Abstract
The host-dependent nature of idiosyncratic drug-induced liver injury (iDILI) suggests that rare genetic polymorphisms may contribute to the disease. Indeed, a few mutations in key genes have already been identified using conventional human genetics approaches. Over 50 commonly used drugs can precipitate iDILI, making this a substantial medical problem. Only recently have human induced pluripotent stem cells been used as a research tool to discover novel iDILI genes and to study the mechanisms of iDILI in vitro. Here we review the current state of stem cell use in the investigation of iDILI, with a special focus on genetics. In addition, the concerns and difficulties associated with genetics and animal model research are discussed. We then present the features of patient-specific pluripotent stem cells (which may be derived from iDILI patients themselves), and explain why these cells may be of great utility. A variety of recent approaches to produce hepatocyte-like cells from pluripotent cells and the associated advantages and limitations of such cells are discussed. Future directions for the use of stem cell science to investigate iDILI include novel ways to identify new iDILI genes, a consideration of epigenetic impacts on iDILI, and the development of new and improved strategies for the production of hepatocytes from human pluripotent cells.
Keywords: DILI, Embryonic stem cell, iPS, Toxicity, Genetics
Introduction
Idiosyncratic drug-induced liver injury (iDILI) is a complex disorder that is typified by the onset of severe (even fatal) liver damage after exposure to drugs that are otherwise well tolerated in the human patient population. The host-dependent (idiosyncratic) nature of iDILI suggests that genetic polymorphisms that cause a predisposition to DILI events likely exist in the population. In these cases, iDILI occurs only when an individual with a predisposing genotype is exposed to the precipitating drug. Put another way, iDILI is a type of drug-genotype interaction, and most individuals with a potential underlying iDILI genotype will never know of their underlying genetic susceptibility because they will fail to encounter the interacting drug.
At present, the only way to “find” an iDILI genotype is for an individual with a predisposing genotype to be treated (unintentionally) with an iDIL-precipitating drug. Current medical practice dictates that all patients are assumed to be normal, and they are treated routinely with potentially DILI-causing drugs. Importantly, these patients are monitored for the onset of liver damage only after the onset of drug treatment, usually by monitoring levels of liver enzymes, such as ALT and others in the blood. Currently, there is no effective way to detect potential iDILI responders prior to drug treatment. As more iDILI polymorphisms in the genome become known, it may be possible to devise genetic tests that identify at-risk individuals. Such a goal will require the comprehensive identification of iDILI risk alleles for each of the 50 or more highly problematic drugs, and an expedient way to gather this genetic information is needed. One way that such information could be gathered is via genome-wide association studies (GWAS). However, about 50 commonly used drugs can cause iDILI, often by completely independent mechanisms, and it is likely that 50 complete GWAS studies would be required. Although GWAS, a recently developed and powerful approach, would work in theory, practicalities of scale and relatively low numbers of iDILI patients for specific drugs prohibit the widespread use of GWAS to comprehensively identify iDILI genes and polymorphisms.
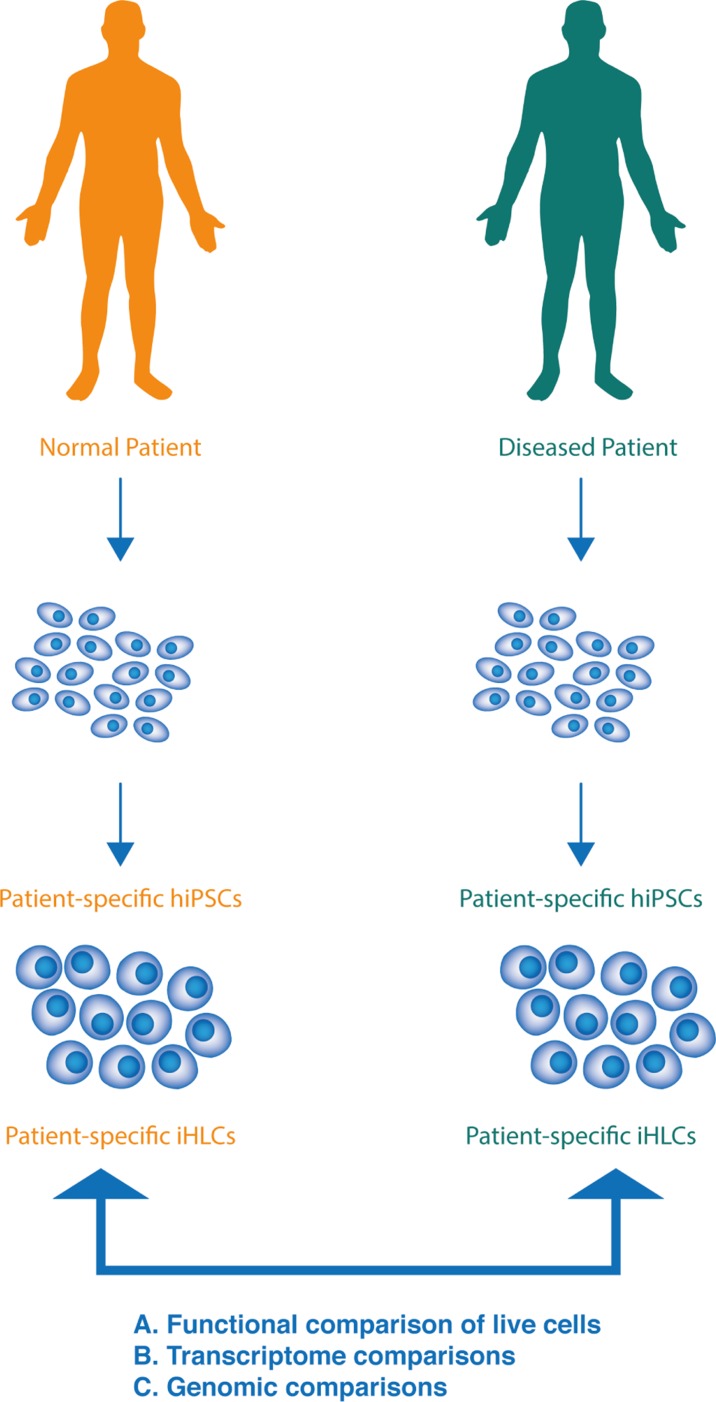
Recently, an entirely different and new approach uses human induced pluripotent stem cells (hiPSCs) that are exact genetic matches to iDILI patients (Fig. 1). These are obtained by a cellular reprogramming approach that takes relatively non-invasive biopsies (either blood or a small skin sample) and reprograms them to an early developmental state that is functionally equivalent to embryonic stem cells. This is achieved by expressing key embryonic transcription factors that induce the state of pluripotency (i.e. the potential to differentiate into any desired cell type, including hepatocytes, present in the adult body). To do this, a patient who has suffered iDILI donates blood or skin that can be derived into hiPSCs and then hepatocytes. These induced hepatocyte-like cells (iHLCs) are subjected to functional toxicity studies, and more importantly, iDILI-causing mutations can be identified in such cells using strategies that are described below. Therefore, it may be possible to identify the majority of iDIL-predisposing genes using at most a few hundred patients rather than several hundred thousand, as would be required for GWAS. In addition, disease mechanisms can never be addressed with GWAS, but iHLCs can be easily studied in vitro, thus providing insight into cellular and molecular mechanisms that govern iDILI.
Fig. 1. Strategy for using iPS reprogramming to investigate iDILI.
Somatic cell samples such as skin or blood are collected from iDILI and control patients. These are used to derive hiPSCs that are exact genetic matches to the donor patients. Such hiPSCs can be indefinitely expanded and passaged, so long as they are maintained under pluripotent conditions. hiPSCs can then be differentiated to iHLCs (hepatocyte-like cells). iHLCs from iDILI and control patients can then be compared using functional tests (A., such as exposure to the iDILI-precipitating drug) or using transcriptome comparisons (B.), which may lead to identification of iDILI genes. Subsequently, causative mutations (C.) can be identified, using transcriptome alterations as a guide.
