNAs contained two additional proteins, which were not present in individual ribosomes or pre-TCs assembled in an in vitro reconstituted system on MVHL-STOP mRNA (a derivative of β-globin mRNA, encoding an MVHL tetrapeptide followed by a UAA stop codon; Alkalaeva et al., 2006) (Fig. 2A; data not shown). These proteins were not specific to RHDV and NV mRNAs, since pre-TCs assembled in RRL on MVHL-STOP mRNA contained identical protein bands (Fig. 2A). Mass spectrometry identified them as eEF2 and serpine1 mRNA-binding protein 1 (SERBP1) (Tables S1–2). Since Stm1, the yeast SERBP1 homologue, was suggested to arrest translation downstream of 80S complex formation (Balagopal and Parker, 2011), we investigated SERBP1’s influence on different stages in protein synthesis. SERBP1 was purified from RRL and also expressed in E. coli as a His-tagged protein (Fig. 2B). The activities of both forms were identical.
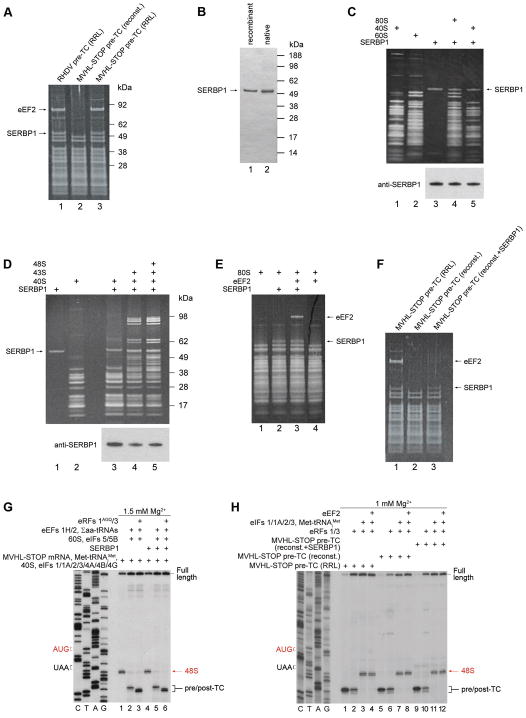
Figure 2. Ribosome-binding properties of SERBP1 and its influence on initiation, elongation, termination and reinitiation.
(A) Protein composition of SDG-purified pre-TCs assembled on RHDV and MVHL-STOP mRNAs in RRL or in an in vitro reconstituted system, assayed by SDS-PAGE followed by SYPRO staining. (B) Purified native and recombinant SERBP1 resolved by SDS-PAGE. (C–F) Association of SERBP1 and eEF2 with 40S subunits, 80S ribosomes, 43S and 48S complexes, and pre-TCs. Ribosomal complexes were separated by SDG centrifugation, and SERPB1’s and eEF2’s presence in ribosomal peak fractions was analyzed by SDS-PAGE followed by (C–F) SYPRO staining and (C, E) western blotting using SERBP1 antibodies. (G) Toe-printing analysis of SERBP1’s influence on initiation, elongation, and association of pre-TCs with eRFs on MVHL-STOP mRNA. (H) Toe-printing analysis of ABCE1-independent reinitiation by eIFs in the presence/absence of eEF2 on pre-TCs assembled on MVHL-STOP mRNA in an in vitro reconstituted system (with/without SERBP1) and in RRL. (G, H) Positions of 48S complexes (red arrows) and pre/post-TCs (black brackets) are shown on the right. Free [Mg2+] is indicated on each panel. See also Tables S1–2.
SERBP1 efficiently bound to vacant 80S ribosomes, 40S subunits, 43S and 48S complexes, and pre-TCs (Figs. 2C–F). It also stabilized ribosomal association of eEF2 with vacant ribosomes, but not with pre-TCs (Figs. 2E–F), consistent with the non-rotated conformation of the ribosome in the latter (des Georges et al., 2014). Taking the ribosome-binding properties of eEF2 and SERBP1 into account, we conclude that preparations of RRL-derived pre-TCs contained true pre-TCs that could be associated with SERBP1, and un-programmed 80S ribosomes bound to both proteins. Importantly, SERBP1 did not affect initiation or elongation on MVHL-STOP mRNA in an in vitro reconstituted system, and pre-TCs assembled in its presence retained the ability to interact with eRFs, evidenced by the appearance of the characteristic +2nt toe-print shift (Fig. 2G; Alkalaeva et al., 2006). To investigate the influence of SERBP1 and eEF2 on reinitiation, we compared this process on pre-TCs assembled on MVHL-STOP mRNA in an in vitro reconstituted system (with/without SERBP1) and in RRL. At 1 mM free Mg2+, ABCE1-independent renitiation by eIFs 2/3/1/1A was equally efficient on reconstituted and RRL-derived pre-TCs, and was not affected by eEF2: in all instances, recycled 40S subunits efficiently scanned mRNA in the 5′-direction, forming 48S complexes at the AUG of the MVHL ORF (Fig. 2H). Analogous results were obtained for reinitiation following ABCE1-mediated splitting of post-TCs at higher [Mg2+] (data not shown). Thus, SERBP1 and eEF2 did not affect reinitiation.
Taken together, these data indicated that RRL-derived pre-TCs assembled on synthetic RHDV and NV mRNAs constitute a valid experimental model for studying calicivirus reinitiation.
Reinitiation by post-termination ribosomes on model RHDV and NV mRNAs
To determine whether reinitiation on calicivirus mRNAs can be executed by post-termination ribosomes, pre-TCs were treated with eRFs in the presence/absence of Met-tRNAiMet, and the position of ribosomes in the resulting complexes was mapped by toe-printing. At 1.5 mM free Mg2+, incubation with eRFs of pre-TCs assembled on RHDV mRNA yielded toe-prints corresponding to eRF1-associated post-TCs (characterized by the +2nt shift), full-length cDNA (indicative of dissociation of deacylated tRNA), and weak toe-prints +16–17nt from the ORF2 AUG, which were strongly enhanced by Met-tRNAiMet (Fig. 3A), indicating that they corresponded to programmed ribosomes assembled on the ORF2 AUG. Thus, on RHDV mRNA, post-termination ribosomes could efficiently rebind initiator tRNA and move 2nt upstream to reinitiate at the ORF2 AUG. Low-level reinitiation in the presence of eRFs alone could be explained by trace contamination of RRL-derived pre-TCs by tRNAiMet (Fig. S1).
