Summary
The Hedgehog (Hh) pathway regulates cell differentiation and proliferation during development by controlling the Gli transcription factors. Cell fate decisions and progression toward organ and tissue maturity must be coordinated and how energy sensor regulates Hh pathway is not clear. AMP-activated Protein Kinase (AMPK) is an important sensor of energy stores that controls protein synthesis and other energy-intensive processes. AMPK is directly responsive to intracellular AMP levels, inhibiting a wide range of cell activities if ATP is low and AMP is high. Thus, AMPK can affect development by influencing protein synthesis and other processes needed for growth and differentiation. Activation of AMPK reduces GLI1 protein levels and stability, thus blocking Sonic hedgehog-induced transcriptional activity. AMPK phosphorylates GLI1 at serines 102 and 408 and threonine 1074. Mutation of these three sites into alanine prevents phosphorylation by AMPK. This in turn leads to increased GLI1 protein stability, transcriptional activity, and oncogenic potency.
Introduction
Regulation of energy production and storage is necessary for living organisms, especially during stages of development that involve substantial growth. Otherwise embryos may invest precious energy in starting organogenesis that cannot be completed. The problem is exemplified by the crucial function of mitochondria, which are the major source of ATP during human pre-implantation development (Wilding et al., 2009). Therefore, developmental control systems that guide the growth of organs and tissues must be coordinated with energy supply.
The Hedgehog (Hh) pathway is essential for development of most organs and tissues. Loss of control of the pathway is oncogenic in tissues where a normal role of the Hh signal is to promote growth. Mutations that deregulate Hh signaling are associated with sporadic and familial skin cancer (basal cell carcinoma) and brain tumors (medulloblastoma). For example, Gorlin Syndrome is due to loss-of-function mutations in the PTCH gene, which encodes the receptor protein Patched1 (Ptch) that binds Hh ligand. Normally Ptch protein restrains Hh transduction, and therefore growth of the skin and cerebellum, until it is inactivated by the Hh ligand, but the tumor cells sense the loss of Ptch function and divide without the need for Hh signals. Given the fine line between mitogenesis and oncogenesis, appropriate regulation of developmental pathways is critical.
Hh signaling controls transcription of target genes by regulating activities of the three Glioma-associated oncogene (Gli1-3) transcription factors. When Hh ligand binds to the Ptch receptor, a 12-pass transmembrane protein, Ptch no longer inhibits the 7-transmembrane domain transducer Smoothened (Smo). In cells not exposed to Hh ligand, Ptch is resident in the plasma membrane overlying primary cilia (Rohatgi et al., 2007); Ptch moves into the cell and is degraded upon binding Hh. Activated Smo then accumulates in primary cilia, which are non-motile solitary appendages on many cell types and serve as transduction centers for Hh signals (Rohatgi et al., 2007). Activation of Smo antagonizes Sufu, Gli1 negative regulator, to promote nuclear translocation of active Gli proteins and induction of genes that control cell proliferation or differentiation during development.
Embryos devote specific regulatory systems to conserving or increasing the energy supply during times of need. One crucial energy-sensing molecule is AMP-activated Protein Kinase (AMPK). AMPK monitors cellular energy status by responding to AMP/ATP ratios, as well as AMP and ATP concentrations (Scott et al., 2009; Steinberg and Kemp, 2009). The levels of AMP and ATP reflect environmental nutrient supply and uptake. High AMP activates AMPK, which then inhibits energy-consuming processes such as protein synthesis, and boosts energy production by increasing glucose uptake and glycolysis (Hardie et al., 2012).
AMPK is a heterotrimer consisting of α, β, and γ subunits. AMPK is activated approximately 1000-fold by phosphorylation of a conserved threonine (Thr172) in the activation loop of the KD by upstream protein kinases such as serine/threonine kinase 11 (STK11; also known as Liver Kinase B1, LKB1) (Jishage et al., 2002). When AMP or ADP concentrations are high, their increased binding to the γ subunit causes a conformational change that promotes phosphorylation of Thr172 by LKB1 and inhibits dephosphorylation (Xiao et al., 2011). Genetic experiments show that zebrafish embryos do not require LKB1 if energy is abundant, but in conditions of energy stress, LKB1 is essential for life (van der Velden et al., 2011). The kinase activity of mammalian phosphorylated AMPK can be enhanced 2 to 5-fold by the binding of AMP to its γ subunit (Sanders et al., 2007; Suter et al., 2006). ATP is an antagonist of AMPK activation, acting by binding to the γ subunit and competing with AMP or ADP binding (Hardie et al., 2012; Xiao et al., 2007).
Development cannot proceed if energy stores are inadequate. Slowing or postponing developmental steps may save the life of a growing animal. Hh signaling has recently been shown to trigger rapid glycolysis in adipocytes by modifying Smo activity, Ca2+ levels, and AMPK activity (Teperino et al., 2012). Thus the Hh developmental pathway alters production of ATP. We have been investigating the complementary possibility, that transduction through the Hh pathway is modulated by energy stores. When energy is scarce and cell division or cell differentiation should be slowed to conserve remaining stores, activated AMPK may indirectly affect developmental pathways like Hh by reducing protein synthesis or other basic gene expression functions. A second possibility is that a more direct regulatory connection links energy sensing with developmental regulators. We have investigated this second possibility using cultured cells that respond to Hh signaling proteins.
Results
Control of Gli1 protein level and stability by AMPK
To investigate whether AMPK regulates the Hh pathway, we used the NIH-3T3 cell line, a well-known Hh-responsive cell line. Cells were treated with the AMPK activator twodeoxyglucose (2DG), a glucose analog that blocks ATP production by inhibiting glycolysis and thereby induces AMPK activation (Wick et al., 1957). Activation of AMPK reduced Gli1 protein levels progressively over time (Figure 1A). As NIH3T3 cells were treated for increasing numbers of hours with 2DG, the amount of activated AMPK (p-AMPK) increased. The amount of a direct AMPK substrate that is often used to measure AMPK activity (Henin et al., 1995), phospho-Acetyl-CoA Carboxylase (p-ACC), also increased for at least four hrs. In addition to 2DG, similar effects were observed when two other AMPK activators (A769662 and AICAR) were applied to NIH 3T3 and pZp53Med1 (Med1) medulloblastoma cells (Figure 1B). In addition, protein levels of Gli2 and Gli3 (FL and R) did not change while AMPK was activated (Figure S1A). AMPK knockout MEF cells had increasing 1.71 fold of Gli1 protein as wild-type MEF and the amount of Gli1 protein was low in AMPK+/+ cells in the presence of 2DG, and did not change in the AMPK-/- cells (Figure 1C). Therefore the low level of Gli1 protein caused by 2DG administration was due to AMPK-dependent action.