Drug-induced liver injury
Idiosyncratic drug-induced liver injury
iDILI is a rare form of iatrogenic liver injury that manifests itself as a wide spectrum of symptoms, ranging from mild and transient increases in plasma markers of hepatocellular injury to more severe forms that present with abdominal symptoms, cholestasis, and, in rare cases, the development of liver necrosis and fulminant hepatic failure. Despite its relatively low frequency (for individual drugs ranging from 1:100,000, e.g., for amoxicillin/clavulanic acid, to 1:100, e.g., for isoniazid),1 iDILI has become a major clinical concern for a number of reasons. First, an increasing number of drugs (>600), belonging to a variety of therapeutic classes, have been implicated in iDILI, with approximately 50 drugs featuring a strong causality. Therefore, the absolute number of all iDILI cases has become impressive when iDILI cases from over 600 drugs are considered in aggregate. Second, the occurrence of iDILI is unpredictable, both for a new drug on the market and for an individual patient. Third, iDILI has been associated with high morbidity and mortality, particularly when combined with jaundice, that can lead to acute liver failure and the need for liver transplants in the most severe cases.2 Finally, iDILI is one of the major reasons for the withdrawal of successfully launched drugs from the market, which may result in the loss of an otherwise important drug.3 Numerous reviews have addressed causality criteria, clinical hallmarks, underreported incidence, potential mechanisms, and prediction of iDILI.2,4–12
Determinants of susceptibility to iDILI
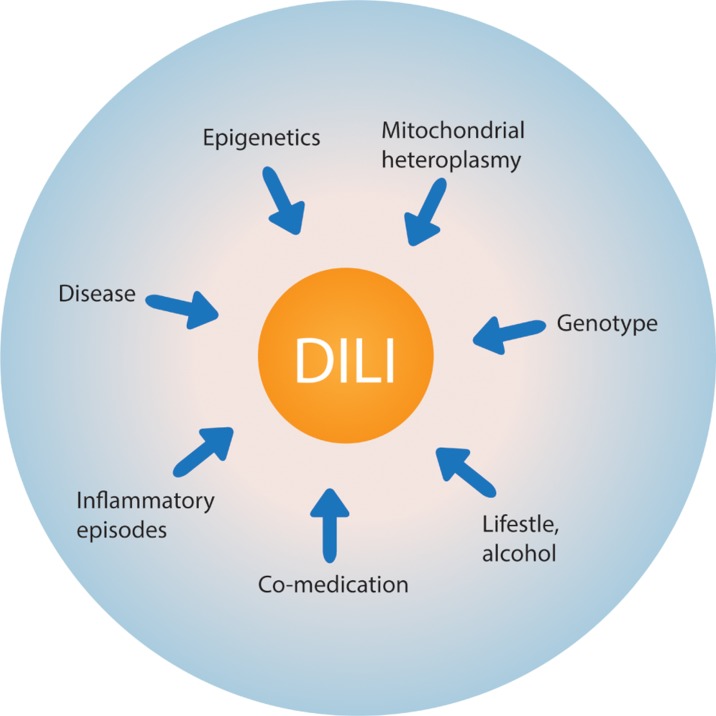
By definition, iDILI is “idiosyncratic”, i.e., dependent on the specific characteristics of the recipient (patient). Thus, besides the toxicodynamic properties of the drug itself (chemotype) and its potential to cause toxicity to hepatocytes by a number of mechanisms, host-specific factors also greatly contribute to the precipitation of toxicity (Fig. 2). There are two major types of host-specific factors: acquired/environmental factors and genotype. Regarding acquired/environmental factors, the underlying disease (indication) against which the patient is being treated,13 underlying infections, and episodes of inflammatory reactions14 can influence the risk. Coadministration with other potentially hepatotoxic drugs or alcohol is another important factor. Taken together, acquired/environmental and genetic determinants of susceptibility to iDILI are responsible for increasing the penetrance and for modulating the expressivity of drug toxicity.10 For these reasons, it is understandable that, despite many attempts with limited success, it is extremely difficult to develop an animal model for iDILI, using, e.g., inbred, virtually genetically identical mice, that are expected to recapitulate the clinical situation in individual patients.
Fig. 2. Determinants of susceptibility to drug-induced liver injury (DILI).
A number of patient-specific (idiosyncratic) factors greatly modulate the sensitivity to the potential hepatotoxic effects of certain drugs.
Role of the genotype
Mutations and/or rare single nucleotide polymorphisms (SNPs) in critical genes have been implicated in modulating the risk for iDILI.15,16 These specific haplotypes or mutations could be responsible for modulating the degree of toxicity at different levels. For example, they could enhance the pro-toxicant pathways in the liver, they could interfere with mechanisms involved in adaptation to the insult or impair defense mechanisms, or they could weaken immune tolerance. While traditional studies have focused on polymorphisms in cytochrome P450 forms that modulate metabolic bioactivation/hepatic clearance of drugs, more recent approaches have addressed the role of genetic variation in genes involved in the adaptive immune system, in detoxification pathways, or in mitochondrial function. For example, the human leukocyte antigen (HLA)-B*5701 haplotype of the major histocompatibility complex (MHC) has been associated with a >80-fold higher risk for developing liver injury from flucloxacillin treatment than individuals with other MHC haplotypes.17 Similarly, carriers of the HLA-A*3303 haplotype have a 36-fold higher risk for iDILI from ticlopidine treatment than other patients.18 These risk factors are statistically robust and point to a key role of these genes in the susceptibility to liver injury. Another example consists of mutations in certain forms of glutathione-S-transferase (GST), which have been implicated in the conjugation of reactive intermediates of many drugs including isoniazid or troglitazone. Phenotypes from null mutations are also quite frequent; specifically, homozygous null mutations encoding the GSTT1 form and the GSTM1 form of GST are present in 50% and 10–25% of Caucasians, respectively. In a study involving 154 patients with a diagnosis of iDILI, 18.2% of patients had the GSTT1/GSTM1 double-null genotype, while only 7.6% of control patients carried this genotype.19 Finally, other studies have analyzed the role of polymorphisms in the mitochondrial polymerase γ (POLG) gene that is involved in mtDNA replication. It was found that the risk for valproic acid-induced DILI was >20-fold increased in individuals carrying the pQ1236H and pE1143G mutations in the POLG gene as compared to wild-type.20
One important point is that these polymorphic risk alleles are quite common in the general population, which raises the question why iDILI is not more common. Most likely, these haplotypes become dangerous only in specific contexts, e.g., exposure to a particular drug. Furthermore, these analyses showed, at best, only a strong correlation with iDILI episodes, and there must undoubtedly be other risk factors. While the identification of such “risky” mutations does not necessarily explain the underlying mechanism of toxicity, it can be used to identify certain individuals or patient subsets who are predisposed to higher risk of iDILI. For the vast majority of drugs, however, the genetic determinants of susceptibility are not known. More recent approaches to establish a link between iDILI and underlying genetic risk factors have aimed at identifying entire functional pathways rather than single genes.
Lessons from current models and the need for novel patient-specific models
Currently there are no fully validated animal models that recapitulate the clinical features of iDILI. It makes sense that normal healthy inbred animals cannot model a disease that is driven by a variety of underlying patient-specific genotypes.21 While certain environmental factors can be modeled in vitro or in vivo, genetic variations are more difficult to adapt in animal models because in most cases, the most relevant “risk genes” are not known. For example, the lipopolysaccharide (LPS) rat or mouse model22 can mimic the underlying presence of sporadic inflammatory episodes, and the heterozygous Sod2 mouse model23 can emulate increased mitochondrial oxidant stress as it occurs with certain forms of underlying mitochondrial disease. However, applications useful for wide-scale screening platforms have not been possible to date. Hepatic cells can be harvested and cultured from patients who had developed iDILI from a particular drug and compared with cells from unaffected patients. At present, however, it is extremely difficult to obtain viable liver tissue from such patients, and primary hepatocytes cannot be easily cultured for extended periods of time. One promising approach is the use and study of stem cells derived from both diseased and healthy patients.