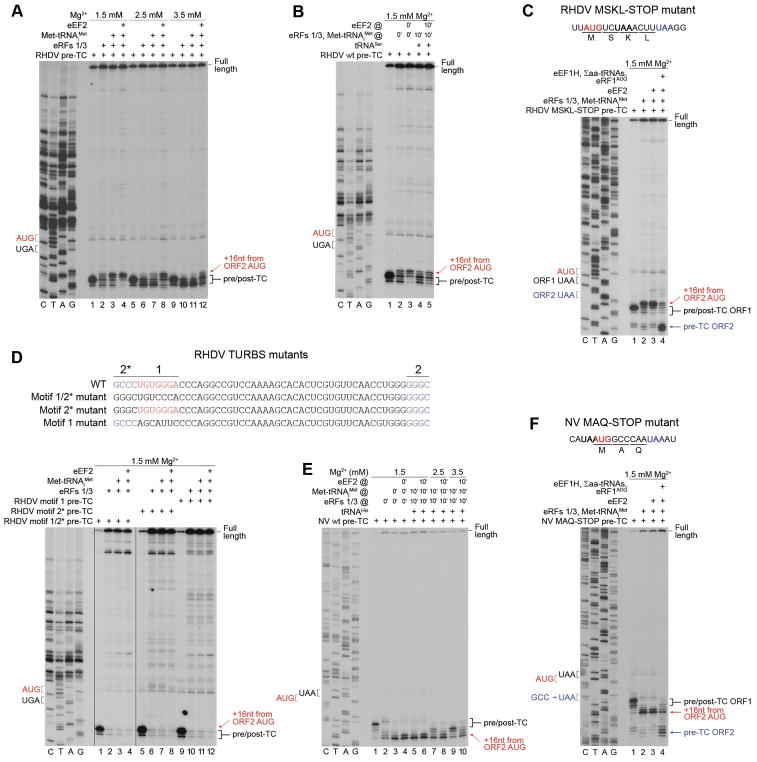
Figure 3. Reinitiation by post-termination ribosomes on model RHDV and NV mRNAs.
(A) Toe-printing analysis of reinitiation by post-termination ribosomes on RHDV mRNA, depending on [Mg2+] and the presence of eEF2. (B) The influence of preincubation of pre-TCs with deacylated tRNA on reinitiation by post-termination ribosomes on RHDV mRNA. (C) Toe-printing analysis of the ability of reinitiation complexes formed on RHDV mRNA to undergo elongation, assayed using “MSKL-STOP” mutant mRNA. (D) Toe-printing analysis of reinitiation by post-termination ribosomes on RHDV TURBS mutant mRNAs, containing substitutions in motifs 1 and 2*. (E) Toe-printing analysis of reinitiation by post-termination ribosomes on NV mRNA, depending on [Mg2+], the presence of eEF2, and preincubation of pre-TCs with deacylated tRNA. (F) Toe-printing analysis of the ability of reinitiation complexes formed on NV mRNA to undergo elongation, assayed using “MAQ-STOP” mutant mRNA. (A–F) Positions of pre/post-TCs on ORF1 (black brackets), 80S reinitiation complexes on ORF2 AUG (red arrows), and eRF1AGQ-associated pre-TCs on ORF2 (blue arrows) are shown on the right. Positions of stop (black or blue) and restart (red) codons are marked on the left. Free [Mg2+] is indicated on each panel. See also Figure S1.
Consistent with previous findings (Skabkin et al., 2013), eEF2 promoted dissociation of eRF1 and destabilized post-TCs, which modestly stimulated reinitiation (Fig. 3A, lane 4). In contrast, elevation of [Mg2+] progressively stabilized post-TCs and concomitantly reduced reinitiation, so that at 3.5 mM Mg2+, low-level reinitiation was observed only in the presence of eEF2 (Fig. 3A, lanes 5–12). Reinitiation was also reduced by preincubation of pre-TCs with tRNASer, which bound to the vacant E-site of SDG-purified pre-TCs (des Georges et al., 2014) and stabilized ribosomal complexes after peptide release (Fig. 3B).
The elongation competence of ribosomes arrested at the ORF2 AUG was confirmed using RHDV “MSKL” mutant mRNA, in which the fifth codon of ORF2 was replaced by a stop codon. Upon addition of eEF2, eEF1H and Σaa-tRNAs, reinitiation complexes formed at the ORF2 AUG of “MSKL” mRNA underwent elongation, yielding stable pre-TCs at the new stop codon in the presence of eRF1AGQ (Fig. 3C). Importantly, no reinitiation occurred on RHDV mRNAs with mutated motif 1, motif 2* or both; moreover, post-TCs formed on these mRNAs were highly unstable (Fig. 3D).
As in the case of RHDV mRNA, at 1.5 mM Mg2+, incubation with eRFs of pre-TCs formed on NV mRNA yielded toe-prints +16–17nt from the ORF2 AUG that were strongly enhanced on addition of Met-tRNAiMet (Fig. 3E, lanes 1–3), indicating that post-termination ribosomes could efficiently rebind initiator tRNA and move 5nt downstream to reinitiate at the ORF2 AUG. Weak toe-prints 1–2nt upstream of the toe-prints attributed to pre-TCs, which appeared after incubation of pre-TCs with eRFs alone, likely corresponded to ribosomes lacking P-site codon-anticodon base-pairing and stabilized by the TURBS. The main difference between RHDV and NV mRNAs was that reinitiation on the latter was less sensitive to preincubation of pre-TCs with deacylated tRNA and to elevation of [Mg2+]: substantial reinitiation, which was fully restored by eEF2, occurred at 3.5 mM Mg2+ even if pre-TCs were preincubated with deacylated tRNA (Fig. 3E, lanes 5–10). The elongation competency of reinitiation complexes was confirmed using NV “MAQ” mutant mRNA, in which the fourth codon was replaced by a stop codon (Fig. 3F).
Thus, at low [Mg2+], efficient reinitiation on calicivirus mRNAs can be promoted by post-termination ribosomes. The integrity of TURBS is essential for the process.
