Abstract
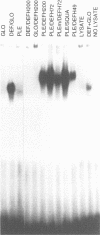
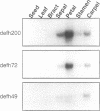
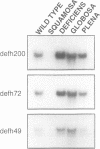
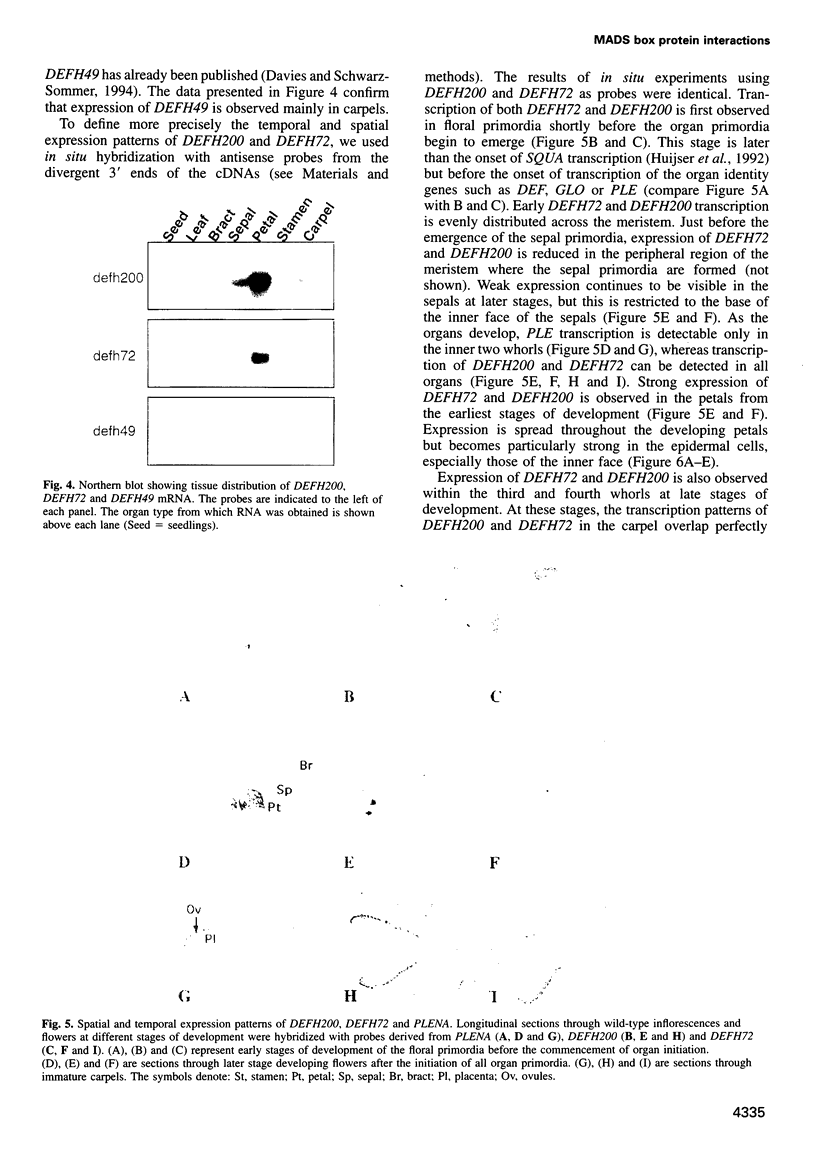
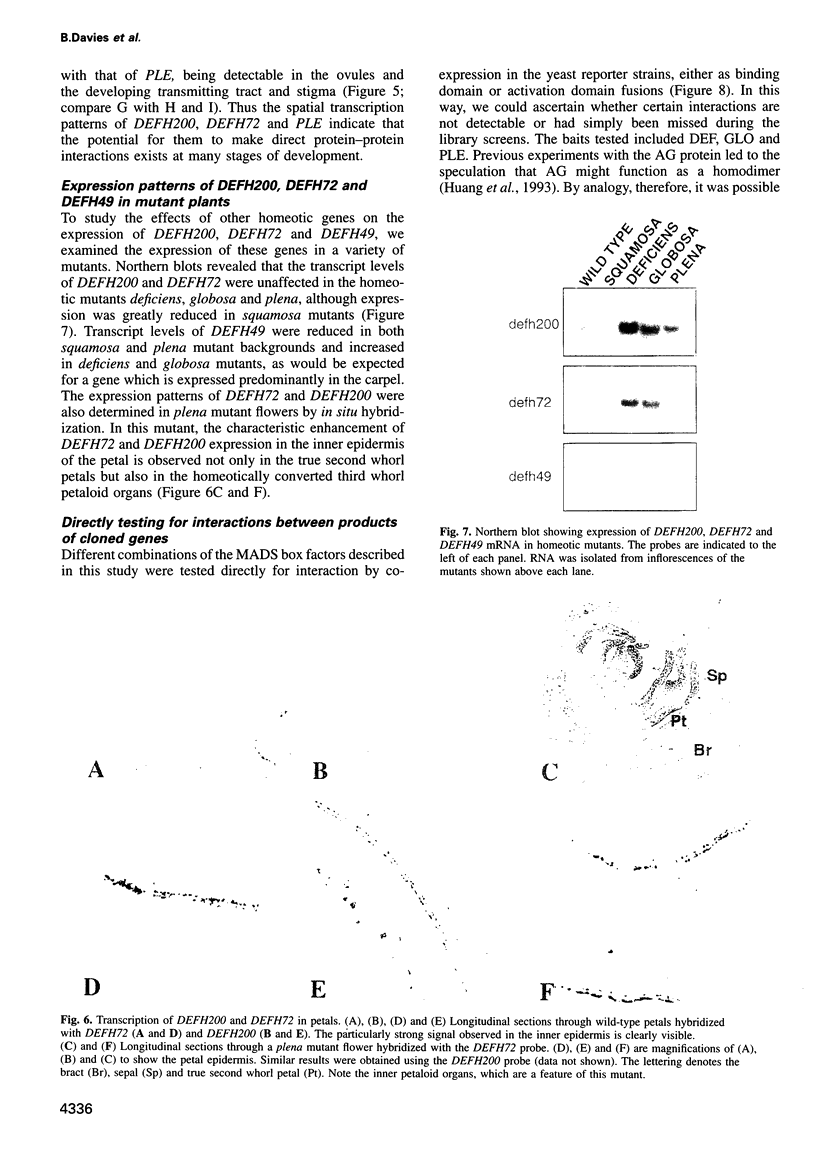
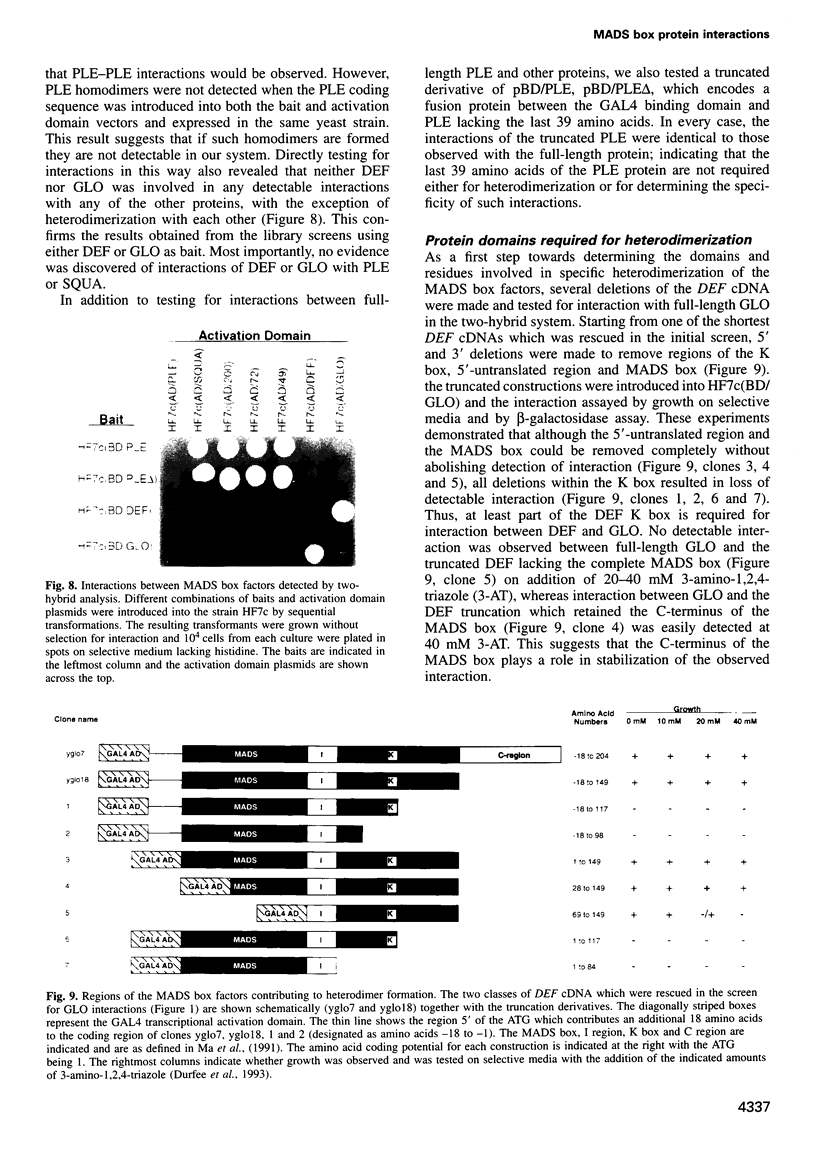
Most known floral homeotic genes belong to the MADS box family and their products act in combination to specify floral organ identity by an unknown mechanism. We have used a yeast two-hybrid system to investigate the network of interactions between the Antirrhinum organ identity gene products. Selective heterodimerization is observed between MADS box factors. Exclusive interactions are detected between two factors, DEFICIENS (DEF) and GLOBOSA (GLO), previously known to heterodimerize and control development of petals and stamens. In contrast, a third factor, PLENA (PLE), which is required for reproductive organ development, can interact with the products of MADS box genes expressed at early, intermediate and late stages. We also demonstrate that heterodimerization of DEF and GLO requires the K box, a domain not found in non-plant MADS box factors, indicating that the plant MADS box factors may have different criteria for interaction. The association of PLENA and the temporally intermediate MADS box factors suggests that part of their function in mediating between the meristem and organ identity genes is accomplished through direct interaction. These data reveal an unexpectedly complex network of interactions between the factors controlling flower development and have implications for the determination of organ identity.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 1990 Feb;4(2):299–312. doi: 10.1101/gad.4.2.299. [DOI] [PubMed] [Google Scholar]
- Angenent G. C., Busscher M., Franken J., Mol J. N., van Tunen A. J. Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell. 1992 Aug;4(8):983–993. doi: 10.1105/tpc.4.8.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent G. C., Franken J., Busscher M., Weiss D., van Tunen A. J. Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J. 1994 Jan;5(1):33–44. doi: 10.1046/j.1365-313x.1994.5010033.x. [DOI] [PubMed] [Google Scholar]
