Abstract
Here, we report an unconventional Chinese pedigree consisting of three branches all segregating prelingual hearing loss (HL) with unclear inheritance pattern. After identifying the cause of one branch as maternally inherited aminoglycoside-induced HL, targeted next generation sequencing (NGS) was applied to identify the genetic causes for the other two branches. One affected subject from each branch was subject to targeted NGS whose genomic DNA was enriched either by whole-exome capture (Agilent SureSelect All Exon 50 Mb) or by candidate genes capture (Agilent SureSelect custom kit). By NGS analysis, we identified that patients from Branch A were compound heterozygous for p.E1006K and p.D1663V in the CDH23 (DFNB12) gene; and patients from Branch B were homozygous for IVS7-2A>G in the SLC26A4 (DFNB4) gene. Both CDH23 mutations altered conserved calcium binding sites of the extracellular cadherin domains. The co-occurrence of three different genetic causes in this family was exceedingly rare but fully compatible with the mutation spectrum of HL. Our study has also raised several technical and analytical issues when applying the NGS technique to genetic testing.
Introduction
Hearing loss (HL) is the most common sensorineural disorder. Severe–to-profound HL affects one of every 1000 neonates. The prevalence increases to about 0.2% before the age of 5 years when languages have been acquired.1,2 It has been estimated that about two-thirds of these cases have a genetic origin, most of which are monogenic.2 Prelingual HL is typically inherited as an autosomal recessive trait with or without accompanying other syndromic features.3 To date, over 160 loci have been identified for hereditary HL, including more than 90 for autosomal recessive nonsyndromic HL (NSHL) and more than 60 for autosomal dominant NSHL. At least 44 genes for autosomal recessive NSHL and 27 genes for autosomal dominant NSHL have been identified (http://hereditaryhearingloss.org, accessed in October 2013). These genes encode proteins participating in a variety of functions: gap junctions, ion homeostasis, extracellular (EC) matrix, transcription factors, cell adhesion, motor proteins, etc. attesting the complexity of hearing mechanism and the genetic heterogeneity of HL.4
Discovering the causal mutations is crucially important for HL diagnosis, especially for prelingual cases. It allows direct estimates of recurrence risk in relatives and helps family planning. In some circumstances, establishing an early genetic diagnosis can also predict the possible phenotypic outcomes and suggest personalized preventative and therapeutic options. As an example, Usher syndromes, which are characterized by both HL and gradual visual impairment, share the same disease genes as several types of autosomal recessive NSHL but are not readily distinguished from autosomal recessive NSHL in infants and early childhood.5 For newborns diagnosed with Usher syndromes, effective measures can slow down or even prevent the progression into retinitis pigmentosa if implemented in time.6
The extremely high genetic heterogeneity of HL makes genetic testing particularly challenging. The target enrichment (TGE) of exons for specific or all genes related to a disease followed by next generation sequencing (NGS) allows a comprehensive survey of mutations affecting protein-coding genes. A number of recent studies reported the application of NGS targeting either the exons of known HL genes (e.g., references 7, 8, 9, 10, 11) or the whole exome (e.g., Woo et al.12) in molecular diagnosis of HL. Notwithstanding their positive findings, some technical issues like the enrichment performance although thoroughly evaluated for some commercial exome kits13,14 remain under-explored for most custom TGE kits. Analytical issues on distinguishing pathogenic mutations from low frequency polymorphisms also persist and confound the clinical interpretations.
In this study, we report an unconventional Chinese family JX-H016 presenting prelingual HL with unknown inheritance pattern. After identifying the cause of one branch as maternally inherited aminoglycoside-induced HL, targeted NGS was applied to identify the genetic causes for the other two branches. Their genomic DNAs were enriched by a commercial whole-exome kit and a custom designed HL kit (CUHK-HL V1) targeting 252 known and candidate HL genes, respectively. The NGS analysis quickly led to the identification of disease-causing mutations in the SLC26A4 and the CDH23 genes for the two other branches. The significance and implications of the findings from this family was discussed in light of the mutation spectrum of HL. We also compared the performances of two TGE kits and discussed the issues to be considered when designing a custom TGE kit.
Materials and methods
Clinical evaluation
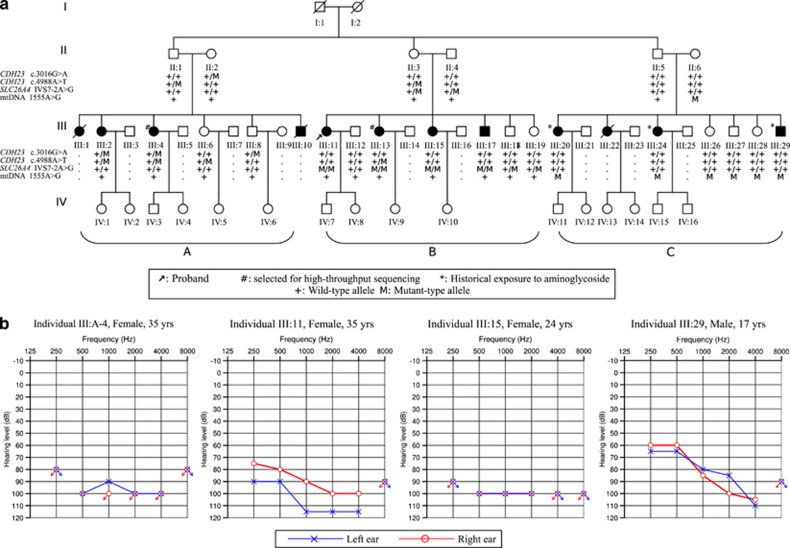
A four-generation Chinese non-consanguineous family, JX-H016, was recruited from an isolated village located in the Jiangxi province in mainland China (Figure 1a). Twenty-two family members including 9 affected subjects and 13 individuals with normal hearing participated in this study. This study was approved by the Ethnic Committee of Chinese PLA General Hospital. Written informed consent was obtained from the adult participants and the guardians on behalf of the children prior to their participation in the study. A medical history was collected using a standard questionnaire, including the age at onset, severity and progression of HL, medication, family history, visual impairment, and other relevant clinical manifestations. All participants underwent audiological examinations including pure-tone audiometry at frequencies 250–8000 Hz, which were found to be consistent with bone conduction values. Immittance testing was applied to evaluate middle-ear pressure, ear canal volumes and tympanic membrane mobility. The degree of HL was evaluated based on the average of audiometric thresholds at 500, 1000 and 2000 Hz.