Reinitiation by recycled 40S subunits on model RHDV mRNA
Efficient reinitiation on NV mRNA by post-termination ribosomes over a wide range of [Mg2+] creates a high background, and reinitiation by recycled 40S subunits was therefore assayed only on RHDV mRNA, where reinitiation by 80S ribosomes was nearly abolished at 3.5 mM Mg2+ (Fig. 3A).
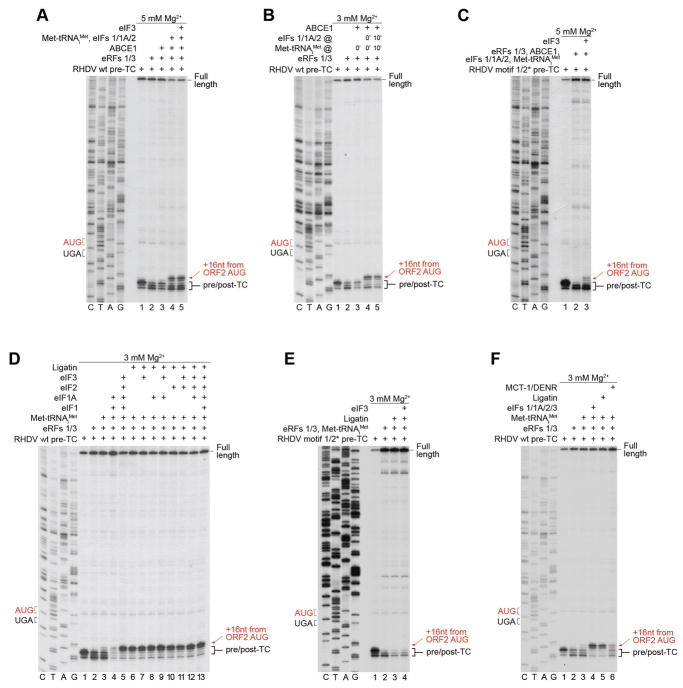
We previously found that at low [Mg2+] (~1 mM), eIFs 3/1/1A can promote efficient splitting of post-TCs assembled on MVHL-STOP mRNA, which in the presence of eIF2-TC is followed by reinitiation (Skabkin et al., 2013). Elevation of [Mg2+] stabilizes post-TCs, making recycling (and in turn reinitiation) dependent on ABCE1. To our surprise, on RHDV mRNA, eIFs promoted efficient ABCE1-independent splitting of post-TCs and subsequent reinitiation at up to 3 mM Mg2+, and even retained ~35% activity at 4 mM (Fig. 4A). Strikingly, eIF3 was not essential, and eIFs 2, 1 and 1A were sufficient for the process (Fig. 4B, lanes 5, 6). The fact that reinitiation required eIF2 (Fig. 4C, lane 7) provided additional confirmation that it was mediated by 40S subunits rather than 80S ribosomes. Further omission of eIF1 impaired recycling, evidenced by the persistence of toe-prints corresponding to post-TCs (Fig. 4C, lane 6). Recycling was also reduced when eIF1 was omitted in the presence of eIF3 (Fig. 4B, lane 4). Importantly, eIF1 could promote efficient recycling in the absence of all other eIFs (Fig. 4C, lane 3). The requirement for reinitiation of eIF1A depended on the presence of eIF3: omission of eIF1A strongly affected the process only in eIF3’s absence (Fig. 4B, lane 3 and Fig. 4C, lane 5). 48S complexes formed with and without eIF3 were equally capable of assembling elongation-competent 80S ribosomes (Fig. 4D).
Figure 4. Reinitiation by 40S subunits following eIF-mediated splitting of post-TCs on model RHDV mRNA.
(A–C) Toe-printing analysis of factor requirements for reinitiation by 40S subunits following ABCE1-independent splitting of post-TCs on model RHDV mRNA. (D) Toe-printing analysis of the ability of 48S complexes formed on the ORF2 AUG of RHDV “MSKL-STOP” mRNA with and without eIF3 to assemble elongation-competent 80S ribosomes. (E) Toe-printing analysis of reinitiation in conditions of delayed addition of eIF2 after preincubation of post-TCs with eIFs 1, 1A and 3, as indicated. (F, G) Toe-printing analysis of the recycling activity of eIF1, individually and in combination with eIF1A on post-TCs assembled on (F) wt and (G) motifs 1/2* mutant RHDV mRNAs. (H) Toe-printing analysis of factor requirements for reinitiation by 40S subunits following ABCE1-independent splitting of post-TCs on RHDV motifs 1/2* mutant mRNA. (A–H) Positions of pre/post-TCs on ORF1 (black brackets), 48S complexes on ORF2 AUG (red arrows), and eRF1AGQ -associated pre-TCs on ORF2 (blue arrows) are shown on the right. Stop (black or blue) and restart (red) codons are marked on the left. Free [Mg2+] is indicated on each panel. See also Figure S2.
Thus, eIF3 was not essential for reinitiation on RHDV mRNA, but had a stimulatory effect if eIF1 and eIF1A were present individually. Moreover, whereas delayed addition of eIF2 after preincubation of post-TCs with eIF1 or eIF1/eIF1A strongly impaired reinitiation, inclusion into preincubation mixtures of eIF3 reduced the inhibition (Fig. 4E, compare lanes 5, 6 with lanes 7, 8). eIF3 may therefore be an auxiliary factor that promotes reinitiation in conditions when the other factors are limited.
To assay whether the ability of post-TCs formed on RHDV mRNA to undergo eIF3-independent recycling and reinitiation is determined by TURBS, we employed pre-TCs assembled on mRNA with mutated motifs 1 and 2* (Fig. 3D, upper panel). Over a wide range of [Mg2+], eIF3-independent recycling on mRNAs with wt and mutated TURBS was similar (Figs. 4F–G), indicating that the process did not depend on the integrity of TURBS. The ability of eIF1 to promote recycling was also not due to the presence of eEF2 and SERBP1, because recycling of RRL-derived pre-TCs formed on MVHL-STOP mRNA (as well as pre-TCs that were reconstituted in vitro in the presence of SERBP1) continued to be eIF3-dependent and inefficient at 3 mM Mg2+ (Fig. S2). In contrast to recycling, reinitiation on mRNA with mutated TURBS strictly required eIF3 and was less efficient than on wt mRNA (Fig. 4H).