Stem cell approaches
Pluripotent cells and induced pluripotent stem cells
Pluripotent cells can in theory give rise to any cell type present in the adult mammalian body plan. The first widely-used pluripotent stem cells were mouse embryonic stem cells (mESCs).24,25 These cells were derived from day 3.5 preimplantation embryos at the blastocyst stage. At this stage of development, there are only a small set of cell types in the embryo. These include trophectoderm cells, which are destined to form the embryonic portion of the placenta, and the inner cell mass (ICM), which subsequently gives rise to all the cells of the embryo proper, and eventually, the adult mouse. It is the ICM cells that can be explanted and used to derive mESCs. Although the ICM exists in the embryo for only a few hours at most, mESCs can be cultured indefinitely in vitro, under conditions that maintain their pluripotency.26 Leukemia inhibitory factor (LIF) is a key cell signaling factor necessary for the maintenance of mESCs in a state of protracted pluripotency.27,28 The LIF receptor signals via Stat3 to the nucleus where key pluripotency transcription factors, including octamer-binding transcription factor 4 (Oct4), sry-related HMG box 2 (Sox2), and Nanog, collude to regulate a large set of genes, whose expression has been aptly described as a pluripotency network.29–32 Human ESCs (hESCs) were first derived from human blastocysts that would otherwise have been discarded from human fertility clinics.33 Unlike mESCs, hESCs depend upon the fibroblast growth factor 2 (FGF2) for maintenance of pluripotency,34 but like mESCs, hESCs contain a pluripotency transcriptional network that is mediated by the master transcription factors OCT4, SOX2, and NANOG. Collectively mESCs and hESCs have been used to model many developmental processes in vitro via directed differentiation experiments. Several dozens of differentiated cell types have been produced in this way, and they have been derived from all three of the principle germ layers, ectoderm, endoderm, and mesoderm. In many cases, these differentiated cells are rather similar to cells from primary organ culture as they become post-mitotic and exhibit gene and protein expression patterns similar to analogous cells in vivo. Such differentiated cells derived from pluripotent cells are far more similar to endogenous cells than traditional immortalized cell lines, which are transformed and usually without a normal complement of chromosomes.
The first example of reprogramming a differentiated vertebrate cell to an early embryonic state was achieved with Xenopus frogs, which were successfully cloned in the early 1960s from intestinal epithelial cells transferred into frog oocytes.35,36 The first time differentiated mammalian cells were successfully reprogrammed was over 40 years later with the advent of Dolly the Sheep.37 In this case, the resulting reprogrammed totipotent cell, achieved by nuclear transfer into an enucleated recipient sheep oocyte, was a one cell embryo that was cultured briefly in vitro to the blastocyst stage and then implanted in a surrogate pseudopregnant female to yield the live-born cloned sheep named Dolly. These successes with animal cloning showed that terminally differentiated vertebrate cells could be reversed to a state of pluripotency, albeit with reprogramming activities only found in the oocyte. In 2006, Shinya Yamanaka succeeded in directly reprogramming cultured adult cells to a state of pluripotency by introducing a set of genes encoding key transcription factors (Oct4, Sox2, kruppel-like factor 4 (Klf4), and proto-oncogene myc (c-Myc)) into mouse fibroblasts using retroviruses.38 The resulting pluripotent cells were deemed induced pluripotent stem cells (iPSCs). After initial viral transduction, the somatic cells were shifted to mESC culture conditions. The mechanism of iPS reprogramming depends upon the forced expression of a defined set of transcription factors, which includes members of the master regulatory transcription factors of the pluripotency network. IPSCs are remarkably similar to ESCs, and most importantly, exhibit a full range of pluripotency. Soon after this achievement, human iPSCs (hiPSCs) were developed.38,39 These, like their mouse counterparts, exhibited nearly complete resemblance to their hESC equivalents. It is crucial to point out that hiPSCs contain exactly the same genetic composition as the human fibroblast donor, thus opening the door to their use as a stem cell platform for personalized medicine. Early mouse iPS cells (miPSC) and hiPSC cell lines were made with integrating retroviruses and lentiviruses, and thus had subtly altered genomes. Since then it has become possible to make iPS cell lines that avoid the use of integrating retrovirus or lentivirus, including the use of messenger RNA encoding the reprogramming factors,40 transposon systems that excise with barely a trace,41,42 and episomes that transiently (but efficiently) express reprogramming factors prior to episomal loss by cell division.43 All of these strategies leave the genome nearly or completely untouched by the iPS reprogramming process.
Disease in a dish: Differentiation of iPS cells to hepatocytes
Most strategies to produce hepatocytes from hESCs or hiPSCs attempt to recapitulate liver development in vivo. In addition, strategies to differentiate hESCs and hiPSCs to human hepatocyte-like cells (hereafter termed “iHLCs” if derived from hiPSCs) have been greatly informed by similar (and earlier) successes with mESCs. Because liver is an endoderm-derived organ, all strategies for production of iHLCs begin with a preculture of hiPSCs. In one set of strategies, hiPSCs are shifted from hiPSC culture medium to an endoderm induction medium containing the signaling factor Activin A with insulin, transferrin, and selenium in trace concentration.44,45 Activin A is a potent inducer of endoderm and rapidly leads to cellular expression of the early endoderm marker SOX17. Cells are then shifted to a medium containing FGF2 and bone morphogenic protein 4 (BMP4), both of which are known to play a role in liver organogenesis in vivo.46–48 These cells are then shifted to a medium containing hepatocyte growth factor (HGF), and then finally to a medium containing oncostatin M and dexamethasone. This typical four stage approach yields hepatocyte-like cells that express both albumin (ALB) and α-fetoprotein (AFP), indicating that these iHLCs are similar to hepatocytes of late embryonic or perinatal origin.44 This same protocol yielded hepatocytes that can also secrete cholesterol and respond to statins, as normal hepatocytes should.44 Human ES and iPS cells seem to form hepatocytes with similar efficiency.49 Fully mature iHLCs are quite difficult to produce with entirely in vitro approaches, but subsequent transplantation into immunocompromised mouse liver seems to affect final stages of maturation.50
Similarly, in one seminal study using analogous approaches to that described above, produced hepatocyte-like cells that expressed a spectrum of mature hepatocyte markers with residual AFP expression.45 Alternate designs for the cell culture have also been tested. For example, one approach yielded cells with at least some level of CYP3A4 expression, the ability to take up indocyanine green, and store glycogen.51 In a different strategy, a cDNA encoding the hepatocyte-specific nuclear receptor/transcription factor HNF4α was ectopically expressed in hiPSCs that were otherwise induced to form hepatocyte-like cells.26 These cells exhibited increased cytochrome P450 (CYP) expression, a finding that is consistent with the fact that many CYP genes contain HNF4α response elements near their promoters. Finally, direct reprogramming of fibroblasts to induced hepatocyte-like cells (iHLC) has been achieved by induced transdifferentiation, in most cases using mouse tail tip fibroblasts as a starting material.52–54
Limitations of stem cell models
iHLCs are a tool not only for the discovery of new iDILI genes but also for the elucidation of the cellular and molecular mechanisms of iDILI. However, iHLCs are not without potential problems. First and foremost, it has been exceedingly difficult to produce iHLCs that completely exhibit normal hepatocellular function. Most notably, iHLCs do not exhibit an adult-like range of CYP expression. Improvements in the maturation of iHLCs will be needed in order to fully realize their potential. Another difficulty (though soluble) is rooted in human genetic variation. Hypothetically, one might reprogram somatic cells from three iDILI patients and three control patients and then compare the two sets of iHLCs. Although the iDILI patients may contain polymorphisms relevant to iDILI, there exists a very large amount of extraneous genetic variation between the two groups. Thus, the confounding factor of genetic background may dilute iDILI phenotypes in vitro. A new tool that partially addresses this problem involves the use of new site-specific recombinase systems such as clustered regularly interspaced short palindromic repeats (CRISPR) or transcription activator-like effector nucleases (TALENS) to correct putative SNPs or other polymorphisms suspected of causing iDILI, thus yielding an isogenic pair of cell lines. This is discussed further below.
Finally, a major limitation of stem cell models should be recognized. Any cells derived from stem cells, even sophisticated co-culture models, cannot fully recapitulate the situation in vivo. For instance, it is likely that immune reactions are important for some (though probably not all) cases of iDILI. It is currently not feasible to combine iHLCs with reconstituted immune systems in vitro. Placing iHLCs into mice is also not a reasonable option to assess the contribution of immune functions since the mouse immune system differs in fundamental ways from the human immune system.