Figure 1.
The pedigree and typical audiograms of patients. (a) The four-generation pedigree of the Chinese family presenting prelingual HL is comprised of three branches (labeled A~C). The affected individuals could only be found in the third generation. Individuals with available DNAs in the second and the third generation were genotyped for the four pathogenic mutations. Two affected members (III-4 and III-13) from the third generation were selected for sequencing. (b) Typical audiograms of selected patients from each branch are shown. While patients in Branch A and B showed bilateral severe-to-profound hearing loss across all frequencies, patients in Branch C all showed severe hearing losses with down-sloping shaped audiograms (only III-29 is shown).
Designing the custom TGE kit
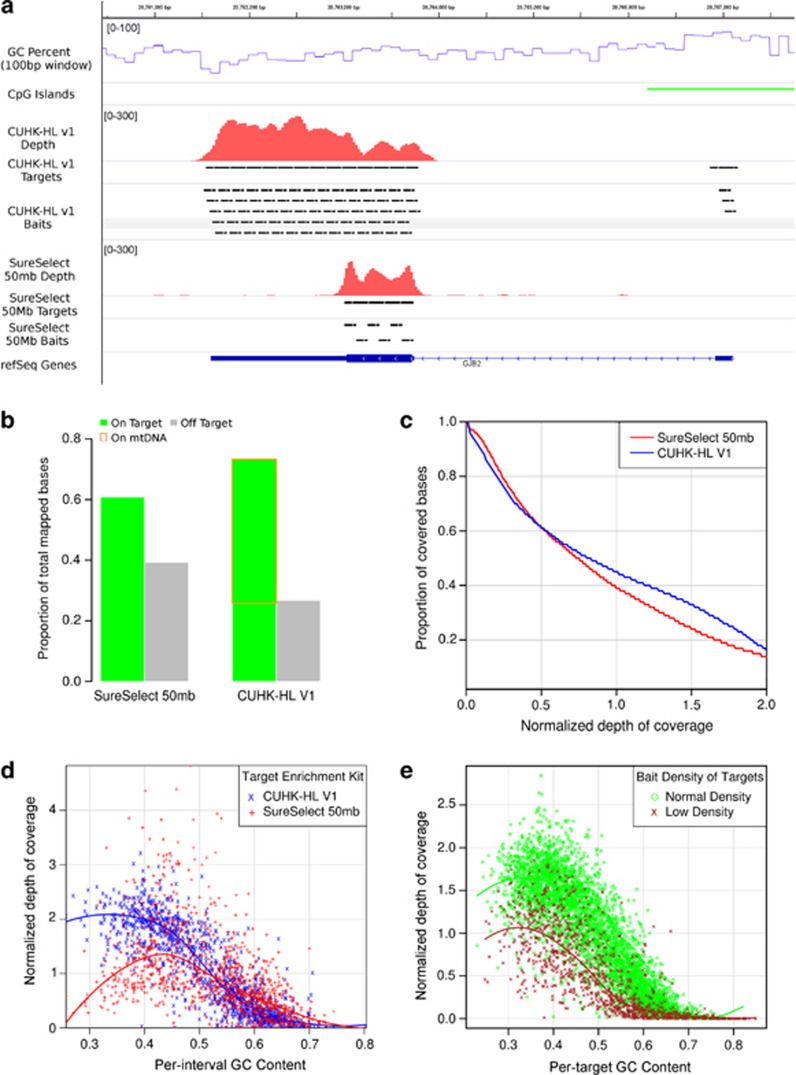
We designed a custom TGE kit (CUHK-HL V1) for the molecular diagnosis of hereditary HL. The kit was designed to target a total of 252 human protein-coding genes related to HL. It included 78 known HL genes (55 nonsyndromic and 23 syndromic HL genes) and 174 candidate HL genes collected based on the functional evidence in knockout mice or from literature survey. The list of 78 known HL genes is given in Supplementary Table S1. We adopted Agilent (Santa Clara, CA, USA) SureSelect TGE technology to manufacture the assay chemistries. As compared with the commercial SureSelect 50 Mb whole-exome kit, it differs in several aspects of the design (summarized in Table 1 and illustrated in Figure 2a). The target regions of the commercial SureSelect All Exon 50 Mb (SureSelect 50 Mb) kit contain all protein-coding exons annotated by the GENCODE project15 as well as 10 bp flanking sequences. In addition, they also include exons of small non-coding RNAs from miRBase and Rfam. In comparison, the CUHK-HL V1 kit was designed to capture only 252 protein-coding genes. All exons including untranslated regions and their 50 base pair (bp) flanking sequences were selected for capture. In addition, the CUHK-HL V1 kit also included the full length mitochondrial DNA (mtDNA) as a single target. We defined the exonic region for a gene as the coding exons plus 10 bp intron–exon boundaries and found that more exonic regions of HL genes are covered by the CUHK-HL V1 kit than SureSelect 50 Mb. About 99.2% of the exonic regions of 55 human NSHL genes are covered by the designed targets of the CUHK-HL V1 kit but only 94.2% by the SureSelect 50 Mb kit. A notable example is the PTPRQ gene, which is virtually not included in the targets of the SureSelect 50Mb kit (Supplementary Table S2). Both kits use 120 bp biotinylated cRNA oligonucleotide baits complementary to the target DNA sequences to hybridize the NGS libraries, but they differ in the bait layouts at targeted regions. While the SureSelect 50 Mb kit contains baits that reside immediately adjacent to each other across the target intervals, the CUHK-HL V1 kit contains densely overlapping baits that cover each target base four times on average (fourfold tiling).