Figure 1. AMPK reduces GLI1 protein levels and stability.
(A) NIH3T3 cells were treated with 25mM 2-deoxyglucose (2DG) for the number of hours indicated to activate AMPK. Cell lysates were analyzed via immunoblot with the indicated antibodies. (B) NIH3T3 and pZp53Med1 (Med1) cells were treated with 150 M A769662 and 0.75 mM AICAR for 6 hours to activate AMPK. Cell lysates were analyzed as shown in (A). (C) WT (AMPK+/+) and KO (AMPK -/-) MEF cells were treated with or without 2DG for four hours and lysed and analyzed as shown in (A). The Gli1 lanes were quantitated using Image J to determine the relative intensity to the control band, and were normalized to the internal loading control, β-Actin, giving the ratio of 1.98 to 1 as indicated. (D) HEK-293 cells were transfected with genes encoding the wild-type (wt) and kinase-dead mutant (DN) forms of AMPK. Cell lysates were analyzed by immunoblot. (E) NIH3T3 cells were serum-starved in Dulbecco's Modified Eagle's Medium (DMEM) (0.5% bovine calf serum) overnight, stimulated with Hh for the indicated hours, and treated with 2DG (25mM) for the number of hours indicated and lysates were analyzed by immunoblot. The western blot was measured using image J to determine the relative intensities of the Gli1 bands, which were normalized using the internal loading control tubulin protein; the numbers are shown. (F) Med1 cells, which have constitutively active Hh target gene expression, were treated with 25mM 2-deoxyglucose (2DG) for the indicated numbers of hours to activate AMPK. Cell lysates were analyzed by immunoblotting with the indicated antibodies. (G) AMPK+/+ and AMPK -/- MEFs were treated with cycloheximide (CHX, 1 μg ml−1) for the indicated times and cell lysates were analyzed by immunoblot with the indicated antibodies. (H) AMPK+/+ and AMPK -/- MEFs were co-treated with cycloheximide (CHX, 1 μg ml−1) and with or without 2DG (25mM) for the indicated times and cell lysates were analyzed by immunoblot with the indicated antibodies.
Knocking down Lkb1, dysregulates the Hh pathway by affecting Gli3 (Jacob et al., 2011). We found that two different Lkb1 shRNAs reduced the level of Lkb1 protein by half in Med1 cells, while the level of Gli1 protein was not affected (Figure S1B). In Lkb1-/- myoblast cells, both Gli1 protein and mRNA levels were not much different in comparison with Lkb1+/+ cells (Figures S1C and S1D). When Lkb1+/+ and Lkb1-/- cells were treated with AICAR, Gli1 protein level was reduced in both (Figure S1C). We conclude that Lkb1 is not involved in the AMPK-mediated reduction of the Gli1 protein level. HEK-293 cells were transfected with a kinase-dead AMPK mutant (Banko et al., 2011) that is unable to phosphorylate proteins. The result was an increase in the Gli1 protein level compared to cells transfected with wild-type AMPK (Figure 1D).
Gli1 is transcriptionally activated by Shh. Activating AMPK reduced Gli1 protein levels in 3T3 cells despite stimulation with Shh ligand (Figure 1E). Co-treatment of the cells with 2DG and Shh increased the amount of activated AMPK measured as p-AMPK and its target p-ACC, and reduced the amount of Gli1 protein compared to cells treated with Shh alone. Med1 cells, which have constitutively active Hh target gene expression (Berman et al., 2002), also had reduced Gli1 protein levels in the presence of 2DG (Figure 1F). The reduction of Gli1 protein may have been due to protein degradation. Co-treatment with 2DG and proteasome inhibitor (MG132) restored the higher amount of Gli1 protein (Figure S1E). This indicates that even upon stimulation by Shh, which increases the amount of Gli1 protein because Gli1 is a transcriptional target, activated AMPK is able to reduce Gli1 protein levels.
The reduction of the Gli1 protein level by activated AMPK could be due to an effect on Gli1 transcription, translation, or post-translational stability. We find that Gli1 protein stability is altered by the activity state of AMPK. We used cycloheximide (CHX) to block translation of new protein and monitored the stability of pre-existing Gli1 protein. When AMPK was functional (Figure 1G, left panel), the amount of Gli1 protein decreased with a half-life of 4 hours. In cells lacking AMPK (Figure 1G, right panel), Gli1 protein remained stable with a half-life of at least 8 hours. Gli1 instability was accelerated in 2DG and CHX co-treated AMPK+/+ cells (Figure 1H) but was not affected in AMPK-/- cells (Figure 1H). The protein stability of Gli2 and Gli3 were not affected in the 2DG and CHX co-treated med1 cells (Figure S1F). In this experiment AMPK could not have been affecting synthesis of Gli1, which was prevented, but instead affected Gli1 protein stability.