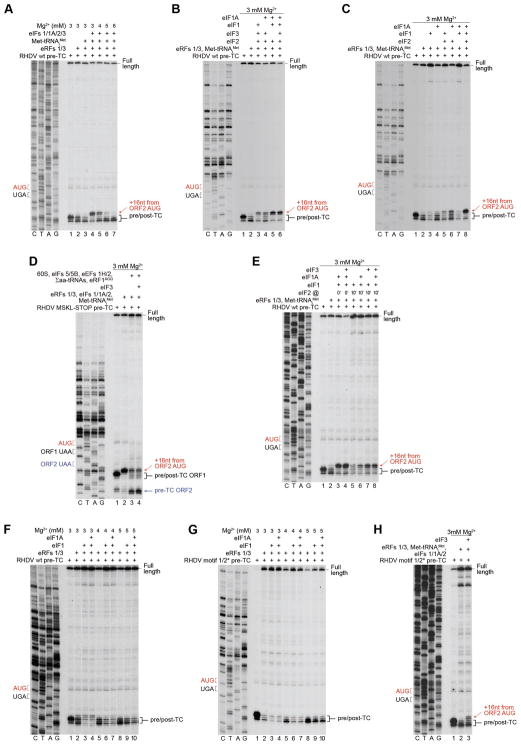
To investigate reinitiation following ABCE1-mediated splitting of post-TCs, [Mg2+] was elevated to 5 mM to minimize ABCE1-independent splitting by eIFs. Again, eIFs 2/1/1A were sufficient for the process (Fig. 5A, lanes 4, 5). The relatively low level of reinitiation was due to the reduced activity of ABCE1 at 5 mM Mg2+ (Pisarev et al., 2010). eIFs 2/1/1A could also promote eIF3-independent reinitiation upon their delayed addition following efficient splitting of post-TCs by ABCE1 at 3 mM Mg2+ (Fig. 5B). In contrast, reinitiation on RHDV mRNA with mutated TURBS was again strictly dependent on eIF3 (Fig. 5C).
Figure 5. Reinitiation by 40S subunits following ABCE1-mediated splitting of post-TCs on model RHDV mRNA. Recycling and reinitiation activities of Ligatin and MCT1-DENR.
(A–C) Toe-printing analysis of factor requirements for reinitiation by 40S subunits following ABCE1-mediated splitting of post-TCs on (A, B) wt and (C) motifs 1/2* mutant RHDV mRNAs. (D–F) Toe-printing analysis of reinitiation by Ligatin and MCT1/DENR on (D, F) wt and (E) motifs 1/2* mutant RHDV mRNAs. (A–F) The positions of pre/post-TCs on ORF1 (black brackets) and 48S complexes on ORF2 AUG (red arrows) are shown on the right. Stop (black) and restart (red) codons are marked on the left. Free [Mg2+] is indicated on each panel.
We have previously found that on MVHL-STOP mRNA, moderate reinitiation could also be promoted by Ligatin and eIF3 following splitting of post-TCs by ABCE1 (Skabkin et al., 2013). Strikingly, on RHDV mRNA, Ligatin alone could mediate efficient ABCE1-independent reinitiation, and its activity was not influenced by other eIFs (Fig. 5D). No reinitiation by Ligatin was observed on mRNA with mutated TURBS (Fig. 5E). Reinitiation could also be promoted by MCT1 and DENR, albeit less efficiently (Fig. 5F).
Thus, irrespective of the mode of splitting of post-TCs and reinitiation, TURBS strongly increases the efficiency of reinitiation by recycled 40S subunits and diminishes its dependence on eIF3.
Reinitiation on non-AUG codons
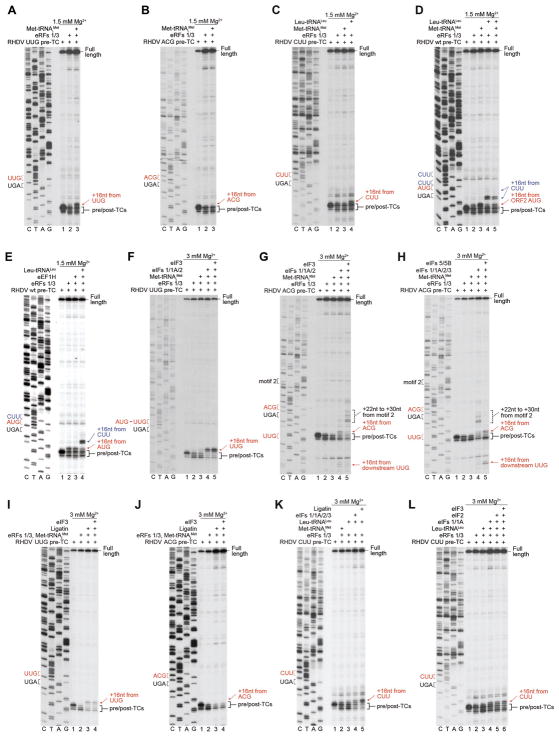
Reinitiation on calicivirus mRNAs is remarkably tolerant of substitutions in the restart codon, so that even changes at two or three positions (to e.g. CUA or UCG) reduced reinitiation by only 3–4 fold (Meyers, 2003; Lütterman and Meyers, 2007, 2014; Napthine et al., 2009). To determine the basis for this plasticity, we investigated reinitiation by post-termination 80S ribosomes and by recycled 40S subunits on mutant RHDV mRNAs, in which the restart AUG was replaced by near-cognate UUG and ACG codons, or by a non-cognate CUU codon.
Wobble reinitiation by 80S ribosomes with Met-tRNAiMet was observed only on the UUG codon with a mismatch in the first position (Figs. 6A–C). However, 80S ribosomes were able to reinitiate at the CUU codon with cognate Leu-tRNALeu (Fig. 6C). Reinitiation by 80S ribosomes with Leu-tRNALeu also occurred on wt RHDV mRNA, in which case complexes primarily formed at the CUU immediately preceding the restart AUG and to a small extent at the next upstream CUU (Fig. 6D). Leu-tRNALeu efficiently competed with Met-tRNAiMet, and when these tRNAs were present in equimolar amounts, formation of complexes on AUG and CUU codons occurred at a ~1:1 ratio (Fig. 6D). Inclusion in reaction mixtures of eEF1H did not alter the position of toe-prints (Fig. 6E), as would have been expected if Leu-tRNALeu had been delivered to the A site, rather than entering the P site on its own. Thus, we did not observe reinitiation primed by eEF1A-mediated delivery of aa-tRNA to the A site.