Table 1. Comparing the design differences between the CUHK-HL V1 and the SureSelect 50Mb target enriched kits.
| Target enrichment kit | CUHK-HL V1 | SureSelect 50 Mb |
|---|---|---|
| Target region length (bp) | 2 062 107 | 51 646 629 |
| Number of baits | 52 049 | 556 569 |
| Baits layout | Overlapping baits, fourfold tiling across the target intervals | Intermediately adjacent, head-to-tail anchored baits across the target intervals |
| Targeted gene groups | 78 known HL genes+174 candidates+entire mtDNA | GENCODE coding genes+noncoding RNAs from miRBase and Rfam |
| Targeted regions for each gene | All exons and UTRs+50 bp flanking sequences | All coding exons+10 bp flanking sequences |
| Proportion of the NSHL gene regionsa covered by the designed targets | 0.992 | 0.942 |
Abbreviations: bp, base pair; UTR, un-translated regions.
Gene regions are defined as all coding exons plus 10 bp flanking sequences at intron–exon junctions.
Figure 2.
Comparing the design and performance of the two target enrichment (TGE) kits. (a) The targeted regions, bait layouts, GC percent and depth of coverage at the GJB2 gene locus. The CUHK-HL V1 kit was targeting at both coding sequences and untranslated regions using fourfold tiling baits, whereas the commercial SureSelect 50 Mb kit was designed to capture only the protein-coding part of the gene using baits that were adjacently riveted to each other. The influence of local GC percent on the read depth is more evident with the CUHK-HL V1 kit: the exon 1 of GJB2 co-localizes with a CpG island on which no reads were mapped; across the exon2, the read depth tended to decrease with increasing GC percent. (b) Enrichment efficiency and the mtDNA effect. Enrichment efficiency can be measured by the proportion of total mapped bases that overlap the designed target regions (on-target proportion). Although the CUHK-HL V1 kit showed a higher on-target proportion than the SureSelect 50 Mb kit (~75% vs ~60%), nearly two-thirds of on-target bases were mapped onto mtDNA which is designed as a single target. (c) Comparing the uniformity of read depths across all NSHL genes. To account for the differences in the designed targets of two TGE kits, the comparison is restricted to the genomic intervals that encompass all coding exons plus 10bp intron–exon boundaries of the NSHL genes (exonic intervals) that overlap the target regions in both TGE kits. To account for the differences in the total sequence amounts, the depth per interval is then normalized by dividing the average depth over the exonic intervals under comparison. The cumulative distributions of the normalized depth on the exonic intervals are shown. The curve can be interpreted as the achieved coverage proportions (y axis) at different normalized depths (x axis). For the normalized depth ranging from 0 to 0.5, we found the SureSelect 50Mb kit consistently but slightly outperformed the CUHK-HL V1 kit on the coverage proportions. (d) The effect of GC content on read depths. Similar to (c), we compared the two TGE kits by using normalized depth over all targeted exonic regions of the NSHL genes. The pattern is quantitatively similar when using all targets. For both kits, regions with very high GC contents (>0.7) had very low depths. While the SureSelect 50Mb kit shows a parabolic relationship between read depth and GC content, the depths on the CUHK-V1 kit targets decrease monotonically with GC content. The difference can most likely be explained by the differences in the bait design. (e) The effect of repeat elements on coverage depths. Because the SureSelect TGE technology tends to avoid placing baits over repeat elements, target regions with low bait density should have higher densities of repeat elements. For the CUHK-HL V1 kit, under the fourfold tiling of 120 bp baits, the expected density should be 1/30; low bait density was defined as <1/50 based on the empirical bait density distribution. After accounting for the GC effect, read depths at the targets of low bait density tend to be shallower than targets with normal bait density. The influence of the bait density on target depth can also be observed for the SureSelect 50 Mb kit (see Table 6).
Targeted NGS
We used the SureSelect 50 Mb and the CUHK-HL V1 kits to capture the genomic DNA of one affected member in Branch A (III-4) and one affected member from Branch B (III-13), respectively (Figure 1a). The experimental procedures were similar for the two kits. Genomic DNA was extracted and purified from peripheral blood leucocytes using QIAamp DNA blood kit (Qiagen, Duesseldorf, Germany). The qualified 3 μg genomic DNA for each sample was randomly sheared into 150~250 bp fragments (Covaris, Woburn, MA, USA) and purified using MinElute PCR purification kit (Qiagen). The fragments were end-repaired, adenylated and ligated to adapters at both ends using NEBNext DNA sample preparation kit (New England Biolabs, Ipswich, MA, USA). The adapter-ligated templates were purified by the Agencourt AMPure SPRI XP beads (Beckman Coulter, Brea, CA, USA); and the fragments with insert size about 250 bp were excised. Extracted DNA was PCR amplified, purified and hybridized to the SureSelect biotinylated RNA library for target capture (Agilent). A total of 500 ng purified amplified library was hybridized to the custom-designed biotinylated cRNA probes for 24 h at 65 °C. Hybridized fragments were enriched using streptavidin-coated magnetic Dynabeads (Invitrogen, Carlsbad, CA, USA), whereas non-hybridized fragments were washed out after 24 h. Captured PCR products were subjected to Agilent 2100 Bioanalyzer to evaluate the magnitude of enrichment. The library enriched by the CUHK-HL V1 kit was sequenced on Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) in 90 bp paired-end (PE) reads. The library enriched by the SureSelect 50 Mb kit was sequenced on GAIIx in 100 bp PE reads using three lanes. Raw image files were processed by Illumina CASAVA Software version 1.7 for base-calling with default parameters.