AMPK reduces Gli1 transcriptional activity
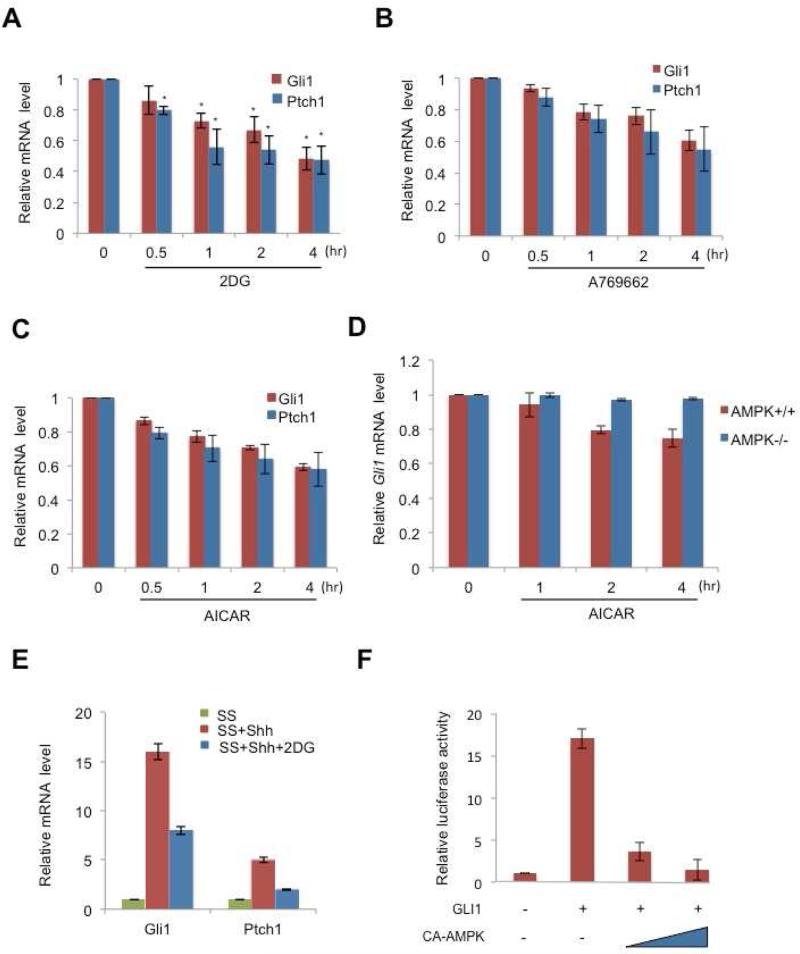
We tested whether AMPK activation affects Gli1 mRNA levels in addition to reducing Gli1 protein stability. In AMPK-/- cells, Gli1 and Ptch1 mRNA were elevated about two-fold compared to AMPK+/+, and consistently, mRNA from the two target genes was higher in DN-AMPK-transfected cells in comparison with WT-AMPK-transfection cells (Figures S2A and S2B). In NIH3T3 cells, treatment with 2DG, A769662 and AICAR led to a time-dependent reduction in the level of mRNA from Gli1 and from another target, Ptch1 (Figures 2A-2C). During the 4 hour time period examined in these experiments, the activation of AMPK by 2DG, A769662, or AICAR shuts down translation and transcription, but the effect is not a general effect on mRNA levels because the control mRNA measured (Gapdh) did not change in amount (Figures 2A-2C). Instead the reduced Gli1 protein, due to its instability and lowered synthesis, causes lower levels of Gli1 and Ptch1 transcripts. In AMPK-/- cells, Gli1 mRNA remained at the same level in the presence of AICAR (Figure 2D). This result is consistent with the results shown in Figure S2A. Gli1 protein level was not affected in the present of 2DG in AMPK-/- cells. In keeping with this view, the addition of AMPK activators together with Shh lowered Gli1 and Ptch1 mRNA levels compared to induction of those targets with Shh alone (Figures 2E and S2C).
Figure 2. AMPK inhibits GLI1 transcriptional activity.
(A) NIH3T3 cells were treated with 25mM 2-deoxyglucose (2DG), (B) 150 M A769662 and (C) 0.75 mM AICAR for the indicated hours and the amount of Gli1 or Ptch1 mRNA was analyzed by qRT-PCR with Gapdh mRNA as the internal control, and normalized to the time zero Gli1 and Ptch1 mRNA levels. The control is provided by time zero, when no chemicals were applied, so the bars indicate Gli1 and Ptch1 mRNA levels relative to those of Gapdh and normalized to levels at time zero. (D) AMPK+/+ and AMPK -/- MEFs were treated with 0.75 mM AICAR for the indicated hours and was analyzed by qRT-PCR as shown as (A). (E) NIH3T3 cells were serum-starved (SS) in DMEM (0.5% bovine calf serum) overnight, stimulated with Shh with or without 25mM 2DG, for 6 hours, and the amount of Gli1 or Ptch1 RNA was analyzed by qRT-PCR and normalized to Gli1 and Ptch1 mRNA levels in serum-starved cells. Three replicate experiments were done with standard deviations. (F) HEK-293 cells were co-transfected with Gli1-RE-Luciferase reporter, GLI1 and constitutively active AMPK (CA-AMPK), and maintained for 36 hours. Cell lysates were analyzed using a luciferase assay to measure reporter-gene transcriptional regulation by GLI1. Representative results from three experiments (n = 3) conducted in duplicate are shown, with standard deviations.
In addition to our in vitro studies with cell lines, we examined whether AMPK controls gli1 mRNA in an in vivo context. In zebrafish embryos, treatment with 2DG led to AMPK activation and inhibition of the activity of its downstream substrate (p-ACC) (Figure S2D). Measured gli1 and ptch1 mRNA levels, we found a reduction in expression levels of these genes in the 2DG treated as compared with the non-treatment group (Figure S2E). Injection of ampk morpholino (MO) into zebrafish embryos led to a reduction of ampk mRNA level in comparison with the control MO (Five base pair mismatches of the ampk MO)(Li et al., 2013) and two-fold elevation of gli1 and ptch1 mRNA levels as compared to the control group (Figure S2F).
AMPK may be acting on Gli1 mRNA levels directly or indirectly, so we used a reporter gene assay to look at direct target gene regulation. We used a synthetic target gene consisting of eight Gli binding sites joined to a luciferase reporter (Sasaki et al., 1997) (Figure 2F). Transfection of 293 cells with the reporter transgene, and another plasmid that encoded constitutively active AMPK, suppressed induction by Gli1. As we show in the following experiments, target gene expression is lower because activated AMPK phosphorylates and destabilizes Gli1 protein, not because AMPK directly reduces Gli1 mRNA.