- Bartel P., Chien C. T., Sternglanz R., Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993 Jun;14(6):920–924. [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993 Jan 15;72(1):85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991 Sep 5;353(6339):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Davies B., Schwarz-Sommer Z. Control of floral organ identity by homeotic MADS-box transcription factors. Results Probl Cell Differ. 1994;20:235–258. doi: 10.1007/978-3-540-48037-2_11. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer K., Chen P. L., Yeh S. H., Yang Y., Kilburn A. E., Lee W. H., Elledge S. J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993 Apr;7(4):555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Feilotter H. E., Hannon G. J., Ruddell C. J., Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994 Apr 25;22(8):1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994 Aug;10(8):286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Flanagan C. A., Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant arabidopsis flowers. Plant Mol Biol. 1994 Oct;26(2):581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E. M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994 Jul 1;8(13):1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C., Savidge B., Yanofsky M. F. Regulation of the arabidopsis floral homeotic gene APETALA1. Cell. 1994 Jan 14;76(1):131–143. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995 Jul;15(7):3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Mizukami Y., Hu Y., Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993 Oct 11;21(20):4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tudor M., Su T., Zhang Y., Hu Y., Ma H. DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell. 1996 Jan;8(1):81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tudor M., Weiss C. A., Hu Y., Ma H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol. 1995 Jun;28(3):549–567. doi: 10.1007/BF00020401. [DOI] [PubMed] [Google Scholar]