To validate and test the segregation pattern of the prioritized variants, primers were designed to amplify the encompassing genomic region. PCR products were sequenced in both forward and reverse directions on an ABI 3100 using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Carlsbad, CA, USA).
Bioinformatics analysis
Raw sequence reads were aligned to the reference genome (NCBI Build 37) using Burrows-Wheeler Aligner (BWA, v0.5.9). The sequence alignment files were processed by Picard (v1.55, http://picard.sourceforge.net) to mark up duplicated reads and calculate summary statistics. Genome Analysis Tool Kit (GATK, v.1.4–9) was used to perform realignment around indels and base quality recalibration to produce analysis-ready alignments. Single-nucleotide variants (SNVs) and small insertion deletions (indels) were called using GATK's Unified Genotyper module. High-quality variants were obtained by GATK's recommended filtering parameters under single sample calling mode. The depth of coverage on given target regions was calculated using the DepthOfCoverage module of GATK. Only high-quality bases (Q>=20) on non-duplicated reads with high mapping quality (MAPQ>=17) were included in evaluating the depth of coverage.
To prioritize the disease-causing variants, we excluded the variants whose allele frequencies are greater than 0.01 in both the public databases and the in-house exome database consisting 170 unrelated samples. The threshold of 0.01 was chosen based on the currently known most common HL-causing mutations in China.16 The functional effects were then annotated by ANNOVAR based on the refSeq Gene model. The evolutionary conservation for at each variant position was measured by Genome Evolutionary Rare Profiling (GERP) and PhyloP. We then focused on the functionally interpretable variants that included: SNVs that were evolutionarily conserved (GERP>2.0) and caused missense or nonsense changes; or SNVs that were located within 2 bp of intron–exon boundaries; and indels that caused in-frame or frameshift alternations. To aid the interpretation of missense SNVs, their pathogenic effect predictions were queried from dbNSFP.17
Results
Pedigree description
The pedigree of family JX-H016, which spanned four generations and comprised 53 members, consisted of three branches (A~C) all segregating prelingual HL. Affected subjects with hearing impairment were only found in the third generation (Figure 1a). The inheritance pattern of this family was unclear. However, when we checked each branch, it was consistent with the autosomal recessive mode. The patients from Branch A and B showed prelingual severe–to-profound HL at all frequencies, whereas the patients from Branch C showed prelingual severe HL with down-sloping audiograms (Figure 1b and Table 2). The severity of hearing impairment did not progress with increasing patient age. Tinnitus and vertigo were not reported by this family. Audiologic evaluation demonstrates normal immittance testing and sensorineural hearing impairment. On the basis of the questionnaires, three affected subjects from Branch C, III-20, III-24 and III-29, had historical exposures to gentamicin or streptomycin (dose uncertain) at the age of 0–2 years. Comprehensive family medical histories and clinical examination of these individuals showed no other clinical abnormalities, including diabetes, cardiovascular diseases, visual problems and neurological disorders. The detailed clinical data of affected subjects of family JX-H016 were summarized in Table 2.
Table 2. Summary of clinical data of affected individuals of family JX-H016.
| Patient | Gender |
Age (years) |
Use of aminoglycoside |
Hearing test PTAa
(dB) |
Audiogram shape | Tinnitus | Vertigo | Noise exposure | ||
|---|---|---|---|---|---|---|---|---|---|---|
| At testing | At onset | Left ear | Right ear | |||||||
| III-1 | Female | — | Prelingual | — | — | — | — | — | — | — |
| III-2 | Female | 36 | Prelingual | No | — | — | — | — | — | — |
| III-4 | Female | 35 | Prelingual | No | 97 | 100 | Flat | No | No | No |
| III-10 | Male | — | Prelingual | — | — | — | — | — | — | — |
| III-11 | Female | 35 | Prelingual | No | 100 | 90 | Sloping | No | No | No |
| III-13 | Female | 27 | Prelingual | No | 98 | 98 | Flat | No | No | No |
| III-15 | Female | 24 | Prelingual | No | 100 | 100 | Flat | No | No | No |
| III-17 | Male | 29 | Prelingual | No | 88 | 90 | Flat | No | No | No |
| III-20 | Female | — | Prelingual | Yes | — | — | — | — | — | — |
| III-22 | Female | — | Prelingual | Yes | — | — | — | — | — | — |
| III-24 | Female | 28 | Prelingual | Yes | 82 | 83 | Sloping | No | No | No |
| III-29 | Male | 17 | Prelingual | Yes | 77 | 82 | Sloping | No | No | No |
PTA, pure-tone average.
Identification of mitochondrial 12S rRNA A1555G mutation in family JX-H016
Because of the aminoglycoside exposures of some patients in this family, we firstly conducted the Sanger sequence to detect the mitochondrial 12S rRNA A1555G mutation in all patients of this family. The homoplasmic A1555G mutation was carried by II-6 and all her offsprings, and presumably also carried by the female offsprings in the fourth generation of Branch C. The 12S rRNA A1555G mutation was not carried by any patient from the other two branches. Other mutations in mitochondrial 12S rRNA were also excluded.