Direct regulation of Gli1 protein stability by AMPK
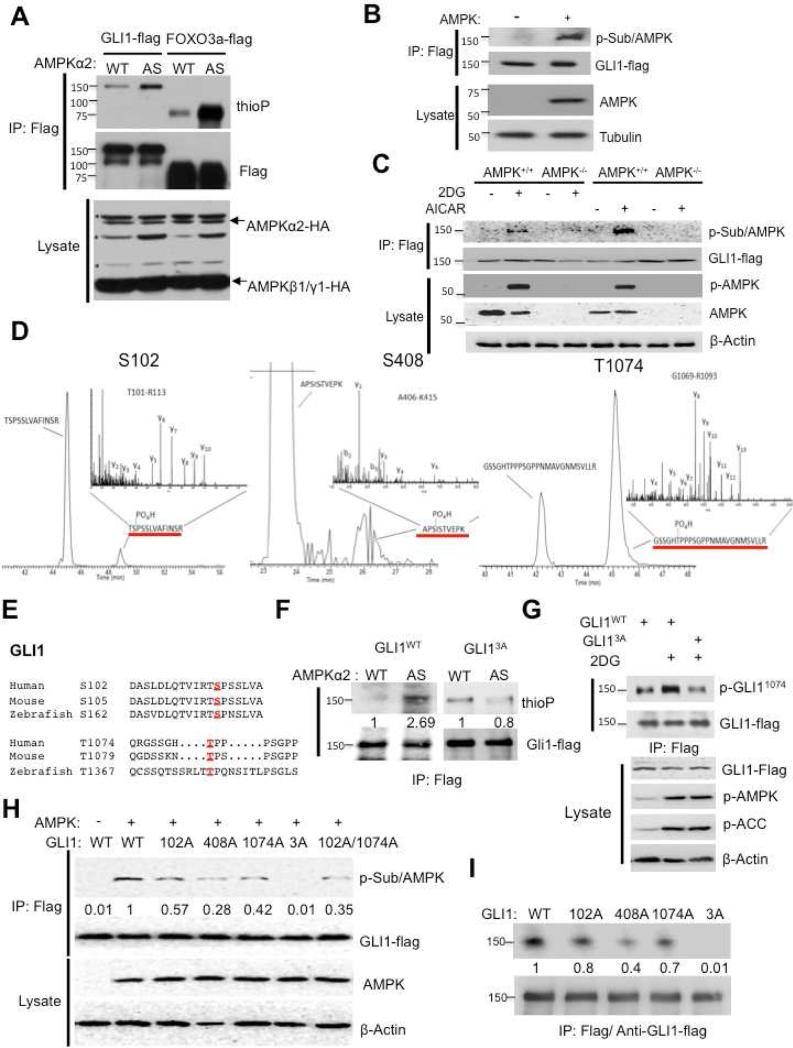
We next examined the mechanism of the influence of AMPK on Gli1 protein stability. To investigate whether AMPK alters Gli1 phosphorylation, we used a chemical genetic approach. The method allows specific labeling of direct substrates of a protein kinase in living cells, thus distinguishing direct from indirect influences of a kinase (Alaimo et al., 2001). ATP-binding pockets of protein kinases contain a conserved “gatekeeper residue” that, during the reaction, is in close contact with the N6 position of the adenine ring of ATP. Substituting a smaller amino acid for this gatekeeper residue enables the mutant protein kinase, which is termed “analog-specific” (AS), to use ATP analogs containing bulky groups at the N6 position (Allen et al., 2007). In contrast, bulky ATP analogs are poor substrates for wild-type (WT) kinases due to steric hindrance by the gatekeeper residue. N6-modified ATPgS nucleotides are accepted by the analog-specific kinase, and the transferred thiophosphate can be alkylated and recognized by a specific monoclonal antibody, thioP antibody (Allen et al., 2005; Allen et al., 2007). The power of the approach is exemplified by an analog-specific version of AMPKα2 (AS-AMPKα2) that was used to identify AMPK substrates in human embryonic kidney 293T cells (Banko et al., 2011).
Using the same protocol, we co-transfected genes encoding HA-tagged WT or AS AMPKα2 with the two other AMPK subunits β1 and γ1, as well as with genes for Flag-tagged GLI1 and FOXO3a into HEK-293 cells. Flag antibody was used to immunoprecipitate (IP) GLI1 and FOXO3a proteins. FOXO3 is AMPK target that served as a positive control. The precipitate was analyzed by immunoblotting with thioP antibody to detect phosphorylated GLI1 and FOXO3a. In AS-AMPKα2-transfected cells treated with 2DG, we found increased phosphorylated GLI1 compared with WT-AMPKα2-transfected cells, as well as increased phosphorylated FOXO3a (Figure 3A).
Figure 3. AMPK directly phosphorylates GLI1.
(A) HEK-293 cells were cotransfected with Flag-tagged GLI1 or Flag-tagged FOXO3, with HA-tagged wild-type or AS-AMPKα2, and with AMPKβ1 and γ1. AS-AMPKα2 phosphorylates the known AMPK substrate FOXO3, which was detected using thioP antibody. FOXO3 and GLI1 were immunoprecipitated with antibodies that recognize the Flag tag and blotted with thioP, Flag, or HA (AMPK) antibody. “ * ” Indicates non-specific bands that were recognized by HA antibody. (B) HEK-293 cells were co-transfected with Flag-tagged GLI1 and HA-tagged AMPK, and cells were lysed in NP40 lysis buffer. GLI1 was immunoprecipitated using an antibody to Flag and the precipitate was analyzed on a protein blot using AMPK phosphorylation-specific substrate antibody (p-Sub/AMPK) and Flag antibody. The introduction of AMPK into the cells causes p-Sub/AMPK to label GLI1-Flag. (C) AMPK+/+ and AMPK -/- MEFs were virus infected with Flag-tagged GLI1 and treated with 25mM 2DG and 0.75mM AICAR for 30 min, and cells were lysed in NP40 lysis buffer and the cells lysates were analysis as shown as (B). (D) HEK-293 cells were co-transfected with Flag-tagged GLI1 and AMPKα2, β1, and γ1. Cells were lysed in NP40 lysis buffer. The lysates were subjected to immunoprecipitation using an antibody to Flag-tag, and the GLI1 band was isolated and subjected to mass spectrometry. Extracted ion chromatograms (EIC) identified phosphorylated peptides S102, S408, and T1074. The inset panels represent the fragment ion spectra determined using high-energy dissociation (HCD). (E) Alignment of two conserved sites (S102 and T1074) in GLI1 that match the optimal AMPK substrate motif and are conserved from human to zebrafish. S408 is present only in the human sequence. (F) HEK-293 cells were co-transfected with Flag-tagged GLI1WT or Flag-tagged GLI13A, either WT-AMPKα2 or AS-AMPK α2, and AMPK β1 and γ1. Cells were lysed in NP40 lysis buffer. Substrates were immunoprecipitated with antibodies to Flag-M2 and blotted with thioP or Flag antibody. (G) 2DG-stimulated phosphorylation of GLI1. Left: HEK293 cells were transfected with Flag-tagged GLI1WT or GLI13A. Thirty-six hours after transfection they were treated with 25mM 2DG for 30 mins, long enough to activate AMPK but not long enough to cause breakdown of Gli1. The lysates were subjected to immunoprecipitation using an antibody to Flag-tag and immunoblotted with phospho-GLI11074 and Flag antibodies. The strong labeling in the center top lane shows that wild-type GLI1, but not GLI13A, is phosphorylated by AMPK. Right: Lysates prepared as in the left panel were immunoblotted with antibodies against Flag, phospho-AMPK (p-AMPK), phospho-ACC (p-ACC), and β-Actin. The p-AMPK and p-ACC lanes show that the AMPK is stimulated by the 2DG. The actin lane controls for loading. The GLI1-Flag lane shows stabilization of GLI1wt and of GLI13A in response to 2DG, though the response to 2DG disappears by 30 minutes. (H) HEK-293 cells were transfected with AMPK and Flag-tagged GLI1WT, GLI1102A, GLI1408A, GLI11074A, GLI1102A/1074A and GLI13A. Gli1 was immunoprecipitated using an antibody to Flag and the precipitate was analyzed on a protein blot using AMPK phosphorylation-specific substrate antibody (p-Sub/AMPK) and Flag antibody. The introduction of AMPK into the cells causes p-Sub/AMPK to label GLI1-Flag. The number indicates the relative intensity of p-Sub/AMPK antibody. (I) HEK-293 cells were transfected with Flag-tagged GLI1WT, GLI1102A, GLI1408A, GLI11074A and GLI13A. Cells were lysed in NP40 lysis buffer. Cell lysates were immunoprecipitated with antibodies to Flag-M2 and the IP GLI1 proteins were subjected to in vitro AMPK kinase assay. The number indicates the relative intensity of AMPK kinase activity.
