Abstract
We present a chemical method to selectively tag and enrich thymine modifications, 5-formyluracil (5-fU) and 5-hydroxymethyluracil (5-hmU), found naturally in DNA. Inherent reactivity differences have enabled us to tag 5-fU chemoselectively over its C modification counterpart, 5-formylcytosine (5-fC). We rationalized the enhanced reactivity of 5-fU compared to 5-fC via ab initio quantum mechanical calculations. We exploited this chemical tagging reaction to provide proof of concept for the enrichment of 5-fU containing DNA from a pool that contains 5-fC or no modification. We further demonstrate that 5-hmU can be chemically oxidized to 5-fU, providing a strategy for the enrichment of 5-hmU. These methods will enable the mapping of 5-fU and 5-hmU in genomic DNA, to provide insights into their functional role and dynamics in biology.
The DNA alphabet includes several modified nucleobases that exist above and beyond the four canonical bases in the genomes of living systems.1 The ability to detect and map such modifications is essential to understand their functional role. Chemical approaches for detecting and mapping cytosine (C) modifications (Figure 1a) have been developed to elucidate their function.2 However, the biological role of the analogous thymine (T) modifications, 5-formyluracil (5-fU) and 5-hydroxymethyluracil (5-hmU) (Figure 1a), has been largely unexplored due to the lack of methods to detect and map these modifications.3 Both 5-fU and 5-hmU are oxidative lesions that occur in mammalian DNA4 and may play an important role in disease. The occurrence of 5-fU has been shown to cause mutagenesis in mammalian cells,5,6 while elevated levels of 5-hmU in the blood are associated with cancer.7,8 Furthermore, the presence of both 5-fU and 5-hmU in synthetic oligomer nucleotides (ODNs) has been shown to perturb DNA–protein interactions and transcription factor binding.9,10 Recently, it has also been shown that 5-hmU is generated by ten-eleven translocation (TET) enzyme mediated oxidation of T in mammalian stem cells and hence could have a potential role in gene regulation.4 Herein, we describe a method to chemically tag 5-fU and 5-hmU to provide a strategy to detect and enrich DNA containing these modifications. Enriched fragments can ultimately be sequenced and aligned against the genome to provide a positional map of where such bases occur, which is critical for understanding their function.11−13
Figure 1.
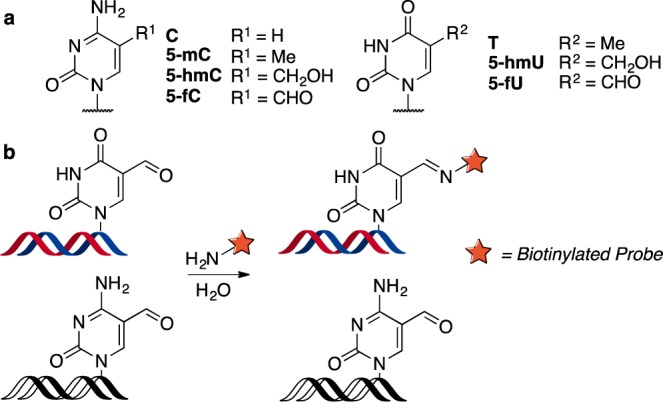
(a) Cytosine and analogous thymine modifications found in DNA. (b) Chemoselective tagging of 5-fU.
We envisaged that the reactive aldehyde functionality on 5-fU could be reacted with a biotinylated probe, analogous to the chemistry previously used to tag 5-fC.11,14 However, to enable the selective enrichment of T modifications, a chemoselective method to distinguish between the aldehyde groups of 5-fU and 5-fC was required (Figure 1b).