Identification of CDH23 and SCL26A4 mutations in family JX-H016 by NGS with two TGE kits
After excluding the mutations in mitochondrial 12S rRNA and GJB2 gene by Sanger sequence, we elected to use the targeted NGS to resolve the genetic causes of Branch A and B. Affected subject III-4 from Branch A and III-13 from Branch B were selected for sequencing. The genomic library of III-4 was enriched by the SureSelect 50 Mb kit; and 19.58 giga base pairs raw sequences were generated using 100 bp PE reads. BWA mapped 95.8% of those reads to the reference genome; and 30.7% of them were marked as duplicates. Not accounting for the duplicated reads and reads with low mapping quality (MAPQ<17), the mean read depth on targets is 131.2 × . The genomic library of III-13 was enriched by the CUHK-HL V1 kit; and a total of 836 Mb raw sequences were generated using 90 bp PE reads. BWA mapped 98.7% of the reads to the genome, of which 15.8% were marked as duplicates. The mean target depth achieved for III-13 is 213.6 × . More than 38 000 and 1700 high-quality variants were called for III-4 and III-13, respectively. After a series of filtering, both patients carried four rare mutations disrupting known NSHL genes (Table 3 and Supplementary Figure S1). In affected subject III-13, we identified the homozygous splicing mutation c.919-2A>G, also known as IVS7-2A>G, of the SLC26A4 (DFNB4) gene. The mutation abolished the splice acceptor of exon 8 and was predicted to skip the entire exon 818 resulting in a truncated protein product. Sanger sequencing confirmed the co-segregation of the homozygote mutation with HL in Branch B (Figure 1a). This mutation did not segregate into the other two branches. In affected subject III-4, we found two heterozygous missense mutations c.3016G>A and c.4988A>T in the CDH23 (DFNB12) gene that resulted in amino acid substitutions p.E1006K and p.D1663V. Sanger sequencing confirmed that each parent contributes one heterozygous allele, and only patients in Branch A carried compound heterozygotes of CDH23 mutations (Figure 1a). Both variants were highly conserved in vertebrates and absent from both public and in-house databases. All rare variants that disrupt known NSHL genes discovered from two sequenced patients are summarized in Table 4.
Table 3. The number of high-quality variants after each step of filtering.
|
III-13 |
III-4 |
|||
|---|---|---|---|---|
| SNVs | Indels | SNV | Indels | |
| All high-quality variants | 1492 | 220 | 35 728 | 2416 |
| After filtering against public databasesa | 50 | 131 | 1172 | 1129 |
| After in-house filtering against in-house exome databaseb | 36 | 22 | 719 | 58 |
| After functional effects filteringc | 11 | 1 | 196 | 14 |
| Variants disrupting known nonsyndromic hearing loss genes | 4 | 0 | 4 | 0 |
| Genes harboring homozygous variants or >=2 heterozygous variants | 1 | 2 | ||
| Recessive hearing loss genes harboring homozygous variants or >=2 heterozygotes | 1 | 1 | ||
Abbreviations: indels, insertions-deletions; SNV, single-nucleotide variants.
Excluding variants having alternative allele frequencies >0.01 in any one of the populations in dbSNP and 1000 genomes.
Excluding variants having alternative allele frequencies >0.01 in 170 other unrelated in-house exomes.
Keeping SNVs that are evolutionarily conserved and cause missense, nonsense changes or potentially disrupt splice sites; and keeping indels that result in inframe or frameshift alternations.
Table 4. All rare variants that disrupt known nonsyndromic hearing loss genes discovered from two sequenced patients.
| Patients | Gene | Variants (dbSNP ID) | Zygosity | Allele frequencya | cDNA change | Protein change | Grantham score | Non-synonymous SNV predictionsb | PhyloPc | GERPc |
|---|---|---|---|---|---|---|---|---|---|---|
| III-4 | GPSM2 | 1:109472416 C>T (rs189033496) | Het | 0.0080 | NM_013296:exon15: c.1909C>T | p.R637W | 101 | +++ + | 3.05 | 5.05 |
| III-4 | MYO6 | 6:76618264 T>C (rs144816202) | Het | 0 | NM_004999:exon32: c.3332T>C | p.V1111A | 64 | +++ + | 4.87 | 6.07 |
| III-4 | CDH23 | 10:73466716 G>A | Het | 0 | NM_022124:exon25: c.3016G>A | p.E1006K | 56 | ? +?? | 5.98 | 5.28 |
| III-4, III-13 | CDH23 | 10:73537579 A>T | Het | 0 | NM_022124:exon37: c.4988A>T | p.D1663V | 152 | ? +?? | 5.12 | 2.18 |
| III-13 | OTOF | 2:26699868 T>A | Het | 0.0011 | NM_004802:exon5: c.326A>T | p.N109I | 149 | +++ + | 3.16 | 3.76 |
| III-13 | SLC26A4 | 7:107323898 A>G (rs111033313) | Hom | 0.0080 | NM_000441:exon8: c.919-2A>G | p.T307Vfs*5d | n.a. | 4.66 | 5.62 | |
| III-13 | MARVELD2 | 5:68715804 G>A | Het | 0.0023 | NM_001038603:exon2: c.592G>A | p.V198M | 21 | ++− − | 2.28 | 5.16 |
Allele frequencies were calculated from the in-house exome database of 440 unrelated samples (Feb 2013; X. Zhou unpublished data).
Results from four non-synonymous SNV effects prediction programs, from left to right: PolyPhen2, SIFT, LRT, MutationTaster. ‘+', deleterious or damaging; ‘−', benign; ‘?', not avaiable.
Measures of evolutionary constraint. PhyloP score is the −log10 of P-value for testing the null hypothesis of neutral evolution, based on 46-way whole-genome alignment of vertebrates. GERP score can be interpreted as the substitutions expected under neutrality minus the number of substitutions observed at the position, which was based on 35-way whole-genome alignment of mammals and had theoretical maximum value of 6.18.
This variant was predicted to abolish splice donor and cause exon 8 skipping.
Comparison of the performance of two TGE kits
To investigate the performance of the SureSelect 50 Mb kit and the CUHK-HL V1 kit, we compared the target coverage of two samples. Of the target bases of III-13 (CUHK-HL V1 kit), 92.8% were covered at least once, 83.3% were covered at >=10 × and 76.5% were at >=20 × . For III-4 (SureSelect 50 Mb kit), at least 96.3% of the target bases were covered at least once, 89.9% were covered at >=10 × and 81.0% at >=20 × (Table 5). The coverage over the exonic regions of the 55 NSHL genes was slightly higher than the overall targets for III-4, but similar or slightly lower than the overall targets for III-13 (Table 5).