In the next experiment we used AMPK phospho-substrate-specific antibody (p-Sub/AMPK (Gwinn et al., 2008), and found that Gli1 was phosphorylated in AMPK-transfected cells. HEK-293 cells were transfected with Flag-tagged Gli1 alone (-) or with AMPK as well (+). Flag-tagged GLI1 was immunoprecipitated with anti-Flag antibodies and separated on a protein gel. The blot was probed with antibodies against AMPK phospho-substrate (p-Sub/AMPK), Flag, AMPK, and tubulin (Figure 3B). The results show that GLI1-Flag was at comparable levels in both extracts, while AMPK was detected only in AMPK-transfected cells; a prominent band was observed with the p-Sub antibody only in cells that had been transfected with AMPK.
We generated MEFs with stable GLI1-Flag expression in AMPK+/+ and AMPK -/- cells to verify that phosphorylation of GLI1 is AMPK-dependent. AMPK was activated using 2DG and AICAR, and Flag-tagged GLI1 was immunoprecipitated with anti-Flag from AMPK+/+ and AMPK -/- cell lines. In the immunoprecipitates, a prominent band was observed with the p-Sub/AMPK antibody in AMPK+/+ MEFs but not in AMPK-/- MEFs (Figure 3C). These results confirm the identity of the AMPK-phosphorylated protein as GLI1.
To discover which GLI1 amino acids were phosphorylated by AMPK, HEK-293 cells were co-transfected either WT-AMPK or DN-AMPK, and GLI1. GLI1 was purified from cell extracts using immunoprecipitation with Flag antibody, further purified by isolating the GLI1 protein from a SDS gel. Mass spectrometry showed that GLI1 was phosphorylated at sites S102, S408, and T1074 (Figures 3D, S3). The same result was obtained in cells transfected with WT AMPK, but not if the cells were transfected with (DN) AMPK. Fourteen potential phosphopeptides were observed after two independent WT AMPK-transfections (Figures S3B-S3D, yellow). Three of these peptides were not phosphorylated in DN-AMPK-transfected cells (Figures S3B-S3D, red circles). In parallel, GLI1 was purified from stable GLI1-Flag expression AMPK+/+ MEFs treated with and without 2DG to modulate endogenous AMPK activity. The same result was found that GLI1 was phosphorylated at sites S102, S408 and T1074 in 2DG treated AMPK+/+ MEFs (Figure S3E). Without 2DG, phosphorylations of S102 and S408 were not detected on GLI1 (Figure S3F). The results indicate that the phosphorylation changes of these sites are in response to metabolic stress in cells.
The sequence for S408 matches the AMPK consensus LRRVXS/TXXXL but not conserved in mouse and zebrafish. S102 and T1074 are conserved in the GLI1 proteins of humans, mice, and zebrafish (Figure 3E) but do not perfectly match the optimal AMPK consensus motif (Table S1).
The three AMPK-phosphorylated amino acids in GLI1 were changed to alanines by mutating the Gli1 gene, i.e. S102A, S408A, and T1074A. This protein, GLI13A, should be immune to AMPK phosphorylation. 293 cells were co-transfected with DNA that encoded either WT-AMPKα2 or AS-AMPKα2, and a construct encoding GLI1WT or GLI13A tagged with a Flag epitope. AS-AMPKa2 no longer phosphorylated the Flag-GLI13A protein (Figure 3F), with the signal dropping to the background levels (0.8 arbitrary units) seen in cells transfected with wild-type GLI1 (2.69 arbitrary units).
An anti-phospho-GLI1 T1074 antibody was prepared that is highly specific. Immunoprecipitated wild-type GLI1-Flag was stained with the p-GLI11074 antibody (Figure 3G, upper panel, top middle lane) while the GLI13A mutant (upper panel, top right lane) was much less. The activation of AMPK is demonstrated here by the phospho-AMPK antibody and by the appearance of p-ACC. In this experiment, 2DG was administered for only 30 min, sufficient to activate AMPK but not long enough to cause loss of GLI1 protein (Figures 1A and 1F). 2DG was necessary for substantial labeling of the WT protein (Figure 3G, top middle lane compared to top left lane). The 2DG-driven phosphorylation of GLI1 was observed with cells transfected with WT GLI1 but not with cells transfected with GLI13A mutant (Figures 3G and S3G). To identify which AMPK phosphorylation site predominates, we repeated the experiment of Figure 2B and cotransfected GLI1WT, GLI13A, the three single mutants, and GLI1102A/1074A with AMPK into 293 cells. In cells transfected with GLI13A, no signal was detected with p-Sub/AMPK, and in cells containing GLI1408A, the signal had 28% of the intensity compared with GLI1WT (Figure 3H). GLI1102A and GLI11074A had 57% and 42% of the control signal, respectively, and the signal of the 102A/1074A double mutant was lower at 35% of the control (Figure 3H).