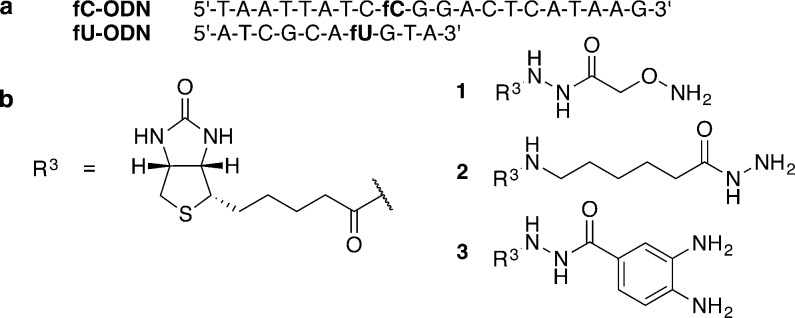
To explore differences in reactivity between the formylated bases, we subjected ODNs that contained either 5-fU (fU-ODN) or 5-fC (fC-ODN) (Figure 2a) to a range of condensation reactions with biotinylated probes 1, 2, or 3 (Figure 2b). Oxyamine 1, used previously for 5-fC enrichment,11,14 can condense on the aldehyde to form an oxime,15,16 while the less nucleophilic acyl hydrazide 2 can react with formyl groups to form a hydrazone.16,17 Furthermore, o-phenylenediamine has been shown to react with the 5-fU mononucleoside to form a stable benzimidazole.18 In order to exploit this chemistry for tagging and subsequent enrichment, we designed a biotinylated o-phenylenediamine linker 3, which was also able to react with 5-fU in a DNA context.
Figure 2.
(a) Sequences of fU-ODN and fC-ODN, and (b) biotinylated oxyamine 1, biotinylated hydrazide linker 2, biotinylated o-phenylenediamine linker 3.
It was found that 5-fU showed greater reactivity than 5-fC (Table 1), providing the basis for selective tagging of T modifications. Previously reported conditions for reacting 5-fC with 1 (Figure 2b)11 require more acidic conditions (pH 5) and the presence of a nucleophilic catalyst p-anisidine (Table 1, entry 1). In contrast, reaction with 5-fU was achievable under less acidic conditions (pH 6) in the absence of catalyst (Table 1, entry 3). Further optimization with a reduced reaction time led to the absence of detectable 5-fC reaction via LC–MS analysis (Table 1, entry 4). Reaction of 5-fU with 2 showed a similar trend in reactivity, although quantitative biotinylation was possible at pH 7 with this probe (Table 1, entries 5–7). Selectivity for 5-fU over 5-fC was also observed when using o-phenylenediamine biotinylated linker 3 (Table 1, entry 8).
Table 1. Reaction Conditions and % of Biotinylation of fU-ODN and fC-ODN with Different Probes.
| probe | conditionsa | fU-ODNb (%) | fC-ODNb (%) | |
|---|---|---|---|---|
| 1 | 1c | p-anisidinef, pH 5, 24 h | >99 | 94 |
| 2 | 1 | pH 5, 24 h | >99 | 56 |
| 3 | 1 | pH 6, 24 h | >99 | 3 |
| 4 | 1 | pH 6, 4 h | 98 | n.d.g |
| 5 | 2d | p-anisidinef, pH 6, 24 h | >99 | 99 |
| 6 | 2 | pH 7, 24 h | >99 | 2 |
| 7 | 2 | pH 7, 4 h | >99 | n.d.g |
| 8 | 3e | pH 7, 4 h | 93 | 1 |
Reactions were carried out in 40 mM NH4OAc (entry 1) or in 40 mM sodium phosphate (entries 2–8).
Conversion was calculated by integration of UV signals of the biotinylated product and the starting material at 260 nm.
Probe 1 was used at 0.4 mM.
Probe 2 was used at 10 mM.
Probe 3 was used at 5 mM.
p-Anisidine was used at 100 mM.
n.d. = not detected.
