Hemophilia A (HA) and B (HB) are estimated to affect 1 in 5,000 male births in the United States each year.[1] Inheritance of mutations in the Factor VIII (F8) gene or Factor IX (F9) gene causes these bleeding disorders. Identification of mutations causing a patient’s hemophilia can lead to better understanding of risk of complications [2], as well as aid in carrier detection in family members [3]. Mutation screening for HA has involved testing for inversions of introns 1 and 22 of F8, as approximately 45% of severe HA patients carry an inversion as their causative mutation [2], and sequencing of the coding regions of F8 to identify point mutations, deletions, or splice-site mutations. Similarly, mutation screening for HB has involved sequencing of the coding regions of F9. However, a subset of patients presenting with hemophilia do not to have a detectable mutation with these methods.[4] Duplication of part of F8 or F9, for example, may not be detected. Also, female family members heterozygous for a large F8 or F9 deletion may not be identified as carriers using these methods, as dosage of the genes is not determined. Recently, Multiplex Ligation-Dependent Probe Amplification (MLPA®, MRC Holland, Amsterdam, Netherlands) has been successfully used to identify large deletions and duplications within F8 and F9.[5-7] This assay quantitatively compares copy numbers of a set of DNA sequences in a patient sample to those in a control sample to screen for the presence of deletions or duplications.[8] The assessment of how this or similar technologies will fit into currently-used mutation screening protocols should be critically evaluated.
We describe here our experience using MLPA® in combination with inversion testing and DNA sequencing for identification of mutations in a large series of hemophilia patients. Additionally, we present our analyses of how this duplication/deletion testing fits into mutation screening algorithms and highlight the importance of careful assay validation and interpretation.
A subset of patients enrolled in the Hemophilia Inhibitor Research Study were studied for this report.[2] At the time of this study, mutation analysis had been completed on 930 HA patients enrolled in the Hemophilia Inhibitor Research Study (250 more patients than in the previous report) and 152 HB patients. Patients with no mutation detected by inversion testing or sequencing and patients with large deletions detected by sequencing were screened with MLPA® SALSA Kits P178 for F8 and P207 for F9 (MRC Holland, Amsterdam, The Netherlands) following the manufacturer’s protocol. MLPA® fragments were separated and sized using the 3730 DNA Analyzer (Applied Biosystems, Carlsbad, California, USA) and analyzed using the GeneMapper® 4.0 (Applied Biosystems) and Coffalyzer® (MRC Holland) software packages. Large duplications were confirmed with TaqMan® CNV assays (Applied Biosystems) custom-designed to assay the region of the duplication following the manufacturer’s protocol.
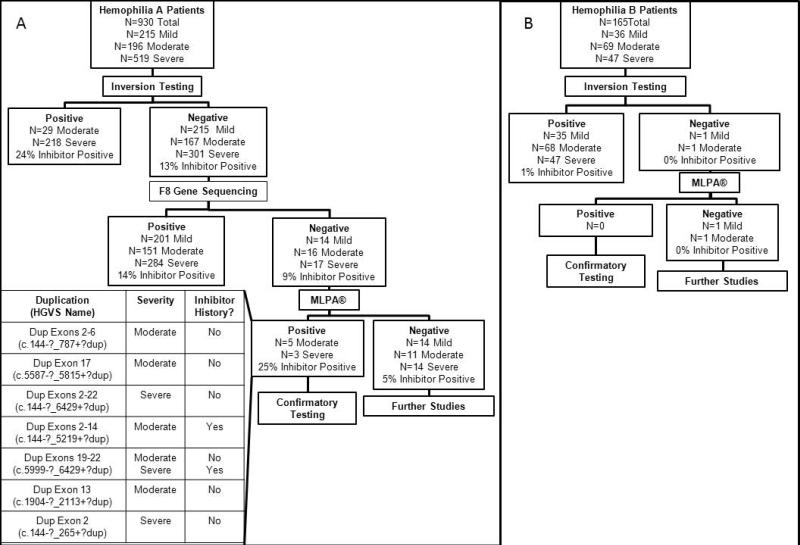
Of the 930 HA patients included in the study, 47 (5%) patients who did not have a mutation detected by inversion testing or F8 sequencing and 28 (3%) HA patients with large deletions within F8 were screened. Large duplications were identified in 8 (17%) patients with no mutation previously detected and were confirmed with TaqMan® CNV assays. All large deletions initially identified by sequencing were confirmed by MLPA®. Figure 1A outlines the algorithm used to identify mutations in this patient population. MLPA® allowed the identification of 8 more mutations in this population, resulting in 96% of patients having an identified mutation.