To prove that AMPK kinase can act directly upon GLI1, an in vitro AMPK kinase assay was performed. In Figure 3I, GLI13A was utterly resistant to AMPK kinase which is similar with results shown in Figure 3H. Each single mutation (102A, 408A, 1074A) moderately reduced the modification by AMPK, from 20% to 60% (Figure 3I). We conclude that S102, S408 and T1074 are all dominant AMPK phosphorylation sites on GLI1.
GLI13A has higher protein stability and transcriptional activity
In this section we show that, compared to GLI1WT, the GLI13A mutant protein has increased stability, is resistant to AMPK-mediated suppression of Gli1 transcriptional activity, and stabilizes Gli1 mRNA at high levels. Using CHX to prevent new protein synthesis as in Figure 1, we found that in transfected 293 cells, GLI13A is considerably more stable than GLI1WT. We produced a phospho-mimic version of GLI1 where each of the three AMPK-phosphorylated residues was replaced by a glutamate; this was called GLI13E and was highly unstable (Figure 4A, right). Similarly, GLI13A was much more stable than GLI13E or GLI1WT in NIH3T3 cell lines in which GLI1 was stably produced (Figure S4A).
Figure 4. GLI13A has higher protein stability and transcriptional activity.
The GLI13A mutant has increased stability and is resistant to AMPK-mediated suppression of Gli1 transcriptional activity. (A) Lysates of HEK-293 cells transfected with Flag–GLI1WT, Flag–GLI13A, or Flag–GLI13E, were harvested at different times after treatment with cycloheximide (CHX, 1 μg ml−1), and analyzed by immunoblot. GLI13E has the two serines and one threonine that are normally phosphorylated by AMPK changed into glutamates to mimic phosphorylated GLI1. (B) HEK-293 cells were co-transfected with Gli1-luciferase reporter, Gli1WT and AMPK for 36 hours. The two amounts of AMPK-expressing plasmid used were 1 and 3ug. Cell lysates were analyzed using a luciferase assay to measure GLI1 transcriptional induction of the introduced Gli1-luciferase target gene. Representative results from three experiments (n = 3), each conducted in duplicate, are shown with standard deviations. (C) HEK-293 cells were co-transfected with Gli1-luciferase reporter, GLI13A, and AMPK and analyzed, as in (B). (D) NIH3T3 cells were transfected with vector control, GLI1WT, or GLI13A for 36 hours, then treated with 2DG (25mM) for 4 hours. RT-PCR was used to measure (D) Gli1 mRNA and (E) Ptch1 mRNA. The experiment was repeated three times. (“ ** ” p<0.01, “ *** ” p<0.001) (F) NIH3T3 vector, GLI1WT and GLI13A producing stable cell lines were treated with AICAR (0.75mM) and A769662 (150 M) for 4 hours. RT-PCR was used to measure (F) Gli1 mRNA and (G) Ptch1 mRNA. The experiment was repeated three times.
To examine regulation of transcription by each version of GLI1, we transfected genes encoding each of the form into 293 cells. Each variant GLI1 was tested with or without co-transfected AMPK. Activation of transcription by GLI1 was measured with a luciferase assay; the cells were also transfected with a plasmid encoding eight Gli consensus sequences in cis to a luciferase gene (Sasaki et al., 1997). Transfected GLI1 activated this target even without added Shh or other agonists. The samples were normalized by comparison to cells transfected with vector alone. Transcriptional induction by wild-type GLI1 was negatively affected by adding AMPK, produced at two levels by transfecting 1 or 3 ug, even without adding 2DG (Figure. 4B). In contrast, GLI13A transcriptional activity was refractory to inhibition by AMPK even at the higher level (Figure 4C). Further activation of AMPK by added 2DG inhibited GLI1 target gene expression if cells contained GLI1WT but not if cells contained GLI13A (Figures 4D, 4E). We repeated the experiments using other AMPK activators (AICAR and A79662) (Banko et al., 2011) in NIH3T3 cell lines stably expressing GLI1 proteins. GLI13A had robust resistance to 2DG-induced lowering of Gli1/Ptch1 mRNA levels (Figures 4F, 4G). Each single mutant had a different degree of moderate to great resistance to 2DG effects (Figures S4B, S4C). These results indicate that mutation of the three residues to alanine did indeed make GLI1 function refractory to inhibition by AMPK.