To rationalize the reactivity differences observed between 5-fU and 5-fC, we performed ab initio quantum mechanical calculations on reduced model systems (Supporting Information, Section 5). We revealed the core electronic factor that defines the increased reactivity of 5-fU, with respect to 5-fC, to be the decreased stability (by 7.71 kcal/mol) of the C–C bonding orbital between the carbons of the aldehyde group and the nucleobase ring. This facilitates the pyramidalization of the aldehyde carbon and formation of the transient C–N bond upon hemiaminal formation (Figure 3a). Furthermore, when considering the transient structures involving the 5-fU model, we located an energy minimum upon the interaction with the incoming nucleophile and a water molecule (Figure 3b and Supporting Information, Section 5). The identified intermediate structure is more stable (ΔE = −13.79 kcal/mol) than the system of non-interacting individual molecules, which may further explain the enhanced reactivity for 5-fU observed over its cytosine counterpart.
Figure 3.
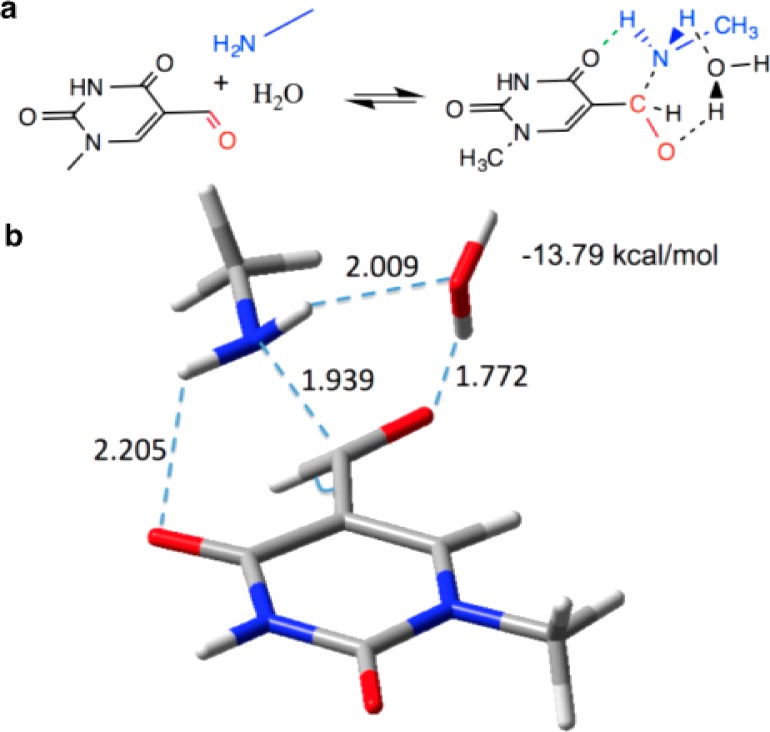
(a) Reduced model systems of 5-formyl-1-methyl uracil (5-fUm), methylamine, and water used in this study. (b) The identified intermediate state along the pathway of hemiaminal formation with 5-fUm. A 6-membered hydrogen transfer transient ring is formed, with the carbon of the aldehyde group partially becoming tetrahedral upon the C–N bond formation. All the outlined distances are in Å.
To demonstrate that DNA containing 5-fU can be selectively enriched, we carried out experiments exploiting the selective reactions developed for probes 1, 2, and 3. A double-stranded 80-mer bearing two modifications per strand was used as a model for 5-fU (fU-DNA), while an analogous ODN containing 5-fC (fC-DNA) and a non-modified ODN (GCAT-DNA) were used as controls (Supporting Information, Table S1). These ODNs were subjected to the biotinylation reaction followed by affinity enrichment using streptavidin-coated magnetic beads. fU-DNA was enriched over fC-DNA by ∼150-fold with each probe, defined by qPCR quantitation (Supporting Information, Figure S4). A similar level of enrichment was observed for fU-DNA over non-modified GCAT-DNA, indicating that captured fC-DNA was at the background level and hence unlikely to be due to covalent reactivity of 5-fC with the biotinylated probes.