In the future, improvements will need to occur if stem cell approaches are to more faithfully model iDILI. Perhaps the area most in need of improvement is the differentiation of stem cells into fully functional hepatocytes that express a normal range of CYP genes.55 Progress is being made in this area through the use of three-dimensional differentiation methods that more closely model cellular positions within organs. Furthermore, new approaches that yield “organoids” (small three-dimensional multicellular structures with organized cells) may lead to better expression of CYP genes.56
Current applications and future directions
iDILI is a disorder that can arise from mutations in at least several key genes, and it likely involves multiple cell types, including hepatocytes and immune cells. It is also possible (though little investigated) that epigenetic alterations in the hepatocyte may lead to changes in gene expression and contribution to the onset of iDILI. The advent of iHLCs constitutes a novel step toward identifying genetic and epigenetic mechanisms leading to predisposition to iDILI and mechanisms involved in the acute phase of iDILI. A number of useful attributes of these cells make them useful for toxicological applications. Foremost, iHLCs are genetically identical to the donor subject who donated somatic cells to produce hiPSCs. In addition, the iPS reprogramming process erases epigenetic modifications that accumulate in primary cells in response to life exposure to toxicants.57–60 Finally, iPSCs and iHLCs, can be produced in unlimited quantities, whereas primary hepatocytes cannot be readily expanded. Hence, the need for repeated liver biopsies from single donors, necessary for conventional experimentation, can be avoided. 61–65
Identification and study of genetic variants causing iDILI
An obvious way to use iPS strategies to investigate iDILI is to collect somatic cell samples (such as skin or nucleated blood cells) and subject them to iPS-mediated reprogramming. The use of iPSCs to investigate iDILI is only now coming to fruition, but here is a reasonable workflow: Cells from at least several subjects who have suffered from iDILI and normal subjects are reprogrammed to yield a set of wild-type and affected (iDILI) iPSCs. Since episodes of DILI affect the liver by definition, it is quite possible that liver biopsies from subjects who have survived (or succumbed) to DILI might be abnormal in terms of their mutational load, gene expression, or even epigenetic content. Since iPSCs are derived from non-liver tissues, these cells are less likely to have undergone drug-induced mutations or other alterations. Sets of iHLCs derived from multiple normal and iDILI patients can then be prepared and investigated to identify changes in gene expression pathways that are potentially involved in iDILI. The iPS approach can yield cells that can be useful to actually test specific mutations in putative iDILI genes such as GSTT1, GSTM1, POLG, and others, as described above.
A notable advantage of the iHLC-based iDILI cell culture model stems from the ability to rescue causative mutations through genome editing.66,67 Using TALENs 68,69 or CRISPR70 homologous recombination genome editing strategies, putative iDILI associated mutations can be repaired in iPSCs. The resulting cells are isogenic with respect to the unedited iPSC cells except for the mutation site. Subsequent differentiation of the repaired and unrepaired iPSCs (which can differ by only a single SNP of interest) to iHLCs yields a powerful pair of experiment and control cell cultures, which may then be interrogated for their response to iDILI-causing drugs. In analogous fashion, using the same recombinase strategies, iDILI associated mutations can also be introduced into the genome of wild-type iPSCs to yield an equivalent pair of wild type and mutant cell lines that are isogenic except for the mutation under investigation. In addition to drug assays, transcriptome and epigenome analyses can be utilized in such paired cell culture approaches to learn more about the role of specific iDILI-promoting mutations and the genes that host such mutations.
Assessment of the contribution of epigenetic regulation to iDILI
Exposure to iDILI-causing drugs may lead to the onset of iDILI in response to epigenetic alterations in hepatocytes. Such alterations could, in theory, be a direct result of exposure to the drug. Alternatively, epigenetic changes might occur over time due to indirect (but consequential) responses71–73 that result in some patients in the deregulation of the expression of proteins that are protective for iDILI. Such genes may be epigenetically silenced more permanently through trimethylation of lysine 27 of histone 3 and methylation of cytosine residues in CpG dinucleotides in gene regulatory regions.74 Similar to perturbed histone acetylation homeostasis in cancer,75 further histone deacetylation may then compact the associated chromatin domains and render the respective genes difficult to transcribe upon exposure to subsequent treatment with an iDILI causing drug. It is likely that candidate genes include not only those encoding proteins but also microRNAs that are protective for iDILI. Certain haplotypes may favor the acquisition of epigenetic marks leading to silencing of iDILI protective genes, adding an additional genetic component that may only be identifiable through whole genome sequencing. In addition, insertion/deletions (indels) and copy number variation for iDILI protective genes may also have a role in the pathogenesis of iDILI by affecting transcription levels and dosage of iDILI protective genes. At present, little is known about an epigenetic influence on iDILI and future research will need to address this issue.
Transcriptome profiling provides a readout that integrates both epigenetic and genetic contributions to gene regulation.76,77 During diagnosis of iDILI in the clinic, it is commonplace to collect a small number of liver cells by needle biopsy.78 From these, it is possible to isolate hepatocytes by laser capture or magnetic bead purification approaches. RNA can be extracted from the iDILI patient hepatocytes and iHLCs are derived from the same individual. Since iPS reprogramming erases only epigenetic information, differences in transcriptome profiles between the patient hepatocytes and iHLCs could be due to either iDILI-caused epigenetic changes in the patient hepatocytes, or incomplete accumulation of proper epigenetic chromatin modification patterns due to infidelities that arise during iHLC differentiation. Future research will be needed to determine if iDILI epigenetic changes can be detected by such strategies. However, such experiments may provide candidate genomic targets for further evaluation using targeted chromatin-immunoprecipitation (ChIP). The alternate approach using ChIP-Seq instead of ChIP based comparisons may currently be quite challenging due to the limited availability of primary cells. In addition, iHLCs are exquisitely well suited for “omics” approaches since their nearly unlimited numbers allow for sufficient transcriptional profiling experiments to achieve a level of experimental replication that is sufficient for the minimization of type one and type two errors.79 Such replication cannot easily be carried out with primary cells. Certain limitations, however, are inherent to this approach. For instance, cell type heterogeneity among collected biopsies confounds the interpretation of large datasets. Laser-capture microscopy based approaches may provide a viable alternative to mixed cell populations normally retrieved via biopsies. However, the limitations in the sample amounts inherent to this technology require amplification based strategies for expression profiling that can introduce a bias sufficient to obscure the contribution of low abundance transcripts to the pathogenesis of iDILI.80–82 Magnetic bead capture of hepatocytes may supply a better alternative as cells could be collected en masse. Alternatively, hepatocytes could be expanded from the initial biopsy but primary hepatocytes have only a limited life span in culture, are difficult to expand, and suffer from drifting gene expression that again can introduce bias sufficient to prohibit the identification of transcripts relevant to iDILI. Clearly, iHLC based transcription surveys, though currently not free from drawbacks, are less prone to these limitations.
Possible xenobiotic-induced predisposition to iDILI
It is possible that prior or concomitant exposure to drugs, other xenobiotics, or even high levels of endogenous metabolites might predispose the onset of iDILI upon treatment with iDILI-causing drugs. Little is known, however, about chemical sensitization to iDILI, so this idea must be considered a working hypothesis. Conventional medical studies typically do not control for prior exposure to compounds that might induce a predisposition to iDILI. Likewise, cell culture models using cell lines such as HepG283 or HepaRG84 are limited since these cell lines cannot capture genotype-based predispositions to the disease. In contrast, iHLCs provide an excellent option to study how prior treatment with predisposing compounds can make cells susceptible to iDILI, since these cells are derived from patients that have already been known to have developed iDILI. For instance, two-staged high throughput screens can be developed where iDILI-iHLCs are treated first with a compound known to elevate oxidative stress in liver cells without causing a disease phenotype. Subsequently, either in the presence or absence of the predisposing compound, the iDILI-inducing drug can be administered and toxicity can be monitored by assays for increased cell death, reduced energy metabolism, or induction of proinflammatory cytokine expression.