Table 5. The summary statistics for two sequenced affected subjects.
| Parameters | III-13 | III-4 |
|---|---|---|
| Target enrichment kit | CUHK-HL V1 | SureSelect 50Mb |
| Total yield of raw sequence reads (Gbp) | 0.837 | 19.58 |
| Percent of aligned reads | 98.7% | 96.3% |
| Percent of duplicated reads | 15.8% | 30.7% |
| Mean depth of coverage on the targeted regionsa | 95.6 × | 110.86 × |
| Mean depth of coverage on the mtDNA | 14 479.2 | 131.3 |
| Percent of target bases covered at >=1 × | 92.8% | 96.3% |
| Percent of target bases covered at >=10 × | 83.1% | 89.9% |
| Percent of target bases covered at >= 20 × | 76.3% | 81.0% |
| Percent of target bases covered at >= 30 × | 71.0% | 72.5% |
| Mean depth of coverage on the NSHL gene regions | 83.5 × | 119.9 × |
| Percent of the NSHL gene regions covered at >=10 × | 85.3% | 90.6% |
| Percent of the NSHL gene regions covered at >=20 × | 74.9% | 83.6% |
| Percent of the NSHL gene regions covered at >=30 × | 67.3% | 76.2% |
Abbreviations: Gbp, giga base pair.
We did not include the mtDNA in calculating the average depth. If the mtDNA target of the CUHK-HL V1 kit were included, the mean target depth for B-3 would be 213.6 × ; and the coverage proportions at 10 × , 20 × and 30 × would be slightly increased to 83.3%, 76.5% and 71.3%, respectively.
To evaluate the enrichment efficiencies of the SureSelect 50 Mb kit and CUHK-HL V1 kit, we compared the proportion of on-target bases. We found that although a larger proportion of mapped bases captured by the CUHK-HL V1 kit were mapped onto the designed target regions (73.2% by CUHK-HL V1 vs 60.7% by SureSelect 50 Mb), the mtDNA target alone subsumed 64.7% of the on-target bases or 47.7% of the total mapped bases (Figure 2b). It made the mtDNA of sample III-13 extremely deeply covered (14 497.2 × ). Although the mtDNA is not targeted by the SureSelect 50 Mb kit, we can still observe a mean depth of 131.3 × on mtDNA. After excluding the mtDNA targets, the differences in the coverage at normalized depths were also reduced (Figure 2c).
To investigate the influence of genomic features on per-target depth, we performed multiple linear regression analysis of the normalized per-target depth, GC content and bait density. The two kits showed different normalized depths for low GC targets (0.3~0.4 GC content). Although depths of those targets in the CUHK-HL V1 kit were typically higher than the mean coverage, the depths of similar targets in the SureSelect 50 Mb kit tended to be lower than the mean (Figure 2d). Consistently, we found GC squared was a significant predictor for the target depth of the SureSelect 50 Mb kit but not for the CUHK-HL V1 kit (Table 6). Target regions with low bait densities tended to have shallower depths after accounting for the GC effect (Figure 2e).
Table 6. Evaluating the influence of the genomic features on the per-target depth.
| Target enrichment kit |
CUHK-HL V1 |
SureSelect 50 Mb |
||
|---|---|---|---|---|
| Dependent variables | β | P-value | β | P-value |
| GC percent | −0.514 | <2e-16 | −0.245 | <2e-16 |
| Squared GC percent | 0.00238 | 0.564 | −0.218 | <2e-16 |
| Bait densitya | 17.77 | <2e-16 | 15.05 | <2e-16 |
Bait density reflects the density of repeat element; target regions rich in repeat element tend to have fewer designed baits.
Discussion
In the present study, we have identified three different genetic defects in an unconventional Chinese family segregating prelingual HL with unclear inheritance pattern. Given a high heterogeneity and its allelic spectrum, the co-occurrence of three different genetic causes in our pedigree is a very rare occasion but not unexpected. The heterogeneity within a single family was reported in a number of other HL pedigrees (summarized in Supplementary Table S3). All reported pedigrees were resolved by using candidate gene sequencing, haplotype analysis and sometimes aided by the audio profiles. In each pedigree, at least one population-specific recurrent mutation was involved, similar to the observation made in our pedigree.