Figure 6. Reinitiation on model RHDV mRNAs, in which the ORF2 AUG was substituted by near- or non-cognate codons.
(A–E) Toe-printing analysis of reinitiation by 80S ribosomes with Met-tRNAiMet or Leu-tRNALeu (as indicated) on RHDV wt and “UUG”, “ACG” and “CUU” mutant mRNAs. (F, G) Toe-printing analysis of initiation factor requirements for reinitiation by 40S subunits with Met-tRNAiMet on “UUG” and “ACG” mRNAs. (H) Toe-printing analysis of the influence of subunit joining on reinitiation on “ACG” mRNA. (I, J) Toe-printing analysis of reinitiation mediated by 40S subunits with Ligatin and Met-tRNAiMet on “UUG” and “ACG” mRNAs. (K, L) Toe-printing analysis of reinitiation mediated by 40S subunits and Ligatin or eIFs (with Met-tRNAiMet or Leu-tRNALeu) on “CUU” mRNA. (A–L) Positions of pre/post-TCs on ORF1 (black brackets), 80S and 48S reinitiation complexes (red and blue arrows), and upstream aberrant complexes (dashed brackets) are shown on the right. Stop (black) and restart (red or blue) codons are marked on the left. Free [Mg2+] is indicated on each panel.
eIFs 1/1A/2, on the other hand, were able to promote reinitiation by recycled 40S subunits with Met-tRNAiMet on both near-cognate codons, albeit reinitiation on the UUG was more efficient (lanes 4 in Figs. 6F and 6G). In contrast to reinitiation on the UUG (Fig. 6F, lane 5), reinitiation on the ACG was suppressed by eIF3 (Fig. 6G, lane 5), indicating that eIF3 influences eIF1’s codon-discriminating activity. The appearance on the “ACG” mRNA of weak toe-prints +15–16nt from the UUG located downstream of the stop codon (Fig. 6G, lane 4) suggests that in the absence of a strong restart codon at the native position, a small proportion of 40S subunits can scan and reinitiate downstream. Reinitiation on ACG and downstream UUG codons was enhanced by eIF5 and eIF5B (Fig. 6H, lane 5), likely due to conversion of 48S complexes into more stable 80S ribosomes. On the “ACG” mRNA, additional toe-prints also appeared +22–30nt from motif 2 (Fig. 6G). These toe-prints, which were enhanced by eIF3, disappeared upon addition of eIF5/eIF5B, suggesting that they corresponded to non-productive 40S complexes, lacking codon-anticodon base-pairing and likely stabilized by TURBS. Wobble reinitiation on the “UUG” (but not on the “ACG”) mRNA could also be mediated by Ligatin (Figs. 6I–J), albeit less efficiently than by eIFs (Fig. 6F). Strong reinitiation at the CUU codon required Ligatin and cognate Leu-tRNALeu (Fig. 6K). Low-level reinitiation at the CUU with Leu-tRNALeu could also be mediated by eIF1 and eIF1A, but it was not stimulated further by eIF2 with or without eIF3 (Fig. 6L).
Taken together, these data indicate that on RHDV mRNA, “wobble” reinitiation with Met-tRNAiMet at near-cognate codons can be promoted by post-termination ribosomes or by recycled 40S subunits and eIFs or Ligatin, with reinitiation by recycled 40S subunits and eIFs being more permissive to codon-anticodon mismatches. Reinitiation on non-cognate codons, on the other hand, requires cognate elongator tRNA and can be mediated by post-termination ribosomes or by recycled 40S subunits and Ligatin.
Preference for the position of the restart AUG
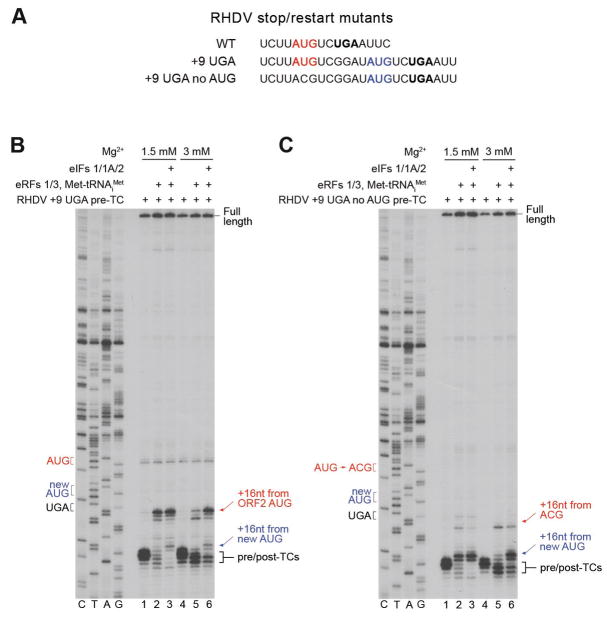
Reinitiation on calicivirus mRNAs does not have a strict requirement for maintenance of the wt spacing between stop and restart codons (Meyers, 2003; Meyers, 2007; Lütterman and Meyers, 2007, 2014). To evaluate the preference for the position of the restart AUG, we therefore moved the stop codon 9nt downstream together with the six upstream nucleotides of its native context, thus creating an additional AUG (Fig. 7A). On this “+9 UGA” mRNA, post-termination ribosomes (Fig. 7B, lanes 2, 5) and the majority of recycled 40S subunits (Fig. 7B, lanes 3, 6) reinitiated on the native AUG. Thus, the position of the native codon was strongly preferred irrespective of the mode of reinitiation. However, when the native AUG was substituted by ACG, efficient reinitiation took place on the newly introduced AUG (Fig. 7C).
Figure 7. The preference for the position of the restart AUG during reinitiation on model RHDV mRNA.