Fig 1.
Mutation screening algorithms incorporating MLPA® a: Mutation screening algorithm for HA patients enrolled in the Hemophilia Inhibitor Research Study b: Mutation screening algorithm for HB patients enrolled in the Hemophilia Inhibitor Research Study
Of the 152 HB patients included in the study, 2 (1%) patients with no mutation detected by F9 sequencing and 6 (4%) patients with large deletions were screened with MLPA®. No large duplications were identified. All large deletions were confirmed by MLPA®. Figure 1B outlines the algorithm used to identify mutations in this patient population. MLPA® did not allow the identification of any more mutations in this population. However, it did allow the confirmation of all of the large deletions detected by sequencing.
In the Hemophilia Inhibitor Research Study 5% of severe HA patients and 13% of severe HB patients carry a large deletion or duplication.[2] Similar distributions have been seen in other population-based studies.[9-11] Inhibitors are estimated to occur in approximately 50% of HA and HB patients with large deletions or duplications.[2, 12] Our experience screening F8 and F9 with MLPA® resulted in the successful confirmation of all 28 deletions in F8 and all 6 deletions in F9 identified by DNA sequencing. Furthermore, 8 duplications were identified in 47 HA patients with no previous mutation identified.
In order to most efficiently identify mutations and classify those at-risk for developing inhibitors as this technology enters the clinical setting, we propose the screening algorithms outlined in Figure 1. Because approximately 40% of HA patients carry an inversion and because inversion carriers are at moderate risk for inhibitor development [12], we propose initially screening for F8 inversions in patients with HA. If no inversion is identified, F8 sequencing would be subsequently conducted. It is estimated this would identify over 95% of mutations leading to HA.[2, 4] If no mutation is identified through sequencing, MLPA® should then be conducted to identify possible large duplications. Similarly, we propose initially sequencing F9, followed by MLPA® if no mutation is identified.
In carrier screening for either HA or HB, the algorithm would be altered depending on the mutation identified in the index case. For example, if a point mutation or small insertion/deletion were identified in the index case, only the amplicon for the gene region surrounding the mutation would need to be sequenced to determine carrier status. Because large deletions and duplications in carriers would result in an altered copy number in the region of the deletion or duplication, we propose using MLPA® to identify carriers of these mutations, as sequencing would not be able to detect these copy number changes.
The proper use of MLPA® or similar technologies to screen for genomic deletions or duplications in hemophilia requires careful attention to several key aspects: choice of control samples; interpretation of results; and choice of DNA extraction methods. Because F8 and F9 are X-linked genes, it is necessary that reference samples be obtained from subjects of the same sex as the tested patient in order to correctly measure the ratio of copy numbers in the samples. It is also important to consider the assay-specific control probe composition when using male reference samples to assay female carriers or, alternatively, using female reference samples to assay male patients. For example, the F8 MLPA® control probe sequences are in X-linked genomic regions, while those in the F9 kit are autosomal. Also, the assay is sensitive to sample impurities and inconsistencies may be introduced by using multiple DNA extraction methods. Finally, due to the complicated nature of quantitative assays to determine copy number variation, assay validation is essential. Initial results must be confirmed by repeated testing with an independent method, such as quantitative PCR.
MLPA® kits surveying the F8 and F9 gene are useful for identifying deletions and duplications causing HA and HB. The kits help to identify previously-undetectable mutations and provide a method for rapid identification of patients most at-risk for complications of inhibitors. However, appropriate caution must be taken to validate results and ensure correct interpretation.