- Huijser P., Klein J., Lönnig W. E., Meijer H., Saedler H., Sommer H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992 Apr;11(4):1239–1249. doi: 10.1002/j.1460-2075.1992.tb05168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T., Brockman L. L., Meyerowitz E. M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992 Feb 21;68(4):683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G. L., Meyerowitz E. M. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994 Feb 25;76(4):703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- Lamb P., McKnight S. L. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991 Nov;16(11):417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- Ma H., Yanofsky M. F., Meyerowitz E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991 Mar;5(3):484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B., Yanofsky M. F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992 Nov 19;360(6401):273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Martin J. F., Miano J. M., Hustad C. M., Copeland N. G., Jenkins N. A., Olson E. N. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol. 1994 Mar;14(3):1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. G., Nordheim A. A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J. 1991 Dec;10(13):4219–4229. doi: 10.1002/j.1460-2075.1991.tb05000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Tan S., Richmond T. J. Structure of serum response factor core bound to DNA. Nature. 1995 Aug 10;376(6540):490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Abu-Abeid M., Zamir D., Nacken W., Schwarz-Sommer Z., Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991 Sep;1(2):255–266. [PubMed] [Google Scholar]
- Pnueli L., Hareven D., Broday L., Hurwitz C., Lifschitz E. The TM5 MADS Box Gene Mediates Organ Differentiation in the Three Inner Whorls of Tomato Flowers. Plant Cell. 1994 Feb;6(2):175–186. doi: 10.1105/tpc.6.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991 Dec;5(12A):2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Purugganan M. D., Rounsley S. D., Schmidt R. J., Yanofsky M. F. Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics. 1995 May;140(1):345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B., Rounsley S. D., Yanofsky M. F. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell. 1995 Jun;7(6):721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P. J., Hansen R., Tetens F., Lönnig W. E., Saedler H., Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992 Jan;11(1):251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science. 1990 Nov 16;250(4983):931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Gille H., Shaw P. E. Identification of amino acids essential for DNA binding and dimerization in p67SRF: implications for a novel DNA-binding motif. Mol Cell Biol. 1993 Jan;13(1):123–132. doi: 10.1128/mcb.13.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi H., Okada K., Shimura Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 1993 Aug;4(2):385–398. doi: 10.1046/j.1365-313x.1993.04020385.x. [DOI] [PubMed] [Google Scholar]
- Shore P., Sharrocks A. D. The MADS-box family of transcription factors. Eur J Biochem. 1995 Apr 1;229(1):1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Sommer H., Beltrán J. P., Huijser P., Pape H., Lönnig W. E., Saedler H., Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990 Mar;9(3):605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H. Thinking about genetic redundancy. Trends Genet. 1993 Nov;9(11):395–399. doi: 10.1016/0168-9525(93)90140-d. [DOI] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W. E., Saedler H., Sommer H., Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992 Dec;11(13):4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990 Jul 5;346(6279):35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zachgo S., Silva E. de A., Motte P., Tröbner W., Saedler H., Schwarz-Sommer Z. Functional analysis of the Antirrhinum floral homeotic DEFICIENS gene in vivo and in vitro by using a temperature-sensitive mutant. Development. 1995 Sep;121(9):2861–2875. doi: 10.1242/dev.121.9.2861. [DOI] [PubMed] [Google Scholar]