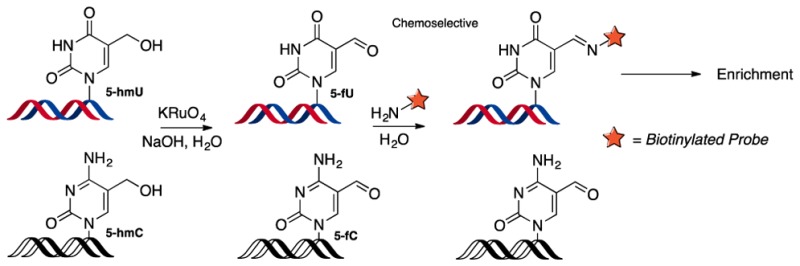
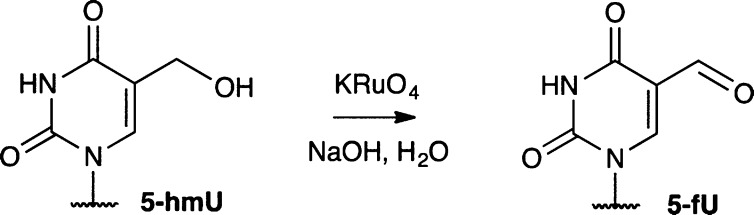
The 5-fU tagging strategy would serve as an important tool to provide a positional map of T modifications, which is required for understanding their biological role. This is particularly useful in mammalian tissue, where 5-fU and 5-fC occur at very similar levels.4 This robust method can therefore be used for the specific enrichment of 5-fU, distinct from 5-fC, to determine its localized position in the genome. In addition, the strategy can also be extended for the mapping of 5-hmU. We demonstrated the clean oxidation of 5-hmU to 5-fU using KRuO4, utilizing conditions originally developed for the oxidation of 5-hmC (Scheme 1).19,20 The completion of oxidation was confirmed by LC–MS analysis of a 5-hmU containing ODN and mononucleoside composition analysis (Supporting Information, Figure S28). The resultant 5-fU can be subsequently tagged by exploiting the same selective chemistry.
Scheme 1. 5-hmU Can Be Chemically Oxidized to 5-fU.
He and co-workers recently reported a method to selectively enrich 5-hmU:G mispairs utilizing a chemoenzymatic glycosylation.21 Our 5-fU tagging strategy following 5-hmU oxidation would be useful to enrich 5-hmU in both matched and mismatched contexts. In particular, this is important for the study of 5-hmU in mammalian cells, where it has been shown that all detectable steady-state 5-hmU exists in a hmU:A base-pair context.4
In conclusion, we demonstrate a method to selectively enrich fragments containing T modifications in DNA by exploiting the chemoselective reactivity of the aldehyde present in 5-fU. Our method will help to further elucidate any potential role, function or consequence of such modified bases in DNA.
Acknowledgments
R.E.H. is supported by The University of Cambridge, F.K. is supported by the Wellcome Trust, and A.B.S. is supported by the Herchel Smith Fund. The Balasubramanian group is core-funded by a Wellcome Trust Senior Investigator Award and by Cancer Research UK. Departmental NMR facilities are supported by EPSRC grant EP/K039520/1. We thank Professor Jonathan Goodman for advice regarding quantum mechanical calculations, and Dr. Pieter Van Delft for assistance with mass spectrometry.
Supporting Information Available
Experimental protocols, the detailed description of the performed calculations, supporting tables, and ODN sequences. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b03730.
The authors declare no competing financial interest.