Certain types of toxicity, e.g., functional hepatic changes, are difficult to assess in cell culture models. For example, cholestasis, which is a frequent hallmark of DILI, has only recently been modeled in vitro. 85,86 Such functional assays have not yet been applied to or validated for iHLCs. Because polymorphisms and rare variants in bile salt transporters have been implicated in the development of drug-induced cholestasis, a genetic analysis of ABCB11 (BSEP), ABCC2 (MRP2), and other genes encoding for canalicular transporters could be analyzed.
Another possibility for xenobiotic-induced predisposition to iDILI is immunological in nature. Since major histocompatibility complex (MHC) genes have already been implicated in iDILI, it follows that dysregulation of the immune system is one route to the onset of iDILI. It may be that iDILI has much in common with an allergic response, where the immune system is first sensitized to a cellular antigen, perhaps by an initial exposure to a DILI drug or a chemically-similar compound. This initial exposure could induce the formation of self-directed (hepatocyte-directed) immune cells, which then become activated and amplified upon later exposure to the iDILI-causing drug. Of note, iHLC approaches can readily be used to study cell-autonomous mechanisms of chemical sensitization but not immunological sensitization mechanisms.
Future goals and directions with iHLC cell culture systems
In order to fully leverage the power of the iHLCs as a cell culture model for iDILI, iHLCs must be produced that more accurately resemble the hepatocytes in vivo. In all current differentiation protocols, iHLCs exhibit a phenotype resembling that of late embryonic or early postnatal primary hepatocytes.87 For instance, current state-of-the-art iHLCs express high levels of α-fetoprotein (AFP) and CYP3A7,44,45,88–90 both markers for embryonic hepatocytes.91,92 Consistent with fairly robust AFP expression, ALB, a marker for adult primary hepatocytes, is expressed at significantly lower levels. Similarly, current iHLCs express low CYP3A4 and CYP2E1.44,45,88–90 It therefore appears that, despite demonstrating other typical liver cellular activities such as indocyanin green uptake, urea production, glycogen storage, LDL uptake,93 and cholesterol secretion,44 current iHLCs may express only low levels of drug metabolizing enzymes that have a key role in the pathogenesis in iDILI. The low expression level of CYPs in particular may lead to a significantly attenuated iDILI phenotype in vitro. Future characterization of iHLCs may require the use of global transcriptome comparisons between fresh primary hepatocytes, but this in itself may not guide the design of improved methods to differentiate detoxification-competent iHLCs from pluripotent cells.
A related technical problem is that primary hepatocytes can be maintained in culture for only a short time, prior to loss of detoxification activities typical of the hepatocyte in vivo. For instance, cultured primary hepatocytes rapidly downregulate many CYPs, including CYP3A4, to nearly undetectable levels.94,95 It is possible that cell culture effects lead to transcriptional deregulation of genes essential for the pathogenesis of iDILI. Although significant improvements in primary hepatocyte culture have been made over the last few years64,96,97 permitting the maintenance of some key hepatocyte enzyme expression for up to several weeks, the length of the drug treatment required for eliciting an iDILI response in culture is unknown and drug dependent. Perhaps of even greater concern is that hepatocytes obtained from iDILI patients by needle biopsy may be abnormal after an episode of iDILI, and thus not suitable for experiments designed to uncover the mechanisms whereby naïve hepatocytes are degraded due to a DILI event.
Based on the above, it is clear that there is much room for improvement in the methodologies leading to the production of iHLCs from human pluripotent cells. Three major strategies to improve iHLCs are described here:
Use of decellularized animal liver matrices for iHLC cultures. Promising recent advances have been made using decellularized liver tissue to produce a native liver extracellular matrix (LEM) as substrate for the growth of primary hepatocytes in subsequent two-dimensional and three-dimensional applications.98 Using LEM, the authors reported significantly improved hepatic functions including elevated albumin secretion and urea synthesis. Moreover, long-term viability was also increased, but a comparison of genome-wide expression profiles between nascent hepatocytes and cells cultured long term was not carried out. Nevertheless, the use of LEM represents an exciting route that may lead to improved iHLC maturation. Likewise, inclusion of sonic hedgehog (SHH) into the stage two (hepatic induction) differentiation cocktail may result in increased transcription of FOXA2,51,99 a transcription factor that is critical at this stage for hepatic induction. Further optimization of growth factor concentrations and timing of addition to the culture medium may be another way to optimize iHLCs.
Three-dimensional and organoid cultures. Three-dimensional cultures represent a family of approaches to produce high quality iHLCs with more adult characteristics. Three-dimensional approaches include the use of bioreactors,100 tethered spheroids,96 three-dimensional-capable substrates such as matrigel sandwich cultures,101–103 caprolactones,104 and alginates.105,106 In addition, three-dimensional co-cultures of mouse embryonic fibroblasts with hepatocytes61 as well as spontaneous organoid formation in co-cultures of hepatocytes, immuneprogenitors, and human umbilical cord vascular endothelial cells107 have given rise to greatly improved primary hepatocyte cultures and are becoming more commonplace in the pharmaceutical industry.
Humanized mouse livers. Transplantation of human hepatocytes into mice constitutes a system that recapitulates many features of the human liver. A major advantage is the ability to produce engrafted human liver tissue that is appropriately vascularized and contains the major cell types present in the human liver.50,108 Moreover, pharmacokinetic studies can be carried out with a dose-response regimen that mimics the treatment protocol in patients and apparent hepatic injury can be monitored over several months, though interim tissue harvesting would be necessary in most cases to assess the state of the graft. Both the immune-compromised FAH and the ALB-HSV-TK mouse models permit the ablation of the host liver in a timed and controlled fashion during which the host liver can be replaced by the human implant.50,108 While in the FAH mouse dietary constraints are required for maintenance of the host liver, in the ALB-HSV-TK mouse, the hepatocytes are ablated in a tissue specific fashion through uptake of dietary gancyclovir where the albumin promoter ensures tissue specific expression of HSV-TK. In addition, these liver replacement models have the advantage that the iDILI-causing drug is not metabolized primarily by the mouse liver, which might result in a greatly attenuated response in the human graft. A particularly attractive variation may be the combination of liver organoid grafts and either the FAH or the ALB-HSV-TK mouse as hosts.
Conclusions
In this review article, we have discussed existing knowledge about iDILI, stem cell approaches to find iDILI genes and to study iDILI mechanisms, and also provided a discussion of current applications and future directions for this field of research. Conventional human genetic approaches are unlikely to yield a comprehensive set of iDILI-associated polymorphisms. New approaches using pluripotent stem cells (especially hiPSCs) promise to provide an expedient way to find new iDILI mutations and to offer a way to study iDILI mechanisms in vitro. Finally, future uses of hiPSCs from iDILI patients were discussed, including the use of iHLCs to study epigenetic contributions to iDILI, and xenobiotic-induced sensitivity for iDILI.
Acknowledgements
This review article was supported by Connecticut Stem Cell Research Grant 11SCDIS02.