(A) The sequence of the ORF2 restart regions in wt and stop/restart mutant RHDV mRNAs. The ORF1 stop codon is in bold black, native ORF2 AUG is in red, newly introduced AUG is in blue. (B, C) Toe-printing analysis of reinitiation by post-termination ribosomes and recycled 40S subunits on RHDV “+9 UGA” and “+9 UGA no AUG” mRNAs. Positions of pre/post-TCs (black brackets), 80S and 48S reinitiation complexes (red and blue arrows) are shown on the right. Stop (black), native restart (red) and newly introduced restart (blue) codons are marked on the left. Free [Mg2+] is indicated on each panel.
DISCUSSION
The preparations of RRL-derived pre-TCs that we employed to recapitulate reinitiation on calicivirus mRNAs contained true pre-TCs that could be associated with SERBP1, and SERBP1/eEF2-bound non-programmed ribosomes. The simultaneous presence of SERBP1 and eEF2 on non-programmed ribosomes has been noted in D. melanogaster embryonic extracts and human peripheral blood mononuclear cells (Anger et al., 2013). SERBP1 and Stm1 have extended structures, and after binding to the head of the 40S subunit, enter the mRNA-binding channel and follow the mRNA-binding path until the P site before contacting the 60S subunit (Ben-Shem et al., 2011; Anger et al., 2013). Interaction with both subunits likely accounts for their ability to promote subunit association (Correia et al., 2004), possibly to sequester ribosomes during stress to protect them from degradation (e.g. van Dyke et al., 2013). Stm1 was also suggested to inhibit translation (Balagopal and Parker, 2011), which would be consistent with the occlusion of the mRNA-binding channel. However, SERBP1 was able to bind to 48S complexes and pre-TCs, both of which contain mRNA in the mRNA-binding cleft. Moreover, it did not interfere with initiation, elongation, association of pre-TCs with eRFs, or canonical reinitiation, making RRL-derived pre-TCs a valid model for studying calicivirus reinitiation despite SERBP1’s presence. These observations also suggest that SERBP1 could have two modes of binding to the 40S subunit, in one of which it might maintain the contact with the head, but be displaced from the mRNA-binding channel.
Recapitulation in vitro of reinitiation on model sg RHDV and NV mRNAs revealed that stabilization of ribosomal association of mRNA by TURBS allows efficient reinitiation by post-termination 80S ribosomes, and diminishes dependence on eIF3 of reinitiation by 40S subunits that can be mediated by eIFs 2/1/1A or Ligatin following ABCE1-dependent or -independent splitting of post-TCs. These advantages were eliminated by substitutions in the essential TURBS motifs, rendering characteristics of the process similar to those of canonical reinitiation, which could be efficiently executed only by recycled 40S subunits and strictly required eIF3 (Skabkin et al., 2013). Since there is no reason to assume that different mechanisms of calicivirus reinitiation cannot all occur in cells, one can speculate that their multiplicity might have additional benefits, such as enabling reinitiation during environmental changes that alter the availability of individual factors, or increasing the probability that the process occurs without delay, which is important because the 40S/TURBS interaction could be disrupted by the next translating ribosome.
Interestingly, in contrast to MVHL-STOP mRNA (Pisarev et al., 2007; 2010), on RHDV mRNA, eIF3 was also not essential for ABCE1-independent recycling, which could be mediated by eIF1 or Ligatin. Ligatin has a C-terminal SUI1/eIF1 domain and competes with eIF1 for binding to the 40S subunit (Skabkin et al., 2010), so mechanisms underlying splitting of post-TCs by both factors might be similar. The difference in eIF3 requirements for splitting of post-TCs on RHDV and MVHL-STOP mRNAs could not be explained by the presence of TURBS or by SERBP1/eEF2 contamination of pre-TCs assembled on RHDV mRNA (Figs. 4F–G, S2). However, it could potentially result from higher affinity to the P/E hybrid state of tRNAVal in post-TCs assembled on RHDV mRNA than of tRNALeu in post-TCs assembled on MVHL-STOP mRNA, which would destabilize post-TCs on the former and also allow their productive interaction with eIF1 or Ligatin. If this explanation is correct, the variety of tRNAs present in pre-TCs formed on ORF1 of different calicivirus sg mRNAs suggests that recycling on some might not be promoted efficiently by eIF1 or Ligatin alone, and would proceed either by the ABCE1-mediated pathway or will involve eIF3. Notably, even though eIF1 was able to promote recycling, eIF1 and eIF2-TC could not mediate efficient reinitiation on RHDV mRNA in the absence of eIF1A. Potential roles of eIF1A include stimulation of ribosomal recruitment of eIF2-TC, stabilization of 40S/mRNA complexes, or cooperation with eIF1 in protecting 40S/mRNA complexes from reassociation with 60S subunits.
Variations in the affinity of specific tRNAs to the P/E hybrid state (Shoji et al., 2009) could also affect the efficiency of reinitiation by post-termination ribosomes on different calicivirus mRNAs and hence, the contribution of this mechanism to the overall level of reinitiation. Thus, post-TCs containing tRNA with high affinity to the P/P orientation will have lower spontaneous dissociation/exchange of tRNA, which would reduce reinitiation by post-termination ribosomes. Here, we also observed differences in reinitiation by post-termination ribosomes on RHDV and NV mRNAs (containing tRNAVal and tRNAArg in post-TCs, respectively): reinitiation on NV mRNA was less sensitive to elevation of [Mg2+] and to the presence of E site tRNA.
The principal difference between TURBS-mediated and conventional reinitiation by recycled 40S subunits is that the former can proceed without eIF3, irrespective of whether recruitment of Met-tRNAiMet is mediated by eIFs 2/1/1A or Ligatin. eIF3-independence is likely the critical factor that allows efficient reinitiation after translation of long ORFs, by which time the transient association of eIF3 with elongating ribosomes would be lost. However, eIF3 did influence TURBS-mediated reinitiation when the other factors were limited, e.g. when eIF1 and eIF1A were present individually or when eIF2-TC was added with a delay, and could therefore stimulate reinitiation in vivo. Consistently, addition of eIF3 enhanced reinitiation on FCV mRNA during translation in RRL (Pöyry et al., 2007). Thus, whereas base-pairing of TURBS with h26 of 18S rRNA allows initial ribosomal retention on the stop/restart region, subsequent binding of eIF3 could provide additional stabilization and enhance recruitment of eIF2-TC. The contribution of eIF3 might vary for different caliciviruses, depending on the strength of the 40S/TURBS interaction. UV cross-linking experiments showed that eIF3 can also directly bind to TURBS (Pöyry et al., 2007). Notably, the eIF3/TURBS interaction on the 40S subunit would not contradict the ribosomal position of eIF3, which binds to rpS13e and rpS27e in the vicinity of h26 (Hashem et al., 2013a) and is thus appropriately positioned to contact TURBS.
