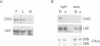Abstract
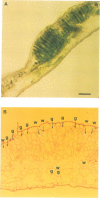
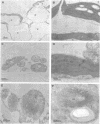
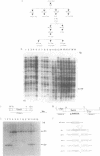
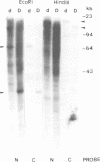
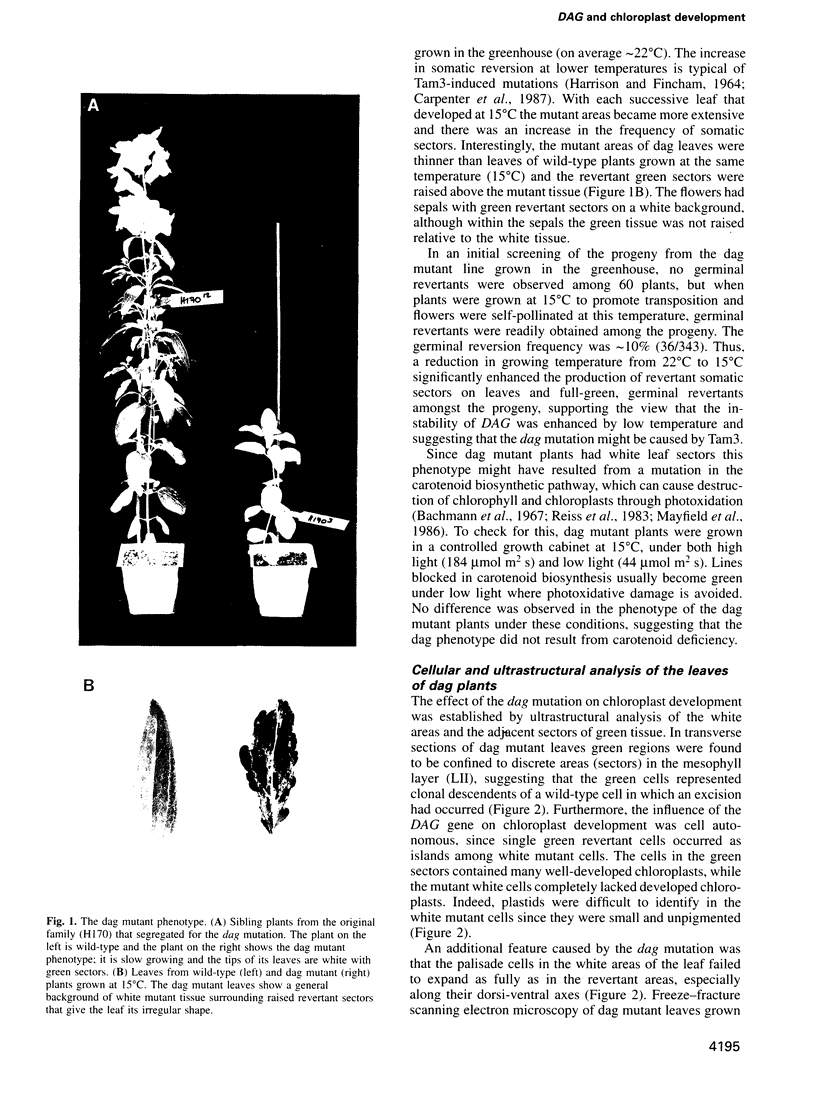
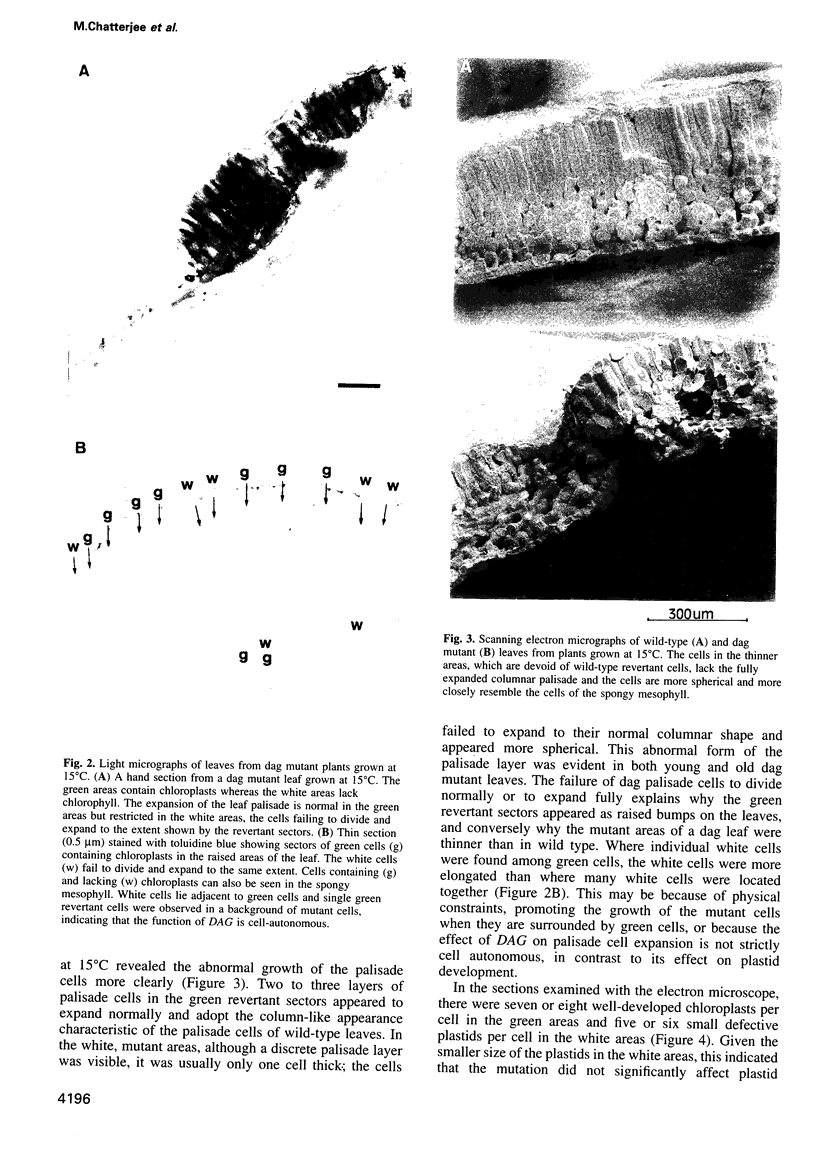
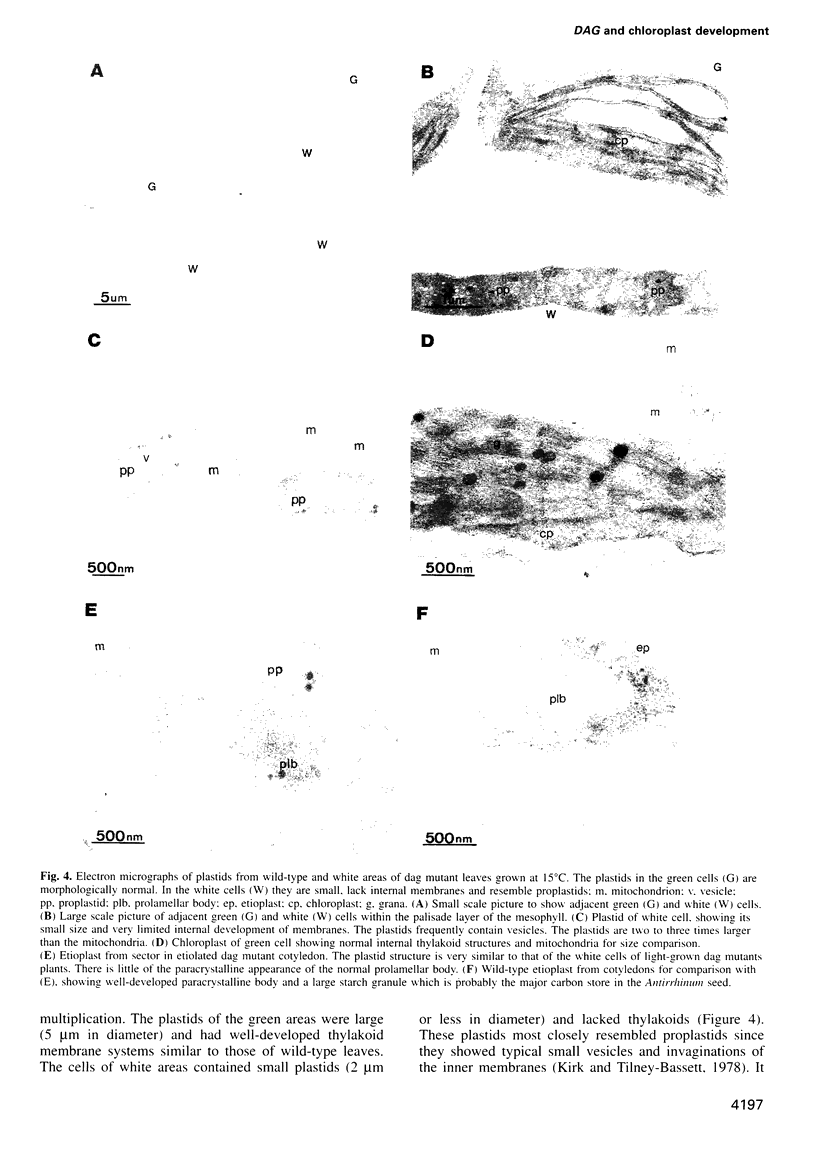
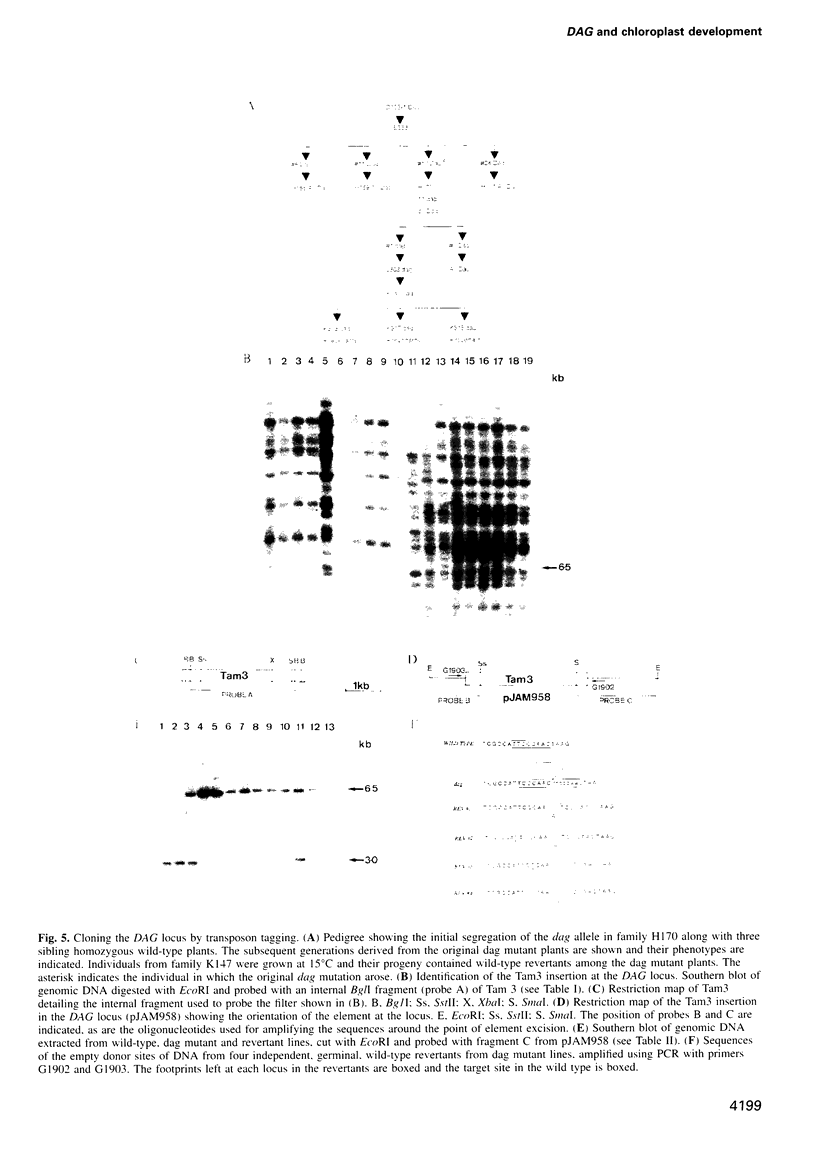
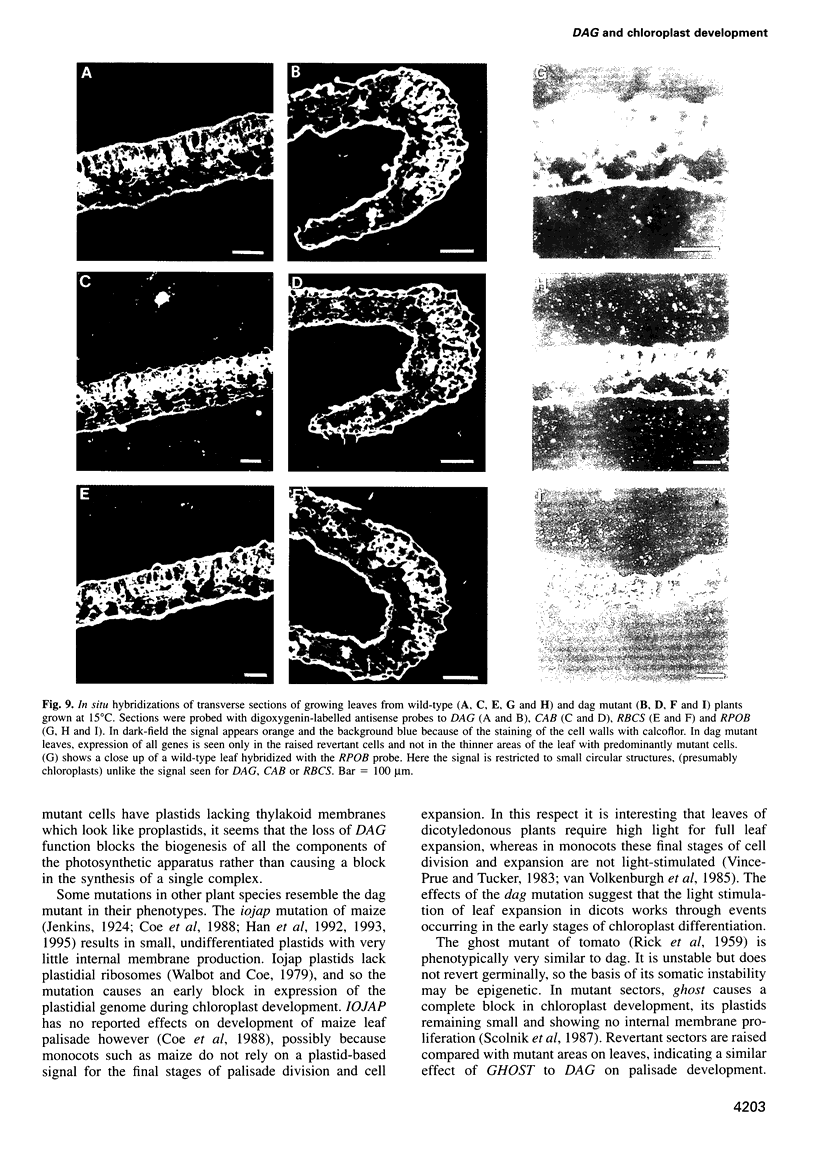
We have identified a mutation at the DAG locus of Antirrhinum majus which blocks the development of chloroplasts to give white leaves with green revertant sectors. The green areas contain normal chloroplasts whereas the white areas have small plastids that resemble proplastids. The cotyledons of dark-grown dag mutant seedlings have plastids which also resemble proplastids. The palisade cells in the white areas of dag mutant leaves also lack their characteristic columnar shape. The DAG locus was cloned by transposon tagging: DAG encodes a novel protein with a predicted Mr of 26k, which is targeted to the plastids. Cleavage of its predicted transit peptide gives a mature protein of Mr 20k. Screening of databases and analysis of Southern blots gave evidence that DAG belongs to a protein family with homology to several