Acknowledgments
This work is supported by the CDC Foundation through grants from Pfizer Inc and Baxter Bioscience. The authors wish to thank the patients who participated in this study, the study coordinators and administrators of the sites, and the members of the Hemophilia Inhibitor Research Study Investigators: Thomas C. Abshire, Amy L. Dunn, Christine L. Kempton, Paula L. Bockenstedt, Doreen B. Brettler, Jorge A. Di Paola, Mohamed Radhi, Steven R. Lentz, Gita Massey, John C. Barrett, Anne T. Neff, Amy D. Shapiro, Michael Tarantino, Brian M. Wicklund, Marilyn J. Manco-Johnson, Christine Knoll, M. Elaine Eyster, Joan C. Gill, Miguel A Escobar, Cindy Leissinger, and Hassan Yaish. Additional contributors were Jennifer Driggers, Dorothy Ellingsen, Heidi Trau, Tengguo Li, Christine De Staerke, and Melissa Creary from the Division of Blood Disorders, National Center on Birth Defects and Developmental Disabilities, Atlanta, GA.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Soucie JM, Evatt B, Jackson D. Occurrence of hemophilia in the United States. The Hemophilia Surveillance System Project Investigators. Am J Hematol. 1998;59:288–94. doi: 10.1002/(sici)1096-8652(199812)59:4<288::aid-ajh4>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 2.Miller CH, Benson J, Ellingsen D, Driggers J, Payne A, Kelly FM, Soucie JM, Craig Hooper W. The Hemophilia Inhibitor Research Study I. F8 and F9 mutations in US haemophilia patients: correlation with history of inhibitor and race/ethnicity. Haemophilia. 2012;18:375–82. doi: 10.1111/j.1365-2516.2011.02700.x. [DOI] [PubMed] [Google Scholar]
- 3.Peyvandi F, Jayandharan G, Chandy M, Srivastava A, Nakaya SM, Johnson MJ, Thompson AR, Goodeve A, Garagiola I, Lavoretano S, Menegatti M, Palla R, Spreafico M, Tagliabue L, Asselta R, Duga S, Mannucci PM. Genetic diagnosis of haemophilia and other inherited bleeding disorders. Haemophilia. 2006;12(Suppl 3):82–9. doi: 10.1111/j.1365-2516.2006.01263.x. [DOI] [PubMed] [Google Scholar]
- 4.Klopp N, Oldenburg J, Uen C, Schneppenheim R, Graw J. 11 hemophilia A patients without mutations in the factor VIII encoding gene. Thromb Haemost. 2002;88:357–60. [PubMed] [Google Scholar]
- 5.Rost S, Loffler S, Pavlova A, Muller CR, Oldenburg J. Detection of large duplications within the factor VIII gene by MLPA. J Thromb Haemost. 2008;6:1996–9. doi: 10.1111/j.1538-7836.2008.03125.x. [DOI] [PubMed] [Google Scholar]
- 6.Kwon MJ, Yoo KY, Kim HJ, Kim SH. Identification of mutations in the F9 gene including exon deletion by multiplex ligation-dependent probe amplification in 33 unrelated Korean patients with haemophilia B. Haemophilia. 2008;14:1069–75. doi: 10.1111/j.1365-2516.2008.01796.x. [DOI] [PubMed] [Google Scholar]
- 7.Acquila M, Pasino M, Di Duca M, Bottini F, Molinari AC, Bicocchi MP. MLPA assay in F8 gene mutation screening. Haemophilia. 2008;14:625–7. doi: 10.1111/j.1365-2516.2008.01659.x. [DOI] [PubMed] [Google Scholar]
- 8.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldenburg J, Pavlova A. Genetic risk factors for inhibitors to factors VIII and IX. Haemophilia. 2006;12(Suppl 6):15–22. doi: 10.1111/j.1365-2516.2006.01361.x. [DOI] [PubMed] [Google Scholar]
- 10.Boekhorst J, Lari GR, D’Oiron R, Costa JM, Novakova IR, Ala FA, Lavergne JM, WL VANH. Factor VIII genotype and inhibitor development in patients with haemophilia A: highest risk in patients with splice site mutations. Haemophilia. 2008;14:729–35. doi: 10.1111/j.1365-2516.2008.01694.x. [DOI] [PubMed] [Google Scholar]
- 11.Margaglione M, Castaman G, Morfini M, Rocino A, Santagostino E, Tagariello G, Tagliaferri AR, Zanon E, Bicocchi MP, Castaldo G, Peyvandi F, Santacroce R, Torricelli F, Grandone E, Mannucci PM, Group AI-GS The Italian AICE-Genetics hemophilia A database: results and correlation with clinical phenotype. Haematologica. 2008;93:722–8. doi: 10.3324/haematol.12427. [DOI] [PubMed] [Google Scholar]
- 12.Coppola A, Santoro C, Tagliaferri A, Franchini M, Di Minno G. Understanding inhibitor development in haemophilia A: towards clinical prediction and prevention strategies. Haemophilia. 2010;16(Suppl 1):13–9. doi: 10.1111/j.1365-2516.2009.02175.x. [DOI] [PubMed] [Google Scholar]