Supplementary Material
References
- Gommersampt J. H.; Borst P. FASEB J. 1995, 9, 1034. [DOI] [PubMed] [Google Scholar]
- Booth M. J.; Raiber E.-A.; Balasubramanian S. Chem. Rev. 2015, 11562240–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Zhao B. S.; He C. Chem. Rev. 2015, 115, 2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T.; Spada F.; Wagner M.; Brandmayr C.; Laube S. K.; Eisen D.; Truss M.; Steinbacher J.; Hackner B.; Kotljarova O.; Schuermann D.; Michalakis S.; Kosmatchev O.; Schiesser S.; Steigenberger B.; Raddaoui N.; Kashiwazaki G.; Mueller U.; Spruijt C. G.; Vermeulen M.; Leonhardt H.; Schaer P.; Mueller M.; Carell T. Nat. Chem. Biol. 2014, 10, 574. [DOI] [PubMed] [Google Scholar]
- Klungland A.; Paulsen R.; Rolseth V.; Yamada Y.; Ueno Y.; Wiik P.; Matsuda A.; Seeberg E.; Bjelland S. Toxicol. Lett. 2001, 119, 71. [DOI] [PubMed] [Google Scholar]
- Kamiya H.; Murata-Kamiya N.; Karino N.; Ueno Y.; Matsuda A.; Kasai H. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2002, 513, 213. [DOI] [PubMed] [Google Scholar]
- Djuric Z.; Heilbrun L. K.; Lababidi S.; Berzinkas E.; Simon M. S.; Kosir M. A. Cancer Epidemiol., Biomarkers Prev. 2001, 10, 147. [PubMed] [Google Scholar]
- Djuric Z.; Heilbrun L. K.; Simon M. S.; Smith D.; Luongo D. A.; LoRusso P. M.; Martino S. Cancer 1996, 77, 691. [PubMed] [Google Scholar]
- Rogstad D. K.; Heo J.; Vaidehi N.; Goddard W. A.; Burdzy A.; Sowers L. C. Biochemistry 2004, 43, 5688. [DOI] [PubMed] [Google Scholar]
- Rogstad D. K.; Liu P.; Burdzy A.; Lin S. S.; Sowers L. C. Biochemistry 2002, 41, 8093. [DOI] [PubMed] [Google Scholar]
- Raiber E.-A.; Beraldi D.; Ficz G.; Burgess H. E.; Branco M. R.; Murat P.; Oxley D.; Booth M. J.; Reik W.; Balasubramanian S. Genome Biol. 2012, 13, R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.-X.; Szulwach K. E.; Fu Y.; Dai Q.; Yi C.; Li X.; Li Y.; Chen C.-H.; Zhang W.; Jian X.; Wang J.; Zhang L.; Looney T. J.; Zhang B.; Godley L. A.; Hicks L. M.; Lahn B. T.; Jin P.; He C. Nat. Biotechnol. 2011, 29, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A.; Huang Y.; Henderson H. R.; Agarwal S.; Rao A. Nat. Protocols 2012, 7, 1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T.; Hackner B.; Truss M.; Muenzel M.; Mueller M.; Deiml C. A.; Hagemeier C.; Carell T. Angew. Chem., Int. Ed. 2011, 50, 7008. [DOI] [PubMed] [Google Scholar]
- Guo P.; Yan S.; Hu J.; Xing X.; Wang C.; Xu X.; Qiu X.; Ma W.; Lu C.; Weng X.; Zhou X. Org. Lett. 2013, 15, 3266. [DOI] [PubMed] [Google Scholar]
- Crisalli P.; Hernandez A. R.; Kool E. T. Bioconjugate Chem. 2012, 23, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raindlová V.; Pohl R.; Sanda M.; Hocek M. Angew. Chem., Int. Ed. 2010, 49, 1064. [DOI] [PubMed] [Google Scholar]
- Guo P.; Xu X.; Qiu X.; Zhou Y.; Yan S.; Wang C.; Lu C.; Ma W.; Weng X.; Zhang X.; Zhou X. Org. Biomol. Chem. 2013, 11, 1610. [DOI] [PubMed] [Google Scholar]
- Booth M. J.; Branco M. R.; Ficz G.; Oxley D.; Krueger F.; Reik W.; Balasubramanian S. Science 2012, 336, 934. [DOI] [PubMed] [Google Scholar]
- Booth M. J.; Ost T. W. B.; Beraldi D.; Bell N. M.; Branco M. R.; Reik W.; Balasubramanian S. Nat. Protocols 2013, 8, 1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.; Song C.-X.; He C. Methods 2015, 72, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.