proteins of unknown function from plants. Expression of DAG is required for expression of nuclear genes affecting the chloroplasts, such as CAB and RBCS, and also for expression of the plastidial gene RPOB encoding the plastidial RNA polymerase beta subunit, indicating that it functions very early in chloroplast development.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachmann M. D., Robertson D. S., Bowen C. C., Anderson I. C. Chloroplast development in pigment deficient mutants of maize. I. Structural anomalies in plastids of allelic mutants at the w3 locus. J Ultrastruct Res. 1967 Nov;21(1):41–60. doi: 10.1016/s0022-5320(67)80005-4. [DOI] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993 Jan 15;72(1):85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Ray S. C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991 Jun 25;19(12):3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Romero J. M., Doyle S., Elliott R., Murphy G., Carpenter R. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell. 1990 Dec 21;63(6):1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988 Nov;7(11):3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987 May 8;49(3):379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990 Feb 26;261(2):455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Giuliano G., Bartley G. E., Scolnik P. A. Regulation of carotenoid biosynthesis during tomato development. Plant Cell. 1993 Apr;5(4):379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C. D., Coe E. H., Jr, Martienssen R. A. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO J. 1992 Nov;11(11):4037–4046. doi: 10.1002/j.1460-2075.1992.tb05497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess W. R., Prombona A., Fieder B., Subramanian A. R., Börner T. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 1993 Feb;12(2):563–571. doi: 10.1002/j.1460-2075.1993.tb05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A., Carpenter R., Doyle S., Coen E. S. Olive: a key gene required for chlorophyll biosynthesis in Antirrhinum majus. EMBO J. 1993 Oct;12(10):3711–3719. doi: 10.1002/j.1460-2075.1993.tb06048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jähnig F. Structure predictions of membrane proteins are not that bad. Trends Biochem Sci. 1990 Mar;15(3):93–95. doi: 10.1016/0968-0004(90)90188-h. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5509–5513. doi: 10.1073/pnas.90.12.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Carpenter R., Sommer H., Saedler H., Coen E. S. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO J. 1985 Jul;4(7):1625–1630. doi: 10.1002/j.1460-2075.1985.tb03829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden C. W., Wolfe K. H., dePamphilis C. W., Palmer J. D. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO J. 1991 Nov;10(11):3281–3288. doi: 10.1002/j.1460-2075.1991.tb04892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J. E. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993 Oct;103(2):309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990 Apr;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. A., Leech R. M. A Genetic Analysis of Chloroplast Division and Expansion in Arabidopsis thaliana. Plant Physiol. 1994 Jan;104(1):201–207. doi: 10.1104/pp.104.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. A., Leech R. M. Chloroplast Division and Expansion Is Radically Altered by Nuclear Mutations in Arabidopsis thaliana. Plant Physiol. 1992 Jul;99(3):1005–1008. doi: 10.1104/pp.99.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. A., Rutherford S. M., Robertson E. J., Leech R. M. arc6, A Fertile Arabidopsis Mutant with Only Two Mesophyll Cell Chloroplasts. Plant Physiol. 1994 Nov;106(3):1169–1177. doi: 10.1104/pp.106.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R. S., Coomber S. A., Bourett T. M., Bartley G. E., Scolnik P. A. Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell. 1994 Sep;6(9):1253–1264. doi: 10.1105/tpc.6.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnie H. M., Reid J., Hohn T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3'-end formation in plants. EMBO J. 1994 May 1;13(9):2200–2210. doi: 10.1002/j.1460-2075.1994.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S., Bauerle C., Hageman J., Keegstra K., Weisbeek P. The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell. 1986 Aug 1;46(3):365–375. doi: 10.1016/0092-8674(86)90657-4. [DOI] [PubMed] [Google Scholar]
- Susek R. E., Ausubel F. M., Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993 Sep 10;74(5):787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Wetzel C. M., Jiang C. Z., Meehan L. J., Voytas D. F., Rodermel S. R. Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994 Aug;6(2):161–175. doi: 10.1046/j.1365-313x.1994.6020161.x. [DOI] [PubMed] [Google Scholar]
- dePamphilis C. W., Palmer J. D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990 Nov 22;348(6299):337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]