The activity of Ligatin in reinitiation on RHDV mRNA is consistent with its ability to stimulate eIF2-independent recruitment of initiator tRNA to 40S/mRNA complexes in which the start codon is placed directly in the P site (Skabkin et al., 2010). On calicivirus mRNAs, such placement is apparently achieved by the TURBS/h26 interaction. Although MCT1-DENR could also mediate reinitiation on RHDV mRNA, their activity was lower than that of Ligatin. MCT1-DENR were recently implicated in reinitiation after short ORFs on a variety of cellular mRNAs in D. melanogaster (Schleich et al., 2014). Although this mechanism is different from reinitiation on calicivirus mRNAs and is more reminiscent of reinitiation on a derivative of β-globin mRNA by Ligatin and eIF3 (Skabkin et al., 2013), it is notable that the activity of MCT1 depended on its phosphorylation at T118/S119 (Schleich et al., 2014). The lower activity of MCT1-DENR in our reconstituted assays could therefore be due to the unphosphorylated state of recombinant MCT1, and the activity of native MCT1-DENR might be closer to that of Ligatin. Notably, the residues that are phosphorylated in MCT1 are not conserved in Ligatin, and its activity might therefore not be dependent on phosphorylation.
The key question concerning the mechanism of Ligatin’s action is how to reconcile its two seemingly opposed activities in promoting tRNA release from recycled 40S subunits and subsequent attachment of initiator tRNA to an AUG codon, which in RHDV mRNA is only 2nt upstream. One likely possibility is that the positions of tRNA on a recycled 40S subunit and on the 40S/mRNA complex following Ligatin-mediated attachment differ. The orientation of tRNA on recycled 40S subunits may not be fully compatible with the presence of Ligatin or with conformational changes that it might induce, so that binding of Ligatin would trigger tRNA’s ejection. Although it is not known whether the ribosomal orientation of Ligatin-attached tRNA is similar to that in 48S or 80S/Met-tRNAiMet/eIF5B complexes (Hussain et al., 2014; Fernández et al., 2013), or differs from them both, it is evidently compatible with binding of 60S subunits and formation of elongation-competent ribosomes (Skabkin et al., 2010, 2013). Although it is possible to cautiously assume that the function and ribosomal location of its SUI1/eIF1 domain are similar to those of eIF1, the function of its N-terminal DUF1947 and PUA domains (which also exist in MCT1) constitutes another unresolved question concerning Ligatin’s mechanism of action. The X-ray structure of MCT1 revealed that DUF1947 contains surface-exposed positively charged patches that could participate in RNA-binding, whereas the PUA domain maintains secondary structure elements that are essential for RNA recognition in PUA domains of other proteins (Tempel et al., 2013). DUF1947 and PUA domains might therefore interact with the 40S subunit, inducing conformational changes in addition to those caused by the SUI1/eIF1 domain, or could directly contact tRNA promoting its recruitment.
Reinitiation on calicivirus mRNAs is remarkably insensitive to substitutions in the restart codon, readily tolerating single substitutions, and retaining substantial activity after substitutions in two or even three positions (Lütterman and Meyers, 2007, 2014; McCormick et al., 2008; Meyers 2003; Pöyry et al., 2007). Although reinitiation on mRNAs with mutated restarts could proceed with Met-tRNAiMet (Lütterman and Meyers, 2007, 2014), it was not apparent whether this accounts for all reinitiation events, nor to which extent neighboring near-cognate codons were used instead of mutated non-cognate restarts. Recapitulation of reinitiation in vitro allowed us to investigate directly whether reinitiation on non-AUG codons proceeds via wobble base-pairing with Met-tRNAiMet or normal base-pairing with cognate elongator tRNAs. We found that on near-cognate codons, reinitiation with Met-tRNAiMet can be executed by all three reinitiation mechanisms: by post-termination ribosomes, by recycled 40S subunits with eIFs 1/1A/2, and by recycled 40S subunits with Ligatin. Reinitiation by 40S with eIFs 1/1A/2 was the least sensitive to the nature of mismatches, likely because of the additional stabilization of tRNA by eIF2. However, on some codons it was inhibited by eIF3, which indicates that eIF3 enhances the codon-discriminating activity of eIF1. The ability of 40S subunits to reinitiate without eIF3 might thus account for efficient reinitiation on those near-cognate codons, which do not function efficiently in the canonical initiation process. Interestingly, reinitiation on codons introduced downstream of the stop codon was restricted to AUG triplets and was sensitive to their nucleotide context (Lüttermann and Meyers, 2014). 3′-directional scanning by reinitiating 40S subunits likely requires eIF4F (Skabkin et al., 2013), implying the involvement of eIF3, which would make eIF1 more selective.
Reinitiation on non-cognate codons strictly required cognate elongator tRNAs, and could be executed by post-termination ribosomes or by 40S subunits and Ligatin. Consequently, there is no reason to assume that reinitiation on near-cognate codons could not utilize cognate elongator tRNAs rather than by Met-tRNAiMet. However, initiation from non-cognate codons raises the question of fidelity. If the position of the restart codon is not strictly determined, and reinitiation can occur within a certain window, then reinitiation by elongator tRNAs could in principle occur in all three reading frames. The reduced reinitiation on mRNAs caused by non-cognate substitutions could in part reflect this problem. When the restart codon is AUG, the possibility of reinitiation with elongator tRNAs from nearby non-cognate codons is likely reduced by the higher affinity of Met-tRNAiMet to the P site. As concerns the preference for the position of the restart site, consistent with a previous report (Pöyry et al., 2007), we also found that when multiple AUGs are present, the position of the native restart is strongly preferred, irrespective of whether reinitiation is executed by post-termination ribosomes or by recycled 40S subunits. However, despite interaction with TURBS, both 80S ribosomes and 40S subunits remain sufficiently mobile to search for a nearby alternative AUG in the absence of the native restart.