Abbreviations
- AFP
α-fetoprotein
- albumin
ALB
- BMP4
bone morphogenic protein 4
- ChIP
chromatin-immunoprecipitation
- CRISPR
clustered regularly interspaced short palindromic repeats
- c-Myc
proto-oncogene myc
- CYP
cytochrome P450
- FGF2
fibroblast growth factor 2
- GWAS
genome-wide association study
- GST
glutathione-S-transferase
- hESC
human embryonic stem cell
- HGF
hepatocyte growth factor
- hiPSC
human induced pluripotent stem cell
- HLA
human leukocyte antigen
- HNF4α
hepatocyte-specific nuclear receptor/transcription factor α
- ICM
inner cell mass
- iDILI
idiosyncratic drug-induced liver injury
- iHLC
induced hepatocyte-like cell
- Indels
insertion/deletions
- iPSCs
induced pluripotent stem cells
- Klf4
kruppel-like factor
- LEM
liver extracellular matrix
- LIF
leukemia inhibitory factor
- LPS
lipopolysaccharide
- mESCs
mouse embryonic stem cells
- MHC
major histocompatibility complex
- Oct4
octamer-binding transcription 4
- POLG
mitochondrial polymerase γ
- SHH
sonic hedgehog
- SNP
single nucleotide polymorphisms
- Sox2
sry-related HMG box 2
- TALENS
transcription activator-like effector nucleases
References
- 1.Bell LN, Chalasani N. Epidemiology of idiosyncratic drug-induced liver injury. Semin Liver Dis. 2009;29:337–347. doi: 10.1055/s-0029-1240002. 10.1055/s-0029-1240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarro VJ, Senior JR. Drug-related hepatotoxicity. New Engl J Med. 2006;354:731–739. doi: 10.1056/NEJMra052270. 10.1056/NEJMra052270. [DOI] [PubMed] [Google Scholar]
- 3.Sakatis MZ, Reese MJ, Harrell AW, Taylor MA, Baines IA, Chen L, et al. Preclinical strategy to reduce clinical hepatotoxicity using in vitro bioactivation data for >200 compounds. Chem Res Toxicol. 2012;25:2067–2082. doi: 10.1021/tx300075j. 10.1021/tx300075j. [DOI] [PubMed] [Google Scholar]
- 4.Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nature Rev Drug Discov. 2005;4:489–499. doi: 10.1038/nrd1750. 10.1038/nrd1750. [DOI] [PubMed] [Google Scholar]
- 5.Watkins P. Idiosyncratic liver injury: Challenges and approaches. Toxicol Pathol. 2005;33:1–5. doi: 10.1080/01926230590888306. 10.1080/01926230590888306. [DOI] [PubMed] [Google Scholar]
- 6.Holt MP, Ju C. Mechanisms of drug-induced liver injury. AAPS J. 2006;8:E48–54. doi: 10.1208/aapsj080106. 10.1208/aapsj080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liguori MJ, Waring J F. Investigations toward enhanced understanding of hepatic idiosyncratic drug reactions. Expert Opin Drug Metab Toxicol. 2006;2:835–846. doi: 10.1517/17425255.2.6.835. 10.1517/17425255.2.6.835. [DOI] [PubMed] [Google Scholar]
- 8.Bleibel W, Kim S, D'Silva K, Lemmer ER. Drug-induced liver injury. Dig Dis Sci. 2007;52:2463–2471. doi: 10.1007/s10620-006-9472-y. 10.1007/s10620-006-9472-y. [DOI] [PubMed] [Google Scholar]
- 9.Hussaini SH, Farrington EA. Idiosyncratic drug-induced liver injury: an overview. Expert Opin Drug Saf. 2007;6:673–684. doi: 10.1517/14740338.6.6.673. 10.1517/14740338.6.6.673. [DOI] [PubMed] [Google Scholar]
- 10.Boelsterli UA, Kashimshetty R. Hepatic Toxicology. In: Roth RA, Ganey PE, editors. Comprehensive Toxicology. Vol. 9. Elsevier; 2010. pp. 383–402. 10.1016/B978-0-08-046884-6.01015-0. [Google Scholar]
- 11.Bjornsson ES, Bergmann OM, Bjornsson HK, Kvaran RB, Olafsson S. Incidence, presentation, and outcomes in patients with drug-induced liver injury in the general population of Iceland. Gastroenterology. 2013;144:1419–1425. doi: 10.1053/j.gastro.2013.02.006. 10.1053/j.gastro.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Yuan L, Kaplowitz N. Mechanisms of drug-induced liver injury. Clin Liver Dis. 2013;17:507–518. doi: 10.1016/j.cld.2013.07.002. 10.1016/j.cld.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boelsterli UA. Disease-related determinants of susceptibility in drug-induced hepatotoxicity. Curr Opin Drug Disc Develop. 2013;6:81–91. [PubMed] [Google Scholar]
- 14.Deng X, Luyendyk JP, Ganey PE, Roth RA. Inflammatory stress and idiosyncratic hepatotoxicity: Hints from animal models. Pharmacol Rev. 2009;61:262–282. doi: 10.1124/pr.109.001727. 10.1124/pr.109.001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russmann S, Jetter A, Kullak-Ublik GA. Pharmacogenetics of drug-induced liver injury. Hepatology. 2010;52:748–761. doi: 10.1002/hep.23720. 10.1002/hep.23720. [DOI] [PubMed] [Google Scholar]
- 16.Daly AK, Day CP. Genetic association studies in drug-induced liver injury. Drug Metab Rev. 2012;44:116–126. doi: 10.3109/03602532.2011.605790. 10.3109/03602532.2011.605790. [DOI] [PubMed] [Google Scholar]
- 17.Daly AK. Molecular basis of polymorphic drug metabolism. J Mol Med (Berl) 1955;73:539–553. doi: 10.1007/BF00195139. [DOI] [PubMed] [Google Scholar]
- 18.Hirata K, Takagi H, Yamamoto M, Matsumoto T, Nishiya T, Mori K, et al. Ticlopidine-induced hepatotoxicity is associated with specific human leukocyte antigen genomic subtypes in Japanese patients: A preliminary case-control study. Pharmacogenom J. 2008;8:29–33. doi: 10.1038/sj.tpj.6500442. 10.1038/sj.tpj.6500442. [DOI] [PubMed] [Google Scholar]
- 19.Lucena M, Andrade RJ, Martínez C, Ulzurrun E, García-Martín E, Borraz Y, et al. Glutathione S-transferase M1 and T1 null genotypes increase susceptibility to idiosyncratic drug-induced liver injury. Hepatology. 2008;48:588–596. doi: 10.1002/hep.22370. 10.1002/hep.22370. [DOI] [PubMed] [Google Scholar]
- 20.Stewart JD, Horvath R, Baruffini E, Ferrero I, Bulst S, Watkins PB, et al. Polymerase g gene POLG determines the risk of sodium valproate-induced liver toxicity. Hepatology. 2010;52:1791–1796. doi: 10.1002/hep.23891. 10.1002/hep.23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixit R, Boelsterli UA. Healthy animals and animal models of human disease(s) in safety assessment of human pharmaceuticals, including therapeutic antibodies. Drug Disc Today. 2007;12:336–342. doi: 10.1016/j.drudis.2007.02.018. 10.1016/j.drudis.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Roth RA, Ganey PE. Animal models of idiosyncratic drug-induced liver injury - Current status. Crit Rev Toxicol. 2011;41:723–739. doi: 10.3109/10408444.2011.575765. 10.3109/10408444.2011.575765. [DOI] [PubMed] [Google Scholar]
- 23.Boelsterli UA, Hsiao CJ. The heterozygous Sod2+/− mouse: Modeling the mitochondrial role in drug toxicity. Drug Discovery Today. 2008;13:982–988. doi: 10.1016/j.drudis.2008.08.002. 10.1016/j.drudis.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 25.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- 27.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. 10.1016/S0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 28.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 29.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136:2311–2322. doi: 10.1242/dev.024398. 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orkin SH. Chipping away at the embryonic stem cell network. Cell. 2005;122:828–830. doi: 10.1016/j.cell.2005.09.002. 10.1016/j.cell.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 34.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 35.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 36.Gurdon JB, Uehlinger V. “Fertile” intestine nuclei. Nature. 1966;210:1240–1241. doi: 10.1038/2101240a0. 10.1038/2101240a0. [DOI] [PubMed] [Google Scholar]