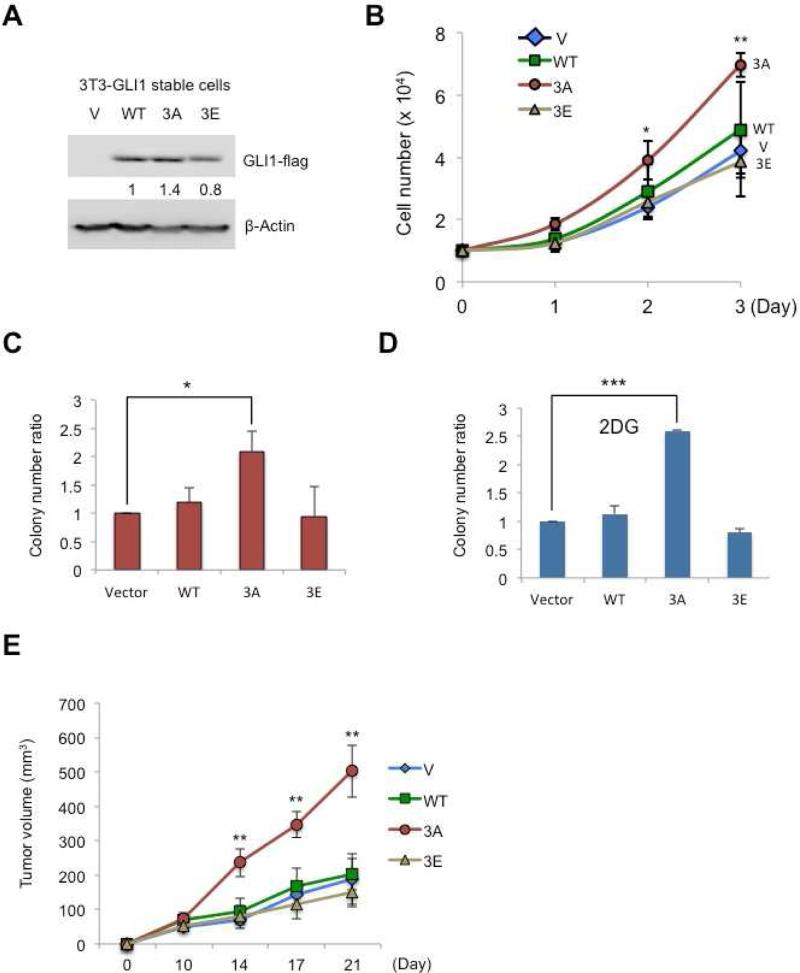
GLI13A has potent cell division-stimulating and oncogenic activity
To further investigate the function of these GLI1 mutants, we generated stable cell lines in 3T3 cells using lentiviral vectors that produced GLI1WT, GLI13A, or GLI13E. The amount of GLI13A protein that accumulated was about 1.4 fold that seen for GLI1WT, normalizing both to actin (Figure 5A). Cell counting with a hemacytometer showed that cells transduced with GLI13A virus had a significantly increased growth rate compared to wild-type and GLI13E cell lines (Figure 5B). Colony formation assays (Figure 5C) showed increased growth of GLI13A-bearing mutant cells compared to other cell lines. To test the effect of activating AMPK, 25mM 2DG was added to the starting cultures and the growth of colonies was measured two weeks later. An even higher colony number difference was observed after treatment with 2DG (Figure 5D) (Yang et al., 2008), but only for cells containing GLI13A. Similar results were obtained using A769662 and AICAR in the colony formation assay (Figure S5). To test the oncogenic impact of GLI1WT in comparison to the mutant proteins, the stably transfected cell lines producing the GLI1 variants were injected subcutaneously into nude mice. Tumor growth was monitored for three weeks. Cells that contained GLI13A grew to a volume about 2.5 times that of the other three cell lines (Figure 5E).
Figure 5.
Tests of cell division, colony formation and oncogenic effects of mutant GLI1 proteins. (A) NIH3T3 cells were infected with vector, GLI1WT, GLI13A, or GLI13E lentivirus, with Flag tags on each protein, and selected for 7 days with puromycin (2.5ugml-1). Cell lysates were analyzed by immunoblotting with Flag antibody to measure the amounts of the expressed proteins. (B) This experiment used NIH3T3 GLI1-stable cell lines (vector control, WT, 3A, and 3E). 5 × 103 cells were seeded into 12-well plates for growth assays, each cell type in triplicate, and cells were counted using a hemocytometer for three consecutive days. The experiment was repeated three times (“ * ” p<0.01, “ ** ” p<0.001). (C) NIH3T3 cells with GLI1WT, GLI13A, or GLI13Estably expressed were seeded into 6-well plates for colony formation assays for 2 weeks. Colonies larger than 1.5 mm were counted. (D) As in (C) the cells were treated with 2DG (25mM) and 2DG-containing medium. The medium in 2DG-treated wells was changed every three days to refresh the 2DG. Colony numbers were counted two weeks later. In (C) and (D), each cell line was seeded in duplicate, with N=3 (“ * ” p<0.05, “ *** ” p<0.0001). (E) NIH3T3 GLI1-stable cell lines (Vector control, WT, 3A, and 3E). 107 cells were injected subcutaneously into the nude mice and tumor growth was monitored for three weeks (“ ** ” p<0.001).
Discussion
The Hh pathway controls cell differentiation and growth in developing embryos and regenerating adult tissues. Most features of the pathway components and their interactions are evolutionarily conserved across many species from Drosophila to humans. In developing and adult animals experience deprivation and stresses, as well as circadian and annual rhythms that demand adaptation of developmental mechanisms to circumstances. One way that this happens is that the pathway itself has built-in feedback controls that buffer the signaling. For example, induction of the Gli1 gene by the pathway creates a positive feedback loop that can maintain expression of target genes including Gli1 itself. A restraining effect is mediated by the induction of ptch by the pathway, since the Ptch protein is a negative regulator. The amount of Ptch will increase in response to increased Hh ligand, so the pathway is buffered. Too much ligand may be reined in by extra Ptch antagonist.
While these sorts of feedback controls are important, they do not cope with the need to coordinate with all relevant aspects of physiology. In particular, dramatic changes in energy stores occur due to the changing abundance of food. Elaborate mechanisms have evolved for cells to adapt to high or low levels of ATP (Hardie et al., 2012; Inoki et al., 2012). In some tissues, such as fly wing discs and mammalian cerebellum, Hh signaling has powerful growth effects. Embarking on that growth when ATP is scarce is unlikely to succeed and may lead to fatal imbalances among tissues and cell types. Thus activation of AMPK in response to high AMP levels shuts down central processes such as protein synthesis and ion transport (Lang and Foller, 2013), while boosting other processes that increase hardiness and allow survival until ATP stores are rebuilt. AMPK has many target proteins that, together, allow coordinated shut down of energy-demanding activities (Hardie et al., 2012). Its activity can be triggered by stimuli such as exercise (Jessen et al., 2014) cytokines, and hypoxia (Evans et al., 2012). Targets have been identified in several ways, such as whole cell proteomics (Banko et al., 2011), in vitro tests (Gwinn et al., 2008), and genetics (Mihaylova and Shaw, 2011). The full range of targets is not known, but among them are transcription factors such as E2F1 (Yang et al., 2014), Msn2 (Petrenko et al., 2013) and FoxO3a (Greer et al., 2007). A recent paper showed that AMPK negatively regulates Gli1 in hepatocellular carcinoma (HCC) but did not address the mechanism (Abi et al.; Xu et al., 2014). They showed that expression of AMPK is negatively correlated with Gli1 in HCC. Their work provides a useful example of the effect of reduced AMPK function in a tumor; as in our experiments, tumor growth is stimulated when Gli1 is not targeted by AMPK.