In future, understanding of the calicivirus reinitiation mechanism could be significantly advanced by structural studies of the 40S/TURBS interaction, which would elucidate details of the contact with h26 (which is particularly interesting because interaction with h26 is also employed by Hepatitis C virus-like IRESs; Hashem et al., 2013b), potential conformational changes in the 40S subunit, and the nature of the eIF3/TURBS interaction. Similar studies on the structure of complexes of the 40S subunit with Ligatin or MCT1-DENR would give valuable insights into how they promote ribosomal recruitment of tRNA during calicivirus reinitiation, noncanonical initiation (Skabkin et al., 2010), and reinitiation after uORFs on cellular mRNAs (Schleich et al., 2014).
EXPERIMENTAL PROCEDURES
Plasmids, purification of factors and ribosomal subunits, and aminoacylation of tRNA are described in the Supplemental Information, which also contains detailed protocols for all experimental procedures.
Assembly of Ribosomal Complexes
On RHDV and NV mRNAs, pre-TCs were formed by translation in RRL in the presence of eRF1AGQ. On MVHL-STOP mRNA, pre-TCs were assembled either in RRL or in an in vitro reconstituted system (Alkalaeva et al., 2006). All pre-TCs were then purified by SDG centrifugation.
Ribosomal association of SERBP1 and eEF2
Ribosomal association of SERBP1 and eEF2 was assayed for pre-TCs formed in RRL or in an in vitro reconstituted system, for individual 40S, 60S subunits and 80S ribosomes after their incubation with SERBP1 and eEF2, and for in vitro reconstituted 43S and 48S complexes. Ribosomal complexes were separated by SDG centrifugation, and ribosomal peak fractions were analyzed by SDS-PAGE followed by SYPRO staining or western blotting using SERBP1 antibodies.
Analysis of reinitiation by toe-printing
To investigate reinitiation, 3.75 nM of SDG-purified pre-TCs were incubated for 10 min at 37°C in 20 μl buffer (20 mM Tris [pH 7.5], 100 mM KCl, 0.25 mM spermidine, 2 mM DTT, 0.3 mM GTP, 0.1 mM ATP and corresponding amounts of MgCl2 to achieve free [Mg2+] indicated on each panel) with different combinations (as indicated in Figures) of the following factors and tRNAs: 50 nM eRF1, 100 nM eRF3, 9 nM Met-tRNAiMet, 9 nM Leu-tRNALeu, 40 nM tRNASer, 40 nM tRNAHis, 50 nM eEF2, 100 nM eIF1, 100 nM eIF1A, 40 nM eIF2, 30 nM eIF3, 100 nM ABCE1, 50 nM Ligatin, 50 nM MCT1, and 50 nM DENR. To examine elongation competence of 48S and 80S reinitiation complexes, reaction mixtures were supplemented with combinations of 50 nM eEF2 (where not added in a previous step), 50 nM eEF1H, 400 nM eRF1AGQ mutant, Σaa-tRNA (~6 nM of each aa-tRNA), 50 nM 60S, 100 nM eIF5, and 90 nM eIF5B, and incubation was continued for another 10 min. Resulting complexes were analyzed by primer extension using AMV-RT. cDNA products were resolved in 6% sequencing gels followed by autoradiography.
Enzymatic foot-printing
100 nM of RHDV mRNA or 22.5 nM of RHDV pre-TC were treated with 0.01 U/μl or 0.025 U/μl of RNaseT1, respectively, for 10 minutes at 37°C. Cleavage sites were identified by primer extension using AMV-RT. cDNA products were resolved in 6% sequencing gels followed by autoradiography.
Supplementary Material
Acknowledgments
We thank L. Yu. Frolova for eRF expression vectors and J. Heaton and T. Gelehrter for SERBP1 antibodies. This work was supported by NIH grants GM59660 and GM80623 and HFSP grant RGP0062/12 to TVP, and NIH grant AI51340 to CUTH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romão L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Correia H, Medina R, Hernández A, Bustamante E, Chakraburtty K, Herrera F. Similarity between the association factor of ribosomal subunits and the protein Stm1p from Saccharomyces cerevisiae. Mem Inst Oswaldo Cruz. 2004;99:733–737. doi: 10.1590/s0074-02762004000700012. [DOI] [PubMed] [Google Scholar]
- des Georges A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CU, Pestova TV, Frank J. Structure of the mammalian ribosomal pre-termination complex associated with eRF1. eRF3.GDPNP. Nucleic Acids Res. 2014;42:3409–3418. doi: 10.1093/nar/gkt1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SH. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342:1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013a;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013b;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural Changes Enable Start Codon Recognition by the Eukaryotic Translation Initiation Complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttermann C, Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J Biol Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- Lüttermann C, Meyers G. The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev. 2009;23:331–344. doi: 10.1101/gad.507609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttermann C, Meyers G. Two alternative ways of start site selection in human norovirus reinitiation of translation. J Biol Chem. 2014;289:11739–11754. doi: 10.1074/jbc.M114.554030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick CJ, Salim O, Lambden PR, Clarke IN. Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J Virol. 2008;82:8917–8921. doi: 10.1128/JVI.02362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry KG, Walker SE, Wang H, Fredrick K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol Cell. 2005;20:613–622. doi: 10.1016/j.molcel.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J Biol Chem. 2003;278:34051–34060. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S, Lever RA, Powell ML, Jackson RJ, Brown TD, Brierley I. Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS One. 2009;4:e8390. doi: 10.1371/journal.pone.0008390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CUT, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CUT, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Küchler K, Stoecklin G, Duncan KE, Teleman AA. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Abdi NM, Bundschuh R, Fredrick K. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 2009;37:4033–4042. doi: 10.1093/nar/gkp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar A, Hellen CUT, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W, Dimov S, Tong Y, Park HW, Hong BS. Crystal structure of human multiple copies in T-cell lymphoma-1 oncoprotein. Proteins. 2013;81:519–525. doi: 10.1002/prot.24198. [DOI] [PubMed] [Google Scholar]
- Van Dyke N, Chanchorn E, Van Dyke MW. The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochem Biophys Res Commun. 2013;430:745–750. doi: 10.1016/j.bbrc.2012.11.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.