The genetic causes in Branch B and C (SLC26A4: c.919-2A>G, mtDNA: A1555G) represent the most common HL-causing mutations in China,19 with an allele frequency of 0.008 in our in-house database; whereas both of the CDH23 mutations in Branch A are private. The mtDNA A1555G mutation was present in matrilineal relatives of Branch C in this Chinese family, consistent with the clinical findings that the affected subjects in Branch C (III-20, III-24 and III-29) had historical exposures to gentamicin or streptomycin at the age of 0–2 years. The SLC26A4 gene encodes pendrin, which is a sodium-independent chloride/iodide transporter. Mutations in this gene are responsible for HL associated with Pendred syndrome or enlarged vestibular aqueduct. The proband (III-11) was examined by temporal bone computed tomography scan and revealed enlarged vestibular aqueduct. She also underwent standard endocrinology examination and found to have normal thyroid hormone. None of the other patients in Branch B had self-reported goiter either, consistent with the rarity of Pendred syndrome among Chinese patients.19 The CDH23 gene encodes a calcium-dependent cell-adhesion protein (cadherin) with 27 EC cadherin domains. Each EC domain contains cadherin-specific motifs XEX, DXD, LDRE, XDX and DXNDN required for cadherin dimerization and Ca2+ binding.20 Mutations in CDH23 gene cause both USH1D and DFNB12. The p.E1006K mutation was reported before,21 whereas the p.D1663V mutation was novel. Both mutations changed the residue at the Ca2+-binding sites. The p.E1006K mutation substituted the negatively charged glutamic acid (E) of the XEX motif at EC10 domain to a negatively charged lysine residue. The p.D1663V mutation substituted the second aspartic acid (D) of the DXD motif at EC16 domain to a hydrophobic valine residue. It is known that homozygous nonsense, frameshift, splice-site and some missense mutations cause USH1D; and DFNB12 is caused exclusively by the missense mutations that are presumed to retain some residual function for retinal and vestibule but not for cochlear. However, the functional effects of novel missense mutations cannot be easily determined. The conserved motifs within EC domains might facilitate the interpretation for a subset of the missense mutations. Previously, Austo et al.22 noted that most missense mutations in the Ca2+-binding motifs were only observed in DFNB12 patients, which led to the suggestion that the impairment of Ca2+ binding may not diminish cadherin's function in retina. However, many of those patients were compound heterozygous for two disease-causing alleles, which confound the interpretation of the phenotypic consequence of each allele. Later, Schultz et al.21 demonstrated that for patients carrying compound heterozygous mutations, USH1D occurs only when two USH1D alleles were in trans; in contrast, when there are both DFNB12 alleles or one DFNB12 and one USH1D allele in trans, the resulting phenotype is DFNB12. The p.E1006K mutation was reported by Schultz et al.21 as the USH1D allele. Therefore, we can predict that the p.D1663V mutation should be DFNB12 allele, and masked the effect of the USH1D allele p.E1006K carried by the Branch A patients. Further establishing the genotype–phenotype correlations of the CDH23 missense mutations can improve the early molecular diagnosis of USH1D patients.
We applied and compared two targeted NGS approaches in this study. The advantage of using custom TGE kit over off-the-shelf commercial exome kits for molecular diagnosis have been discussed previously,11,23 including cost saving in sequencing, deeper coverage in candidate genes, shorter turnaround time, easier data management, etc. There was a consensus that before the clinical application of a custom TGE kit, its performance should be extensively evaluated and validated. In this regard, we noted that previous studies mainly focused on evaluating the accuracy of variant calls and genotype concordance,10,11,24 although the general problem of variant calling and quality control was already well solved (e.g., DePristo et al.25). Therefore, we focused in this study on the comparative evaluation of the coverage depth, which is the major determinant of the power for variant discovery.
In targeted resequencing projects, the sequencing is commonly considered as completed if 80% of the target regions are covered by >=20 × . For the application in molecular diagnosis, the coverage requirement is higher. For the sequenced samples, we typically observed that the achieved coverage at given depth would reach a plateau at the increase of raw sequences. This efficiency trend was known to be influenced by a number of factors including the library complexity, kit performance and experimental conditions. Per-target depths are known to be highly correlated among different samples enriched and sequenced using the same platform (e.g., Plagnol et al.26). The two TGE kits used in this study were based on the same technology and had almost the same experimental procedures, so the experimental differences should be controlled to the minimal. Although different sequencing protocols were used (90 bp PE for III-13, 100 bp PE for III-4), the results were quantitatively similar after we redid the analyses using 90 bp PE reads (by trimming out 10 bp at the 3' end) of III-4. Therefore, we believe the differences observed on the two samples mainly reflect the differences in the kit performances.
Among samples enriched by the CUHK-HL V1 kit within the same batch, the proportion of bases mapped onto mtDNA varies from 30 to 80%, which is highly correlated with the proportion of total on-target bases and the uniformity over all target regions (data not shown). Although it is intuitive that higher on-target proportions for the samples enriched by the CUHK-HL V1 kit can result from its higher bait density, it can also be influenced by the effect of the mtDNA target.
Our evaluation suggests rooms for improvement for our custom TGE kit, and also illustrates several issues that need to be considered in the kit design. First, the inclusion of the entire mtDNA as a separate target should be treated with caution. Although the deep coverage on mtDNA can have the benefit for detecting structural variants and low-level heteroplasmy (as demonstrated by Calvo et al.27 in diagnosing mitochondrial disorders), it also incurred a great loss in enrichment efficiency. For the genetic diagnosis of HL, which has a very limited mutation spectrum at mtDNA,28 a dedicated target of entire mtDNA may not be necessary. Some recent studies even demonstrated that the mtDNA mutations could well be discovered in exome sequencing in which mtDNA was not specifically targeted (e.g., Dinwiddie et al.29). Second, the calibration and optimization for the TGE kit is a complicated issue. It was shown previously that by using overlapping baits, the NimbleGen SeqCapEZ whole-exome kit showed the highest on-target proportion and most uniform target depths.13 Here, we applied a similar design philosophy to our custom kit using the Agilent's SureSelect technology. Although we found an improvement over the low GC targets (Figure 2d), the overall target uniformity after excluding mtDNA did not improve over the commercial products. Nevertheless, the observed difference is small and already offset by the reduced sequencing amount. We also found that the use of overlapping baits may have a benefit to reduce the reference bias for heterozygous SNVs, because the allele balances of heterozygous SNVs called from the samples enriched by the custom kit were closer to 0.5 and showed less variability than other whole-exome kits based on the same technology (Supplementary Figure S2). Finally, all of the TGE methods based on hybridization suffer from the bias caused by GC content and repeat elements. To fill in those coverage gaps, alternative approaches like PCR-based TGE technology should be considered,30 although they suffer from other problems like variable depths across samples, allele dropouts, etc.
Taken together, three different genetic causes of prelingual HL were identified in this family. The apparently recessive HL in Branch C was indeed caused by the maternally inherited mtDNA A1555G mutation with variable penetrance (induced by ototoxic drugs). The Branch A was diagnosed as DFNB12 caused by the compound heterozygote for two missense mutations; and Branch B was diagnosed as DFNB4 with enlarged vestibular aqueduct because of a homozygous splice-site mutation. We have also evaluated two targeted NGS approaches. Our experiences not only demonstrated the effectiveness of NGS approach in molecular diagnosis, but also underscored the ongoing challenges in the issues like designing the custom enrichment kit, evaluating the pathogenicity of variants and predicting phenotype outcomes from genotypes.