- 37.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 40.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 44.Krueger WH, Tanasijevic B, Barber V, Flamier A, Gu X, Manautou J, et al. Cholesterol-secreting and statin-responsive hepatocytes from human ES and iPS cells to model hepatic involvement in cardiovascular health. PLoS One. 2013;8:67296. doi: 10.1371/journal.pone.0067296. 10.1371/journal.pone.0067296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu J, Sadler KC. New school in liver development: lessons from zebrafish. Hepatology. 2009;50:1656–1663. doi: 10.1002/hep.23157. 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller KM, Themanns M, Friedbichler K, Kornfeld JW, Esterbauer H, Tuckermann JP, et al. Hepatic growth hormone and glucocorticoid receptor signaling in body growth, steatosis and metabolic liver cancer development. Mol Cell Endocrinol. 2012;361:1–11. doi: 10.1016/j.mce.2012.03.026. 10.1016/j.mce.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell research. 2009;19:1233–1242. doi: 10.1038/cr.2009.107. 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 50.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura N, Saeki K, Mitsumoto M, Matsuyama S, Nishio M, Saeki K, et al. Feeder-free and serum-free production of hepatocytes, cholangiocytes, and their proliferating progenitors from human pluripotent stem cells: application to liver-specific functional and cytotoxic assays. Cell Reprogram. 2012;14:171–185. doi: 10.1089/cell.2011.0064. [DOI] [PubMed] [Google Scholar]
- 52.Khurana S, Jaiswal AK, Mukhopadhyay A. Hepatocyte nuclear factor-4alpha induces transdifferentiation of hematopoietic cells into hepatocytes. J Biol Chem. 2010;285:4725–4731. doi: 10.1074/jbc.M109.058198. 10.1074/jbc.M109.058198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 54.Ji S, Zhang L, Hui L. Cell fate conversion: direct induction of hepatocyte-like cells from fibroblasts. J Cell Biochem. 2013;114:256–265. doi: 10.1002/jcb.24380. 10.1002/jcb.24380. [DOI] [PubMed] [Google Scholar]
- 55.Jozefczuk J, Prigione A, Chavez L, Adjaye J. Comparative analysis of human embryonic stem cell and induced pluripotent stem cell-derived hepatocyte-like cells reveals current drawbacks and possible strategies for improved differentiation. Stem Cells Dev. 2011;20:1259–1275. doi: 10.1089/scd.2010.0361. 10.1089/scd.2010.0361. [DOI] [PubMed] [Google Scholar]
- 56.Willenbring H, Soto-Gutierrez A. Transplantable liver organoids made from only three ingredients. Cell stem cell. 2013;13:139–140. doi: 10.1016/j.stem.2013.07.014. 10.1016/j.stem.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, Xue WJ, Li P, Sha ZY, Huang H, Rui L, et al. RASSF1A hypermethylation is associated with aflatoxin B1 and polycyclic aromatic hydrocarbon exposure in hepatocellular carcinoma. Hepatogastroenterology. 2012;59:1883–1888. doi: 10.5754/hge11731. [DOI] [PubMed] [Google Scholar]
- 58.Clarke JD, Cherrington NJ. Genetics or environment in drug transport: the case of organic anion transporting polypeptides and adverse drug reactions. Expert Opin Drug Metab Toxicol. 2012;8:349–360. doi: 10.1517/17425255.2012.656087. 10.1517/17425255.2012.656087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castillo P, Ibáñez F, Guajardo A, Llanos MN, Ronco AM. Impact of cadmium exposure during pregnancy on hepatic glucocorticoid receptor methylation and expression in rat fetus. PLoS One. 2012;7:44139. doi: 10.1371/journal.pone.0044139. 10.1371/journal.pone.0044139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeybel M, Mann DA, Mann J. Epigenetic modifications as new targets for liver disease therapies. J Hepatol. 2013;59:1349–1353. doi: 10.1016/j.jhep.2013.05.039. 10.1016/j.jhep.2013.05.039. [DOI] [PubMed] [Google Scholar]
- 61.Leite SB, Teixeira AP, Miranda JP, Tostões RM, Clemente JJ, Sousa MF, et al. Merging bioreactor technology with 3D hepatocyte-fibroblast culturing approaches: Improved in vitro models for toxicological applications. Toxicol In Vitro. 2011;25:825–832. doi: 10.1016/j.tiv.2011.02.002. 10.1016/j.tiv.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Janorkar AV, Harris LM, Murphey BS, Sowell BL. Use of three-dimensional spheroids of hepatocyte-derived reporter cells to study the effects of intracellular fat accumulation and subsequent cytokine exposure. Biotechnol Bioeng. 2011;108:1171–1180. doi: 10.1002/bit.23025. 10.1002/bit.23025. [DOI] [PubMed] [Google Scholar]
- 63.Goral VN, Hsieh YC, Petzold ON, Clark JS, Yuen PK, Faris RA. Perfusion-based microfluidic device for three-dimensional dynamic primary human hepatocyte cell culture in the absence of biological or synthetic matrices or coagulants. Lab Chip. 2010;10:3380–3386. doi: 10.1039/c0lc00135j. 10.1039/c0lc00135j. [DOI] [PubMed] [Google Scholar]
- 64.Miranda JP, Leite SB, Muller-Vieira U, Rodrigues A, Carrondo MJ, Alves PM. Towards an extended functional hepatocyte in vitro culture. Tissue Eng Part C Methods. 2009;15:157–167. doi: 10.1089/ten.tec.2008.0352. 10.1089/ten.tec.2008.0352. [DOI] [PubMed] [Google Scholar]
- 65.Guillouzo A, Guguen-Guillouzo C. Evolving concepts in liver tissue modeling and implications for in vitro toxicology. Expert Opin Drug Metab Toxicol. 2008;4:1279–1294. doi: 10.1517/17425255.4.10.1279. 10.1517/17425255.4.10.1279. [DOI] [PubMed] [Google Scholar]
- 66.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng LT, Sun LT, Tada T. Genome editing in induced pluripotent stem cells. Genes Cells. 2012;17:431–438. doi: 10.1111/j.1365-2443.2012.01599.x. 10.1111/j.1365-2443.2012.01599.x. [DOI] [PubMed] [Google Scholar]
- 68.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol Bioeng. 2013;110:1811–1821. doi: 10.1002/bit.24890. 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 69.Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Bio. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 71.Avan A, Crea F, Paolicchi E, Funel N, Galvani E, Marquez VE, et al. Molecular mechanisms involved in the synergistic interaction of the EZH2 inhibitor 3-deazaneplanocin A with gemcitabine in pancreatic cancer cells. Mol Cancer Ther. 2012;11:1735–1746. doi: 10.1158/1535-7163.MCT-12-0037. 10.1158/1535-7163.MCT-12-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.D'Addario C, Caputi FF, Ekström TJ, Di Benedetto M, Maccarrone M, Romualdi P, et al. Ethanol induces epigenetic modulation of prodynorphin and pronociceptin gene expression in the rat amygdala complex. J Mol Neurosci. 2013;49:312–319. doi: 10.1007/s12031-012-9829-y. 10.1007/s12031-012-9829-y. [DOI] [PubMed] [Google Scholar]
- 73.Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis. 2010;31:489–495. doi: 10.1093/carcin/bgp305. 10.1093/carcin/bgp305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Binder H, Steiner L, Przybilla J, Rohlf T, Prohaska S, Galle J. Transcriptional regulation by histone modifications: towards a theory of chromatin re-organization during stem cell differentiation. Phys Biol. 2013;10:026006. doi: 10.1088/1478-3975/10/2/026006. 10.1088/1478-3975/10/2/026006. [DOI] [PubMed] [Google Scholar]
- 75.Parbin S, Kar S, Shilpi A, Sengupta D, Deb M, Rath SK, et al. Histone deacetylases: a saga of perturbed acetylation homeostasis in cancer. J Histochem Cytochem. 2014;62:11–33. doi: 10.1369/0022155413506582. 10.1369/0022155413506582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi JK. Systems biology and epigenetic gene regulation. IET Syst Biol. 2010;4:289–295. doi: 10.1049/iet-syb.2010.0008. 10.1049/iet-syb.2010.0008. [DOI] [PubMed] [Google Scholar]
- 77.Hoheisel JD. Microarray technology: beyond transcript profiling and genotype analysis. Nat Rev Genet. 2006;7:200–210. doi: 10.1038/nrg1809. 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 78.Kleiner DE, Chalasani NP, Lee WM, Fontana RJ, Bonkovsky HL, Watkins PB, et al. Hepatic histological findings in suspected drug-induced liver injury: systematic evaluation and clinical associations. Hepatology. 2014;59:661–670. doi: 10.1002/hep.26709. 10.1002/hep.26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin LJ, Woo JG, Avery CL, Chen HS, North KE, Au K, et al. Multiple testing in the genomics era: findings from Genetic Analysis Workshop 15, Group 15. Genet Epidemiol. 2007;31(Suppl 1):S124–131. doi: 10.1002/gepi.20289. 10.1002/gepi.20289. [DOI] [PubMed] [Google Scholar]