Acknowledgments
We sincerely thank all the family members for their participation and support in this study. These investigations were supported by Key Project of National Natural Science Foundation of China (81030017), National Science Fund for Distinguished Young Scholars (81125008) to HJ Yuan, the National Basic Research Programs (No. 2013CB945402 to DY Han and HJ Yuan; No. 2012CB316504 and 2012CB316503 to XG Zhang and MQ Zhang), and Hong Kong RGC General Research Fund (463811) to KW Choy.
Footnotes
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary Material
References
- Morton N. E. Genetic epidemiology of hearing impairment. Ann. N. Y. Acad. Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- Morton C. C, Nance W. E. Newborn hearing screening—a silent revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- Smith R. J, Bale J. F, Jr., White K. R. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- Petit C. From deafness genes to hearing mechanisms: harmony and counterpoint. Trends Mol. Med. 2006;12:57–64. doi: 10.1016/j.molmed.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Petit C. Usher syndrome: from genetics to pathogenesis. Annu. Rev. Genomics Hum. Genet. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- Musarella M. A, Macdonald I. M. Current concepts in the treatment of retinitis pigmentosa. J. Ophthalmol. 2011;2011:753547. doi: 10.1155/2011/753547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer A. E, DeLuca A. P, Hildebrand M. S, Taylor K. R, Gurrola J, 2nd, Scherer S, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc. Natl Acad. Sci. USA. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman L. M, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Qian D, Ahmad S, Mattox D, Todd N. W, Han H, et al. A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach. Genet. Test. Mol. Biomarkers. 2012;16:536–542. doi: 10.1089/gtmb.2011.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Sommen M, Corneveaux J. J, Reiman R. A, Hackett N. J, Claes C, et al. A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am. J. Med. Genet. A. 2013;161A:145–152. doi: 10.1002/ajmg.a.35737. [DOI] [PubMed] [Google Scholar]
- Shearer A. E, Black-Ziegelbein E. A, Hildebrand M. S, Eppsteiner R. W, Ravi H, Joshi S, et al. Advancing genetic testing for deafness with genomic technology. J. Med. Genet. 2013;50:627–634. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. M, Park H. J, Baek J. I, Park M. H, Kim U. K, Sagong B, et al. Whole-exome sequencing identifies MYO15A mutations as a cause of autosomal recessive nonsyndromic hearing loss in Korean families. BMC Med. Genet. 2013;14:72. doi: 10.1186/1471-2350-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. J, Chen R, Lam H. Y, Karczewski K. J, Euskirchen G, Butte A. J, et al. Performance comparison of exome DNA sequencing technologies. Nat. Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulonen A. M, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S, et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey A. J, Kokocinski F, Calafato M. S, Scott C. E, Palta P, Drury E, et al. The GENCODE exome: sequencing the complete human exome. Eur. J. Hum. Genet. 2011;19:827–831. doi: 10.1038/ejhg.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin A, Liu C, Zhang Y, Wu J, Mai M, Ding H, et al. The carrier rate and mutation spectrum of genes associated with hearing loss in South China hearing female population of childbearing age. BMC Med. Genet. 2013;14:57. doi: 10.1186/1471-2350-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucke P. J, Van Hauwe P, Everett L. A, Demirhan O, Kabakkaya Y, Dietrich N. L, et al. Identification of two different mutations in the PDS gene in an inbred family with Pendred syndrome. J. Med. Genet. 1999;36:475–477. [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, You Y, Huang D, Cui J, Wang Y, Wang Q, et al. Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. J. Transl. Med. 2009;7:79. doi: 10.1186/1479-5876-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Weihofen W. A, Gaudet R, Corey D. P. Structural determinants of cadherin-23 function in hearing and deafness. Neuron. 2010;66:85–100. doi: 10.1016/j.neuron.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. M, Bhatti R, Madeo A. C, Turriff A, Muskett J. A, Zalewski C. K, et al. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J. Med. Genet. 2011;48:767–775. doi: 10.1136/jmedgenet-2011-100262. [DOI] [PubMed] [Google Scholar]
- Astuto L. M, Bork J. M, Weston M. D, Askew J. W, Fields R. R, Orten D. J, et al. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002;71:262–275. doi: 10.1086/341558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Tang W, Ahmad S, Lu J, Colby C. C, Zhu J, et al. Applications of targeted gene capture and next-generation sequencing technologies in studies of human deafness and other genetic disabilities. Hear. Res. 2012;288:67–76. doi: 10.1016/j.heares.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran T. A, Husami A, Kissell D, Zhang W, Keddache M, Black A. P, et al. Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss. Otolaryngol. Head Neck Surg. 2013;148:1007–1016. doi: 10.1177/0194599813482294. [DOI] [PubMed] [Google Scholar]
- DePristo M. A, Banks E, Poplin R, Garimella K. V, Maguire J. R, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Curtis J, Epstein M, Mok K. Y, Stebbings E, Grigoriadou S, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S. E, Compton A. G, Hershman S. G, Lim S. C, Lieber D. S, Tucker E. J, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G, Smith R. J. Maternally inherited hearing impairment. Clin Genet. 2000;57:409–414. doi: 10.1034/j.1399-0004.2000.570601.x. [DOI] [PubMed] [Google Scholar]
- Dinwiddie D. L, Smith L. D, Miller N. A, Atherton A. M, Farrow E. G, Strenk M. E, et al. Diagnosis of mitochondrial disorders by concomitant next-generation sequencing of the exome and mitochondrial genome. Genomics. 2013;102:148–156. doi: 10.1016/j.ygeno.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Warner J. B, Nakano M, Libby B, Medkova M, David P. H, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.