- 80.Adjaye J. Generation of amplified RNAs and cDNA libraries from single mammalian cells. Methods Mol Med. 2007;132:117–124. doi: 10.1007/978-1-59745-298-4_10. 10.1007/978-1-59745-298-4_10. [DOI] [PubMed] [Google Scholar]
- 81.Singh R, Maganti RJ, Jabba SV, Wang M, Deng G, Heath JD, et al. Microarray-based comparison of three amplification methods for nanogram amounts of total RNA. Am J Physiol Cell Physiol. 2005;288:C1179–1189. doi: 10.1152/ajpcell.00258.2004. 10.1152/ajpcell.00258.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neira M, Azen E. Gene discovery with laser capture microscopy. Methods Enzymol. 2002;356:282–289. doi: 10.1016/s0076-6879(02)56941-x. 10.1016/S0076-6879(02)56941-X. [DOI] [PubMed] [Google Scholar]
- 83.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 84.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ansede JH, Smith WR, Perry CH, St Claire RL, 3rd, Brouwer KR. An in vitro assay to assess transporter-based cholestatic hepatotoxicity using sandwich-cultured rat hepatocytes. Drug Metab Dispos. 2010;38:276–280. doi: 10.1124/dmd.109.028407. 10.1124/dmd.109.028407. [DOI] [PubMed] [Google Scholar]
- 86.Greer ML, Barber J, Eakins J, Kenna JG. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268:125–131. doi: 10.1016/j.tox.2009.08.007. 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Gerbal-Chaloin S, Funakoshi N, Caillaud A, Gondeau C, Champon B, Si-Tayeb K. Human induced pluripotent stem cells in hepatology: beyond the proof of concept. Am J Pathol. 2014;184:332–347. doi: 10.1016/j.ajpath.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 88.Kawabata K, Inamura M, Mizuguchi H. Efficient hepatic differentiation from human iPS cells by gene transfer. Methods Mol Biol. 2012;826:115–124. doi: 10.1007/978-1-61779-468-1_10. 10.1007/978-1-61779-468-1_10. [DOI] [PubMed] [Google Scholar]
- 89.Takata A, Otsuka M, Kogiso T, Kojima K, Yoshikawa T, Tateishi R, et al. Direct differentiation of hepatic cells from human induced pluripotent stem cells using a limited number of cytokines. Hepatol Int. 2011;5:890–898. doi: 10.1007/s12072-011-9251-5. [DOI] [PubMed] [Google Scholar]
- 90.Shiraki N, Yamazoe T, Qin Z, Ohgomori K, Mochitate K, Kume K, et al. Efficient differentiation of embryonic stem cells into hepatic cells in vitro using a feeder-free basement membrane substratum. PLoS One. 2011;6:24228. doi: 10.1371/journal.pone.0024228. 10.1371/journal.pone.0024228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nayak NC, Mital I. The dynamics of alpha-fetoprotein and albumin synthesis in human and rat liver during normal ontogeny. Am J Pathol. 1977;86:359–374. [PMC free article] [PubMed] [Google Scholar]
- 92.Greuet J, Pichard L, Bonfils C, Domergue J, Maurel P. The fetal specific gene CYP3A7 is inducible by rifampicin in adult human hepatocytes in primary culture. Biochem Biophys Res Commun. 1996;225:689–694. doi: 10.1006/bbrc.1996.1231. 10.1006/bbrc.1996.1231. [DOI] [PubMed] [Google Scholar]
- 93.Baharvand H, Hashemi SM, Shahsavani M. Differentiation of human embryonic stem cells into functional hepatocyte-like cells in a serum-free adherent culture condition. Differentiation. 2008;76:465–477. doi: 10.1111/j.1432-0436.2007.00252.x. 10.1111/j.1432-0436.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 94.LeCluyse E, Madan A, Hamilton G, Carroll K, DeHaan R, Parkinson A. Expression and regulation of cytochrome P450 enzymes in primary cultures of human hepatocytes. J Biochem Mol Toxicol. 2000;14:177–188. doi: 10.1002/(sici)1099-0461(2000)14:4<177::aid-jbt1>3.0.co;2-4. 10.1002/(SICI)1099-0461(2000)14:4<177::AID-JBT1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 95.Pascussi JM, Drocourt L, Gerbal-Chaloin S, Fabre JM, Maurel P, Vilarem MJ. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem. 2001;268:6346–6358. doi: 10.1046/j.0014-2956.2001.02540.x. 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 96.Xia L, Sakban RB, Qu Y, Hong X, Zhang W, Nugraha B, et al. Tethered spheroids as an in vitro hepatocyte model for drug safety screening. Biomaterials. 2012;33:2165–2176. doi: 10.1016/j.biomaterials.2011.12.006. 10.1016/j.biomaterials.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Tostões RM, Leite SB, Serra M, Jensen J, Björquist P, Carrondo MJ, et al. Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology. 2012;55:1227–1236. doi: 10.1002/hep.24760. 10.1002/hep.24760. [DOI] [PubMed] [Google Scholar]
- 98.Lee JS, Shin J, Park HM, Kim YG, Kim BG, Oh JW, et al. Liver extracellular matrix providing dual functions of two-dimensional substrate coating and three-dimensional injectable hydrogel platform for liver tissue engineering. Biomacromolecules. 2014;15:206–218. doi: 10.1021/bm4015039. 10.1021/bm4015039. [DOI] [PubMed] [Google Scholar]
- 99.Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, et al. Differentiation of human ES and Parkinson's disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci. 2010;45:258–266. doi: 10.1016/j.mcn.2010.06.017. 10.1016/j.mcn.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sivertsson L, Synnergren J, Jensen J, Björquist P, Ingelman-Sundberg M. Hepatic differentiation and maturation of human embryonic stem cells cultured in a perfused three-dimensional bioreactor. Stem Cells Dev. 2013;22:581–594. doi: 10.1089/scd.2012.0202. 10.1089/scd.2012.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang S, Nagrath D, Chen PC, Berthiaume F, Yarmush ML. Three-dimensional primary hepatocyte culture in synthetic self-assembling peptide hydrogel. Tissue Eng Part A. 2008;14:227–236. doi: 10.1089/tea.2007.0143. 10.1089/tea.2007.0143. [DOI] [PubMed] [Google Scholar]
- 102.Chen HL, Wu HL, Fon CC, Chen PJ, Lai MY, Chen DS. Long-term culture of hepatocytes from human adults. J Biomed Sci. 1998;5:435–440. doi: 10.1007/BF02255932. 10.1007/BF02255932. [DOI] [PubMed] [Google Scholar]
- 103.Ferrini JB, Pichard L, Domergue J, Maurel P. Long-term primary cultures of adult human hepatocytes. Chem Biol Interact. 1997;107:31–45. doi: 10.1016/s0009-2797(97)00072-0. 10.1016/S0009-2797(97)00072-0. [DOI] [PubMed] [Google Scholar]
- 104.Chua KN, Lim WS, Zhang P, Lu H, Wen J, Ramakrishna S, et al. Stable immobilization of rat hepatocyte spheroids on galactosylated nanofiber scaffold. Biomaterials. 2005;26:2537–2547. doi: 10.1016/j.biomaterials.2004.07.040. 10.1016/j.biomaterials.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 105.Lau TT, Ho LW, Wang DA. Hepatogenesis of murine induced pluripotent stem cells in 3D micro-cavitary hydrogel system for liver regeneration. Biomaterials. 2013;34:6659–6669. doi: 10.1016/j.biomaterials.2013.05.034. 10.1016/j.biomaterials.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 106.Fang S, Qiu YD, Mao L, Shi XL, Yu DC, Ding YT. Differentiation of embryoid-body cells derived from embryonic stem cells into hepatocytes in alginate microbeads in vitro. Acta Pharmacol Sin. 2007;28:1924–1930. doi: 10.1111/j.1745-7254.2007.00713.x. 10.1111/j.1745-7254.2007.00713.x. [DOI] [PubMed] [Google Scholar]
- 107.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 108.Zhang Y, Huang SZ, Zeng YT. The effect of HSV-tk/GCV on hepatic specific damage driven by murine ALB promoter/enhancer. Yi Chuan Xue Bao. 2004;31:1053–1060. [PubMed] [Google Scholar]