Our research shows that GLI1 can be added to the list of direct AMPK targets. S408 is found in a sequence that perfectly matches the AMPK consensus site, while S102 and T1074 (SP and TP) sequences are not perfectly matched to the traditional AMPK consensus site. We examined other kinases such as p38 and JNK, MAPK-type proline directed kinases, and found that their inhibition did not influence Gli1 protein levels (Data not shown). Mutation of each of the individual sites on Gli1 impacts AMPK-mediated phosphorylation activity, and each mutation site may exert effects on other sites (Figures 3h and S3G). Overall, current results show that two sets of “AMPK-dependent” phosphorylation sites including two SP/TP sites (S102 and T1074, Figures S3E, S3F) with high basal stoichiometry and modest inducibility, and one site matching the AMPK consensus (S408, Figures S3E, S3F) with low basal stoichiometry but very high inducibility. Collectively, these AMPK-regulated sites in GLI1 control its protein stability.
Post-translational modification of Gli proteins by other signaling pathways, contributes to the formation of many cancers with elevated Gli activity (Amakye et al., 2013; Hui and Angers, 2011; Niewiadomski et al., 2014). The mTOR/ S6K kinase pathway was shown to enhance Gli1 transcriptional activity (Wang et al., 2012). Since the mTOR/S6K pathway has been reported to be involved in the development of various tumors, targeting mTOR is becoming one of the major methods for cancer treatment. In addition, AMPK has been shown to suppress mTOR/S6K1 activity through direct phosphorylation of mTOR and Tuberous Sclerosis Complex (Kim et al.), the upstream negative regulator of mTOR (Kahn et al., 2005). Given our findings we now know that AMPK both directly and indirectly suppresses the Hh/Gli1 pathway. AMPK conditional knockout models have been created (Viollet et al., 2009), but the role of AMPK in tumorigenesis has not been extensively studied. Loss of AMPK is insufficient to provoke tumor formation in mice, but genetic ablation of the α1 catalytic subunit of AMPK accelerates the development of lymphomas driven by Myc over-expression (Faubert et al., 2013). In contrast, deletion of the AMPKα2 but not the AMPKα1 subunit of AMPK increases susceptibility to H-RasV12 induced transformation in murine fibroblasts (Phoenix et al., 2012), raising the interesting possibility that the AMPKα2 subunit may contribute to tumor suppression in a way that is independent of, or in addition to, the energy-sensing function of AMPK. This suggests that AMPK activity opposes tumorigenesis. Loss of AMPK function evidently fosters tumor progression, perhaps by heightening activities of pathways that spur cell growth and proliferation.
In summary, we found that AMPK inhibits Gli1 protein levels and transcriptional activity. AMPK phosphorylated GLI1 at three novel sites and induced GLI1 protein degradation. Mutation of these three sites into Alanine prolonged GLI1 protein stability, transcriptional activity and oncogenic function (Figure 6). We report here that an energy sensor, AMPK, directly targets the Hh transcriptional activator, GLI1, and suppresses GLI1 activity. Revealing the detailed molecular mechanism of how AMPK modulates Hh pathway will enable us to further understand the coordination between energy metabolism regulation and Hh pathway during development.
Figure 6. AMPK phosphorylates GLI1 and inhibits Hedgehog pathway.
The diagram shows that activated AMPK directly phosphorylates GLI1 on S102, S408, and T1074 sites. Phosphorylation lowers GLI1 protein stability, thus reducing GLI1 transcriptional activity and mitigates cell growth.
Experimental Procedures
Reagents and plasmids
AMPK activators 2DG and metformin were purchased from Sigma and AICAR from Calbiochem and A-769662 were purchase from Selleckchem. Cycloheximide (CHX) and glucose were purchased from Sigma. The dual-luciferase assay kit was purchased from Promega. The Gli1-luciferase reporter contained eight directly repeated copies of the consensus Gli1 binding site (Sasaki et al., 1997). GLI1 HA-tagged, Flag-tagged and pCDH-CMV-MCS-EF1-Puro constructs were gifts from Dr. Mien-Chie Hung (Wang et al., 2012) (UTMDACC, Houston, TX). LKB1 shRNA plasmids were gifts from Dr. Hui-Kuan Lin (UTMDACC, Houston, TX). Mutated constructs derived from the control HA-tagged and Flag-tagged GLI13A and GLI13E were generated using the Quick Change multisite-directed mutagenesis kit from Stratagene. NheI-forward primer: 5' GGCGAGCTAGCATGGACTACAAAGACCATGAC 3' and BstBI-reverse primer: 5' AGTATTCGACACCCCGGATCCTC 3' were used to subclone the GLI1WT, GLI13A and GLI13E into pCDH-CMV-MCS-EF1-Puro.
Immunoblotting and immunoprecipitation assays
Immunoblotting and immunoprecipitation were performed as previously described (Yang et al., 2008), with the following antibodies: Gli1 and GFP (Santa Cruz Biotechnology); Gli1, AMPK, p-AMPK, ACC, p-ACC, p-AMPK/Sub,LKB1, JNK, p-JNK, p-38 and p-p38 (Cell Signaling); Actin, Tubulin, Flag-M2 (Sigma) and HA (Roche); Thiophosphate Ester Specific Ab (Epitomics). The Gli1 antibody used to generate the data shown of Santa Cruz and Cell Signaling antibodies at 1:1000 dilution in 3% milk.
Quantitative real-time PCR
Total RNA was isolated from NIH3T3 fibroblasts and PZp53MED cells using Trizol reagent (Invitrogen). One microgram of RNA was reverse-transcribed with random hexamer primers using SuperScript III reverse transcriptase (Invitrogen). A fraction (1/20) of the resultant cDNA was used as a template for amplification with TaqMan quantitative PCR probes (Applied Biosystems) on an Applied Biosystems 7500 Fast thermocycler: Gapdh (Mm99999915_g1), Gli1 (Mm00494645_m1) and Ptc1 (Mm00436026_m1).
Cell culture, MTT cell growth, and colony formation assay analysis
NIH 3T3 cells were cultured in DMEM supplemented with 10% BCS at 5%CO2. All other cell cultures were kept in DMEM supplemented with 10% FBS at 5% CO2. The concentrations and time for each chemical treatment were as follows: 2DG (25 mM, 4 h) and CHX (1 μg ml−1), unless otherwise noted. The cell growth rate was determined using MTT and cell counting assays (Yang et al., 2008). For colony formation assays, 5 × 104 cells were placed in 1.5 ml DMEM with 10% FBS and 0.3% agarose and overlaid onto 3 ml DMEM with 10% FBS and 0.6% agarose in each well of a six-well plate and medium with or without 25mM 2DG was applied to each well until the end of the assay. After 2–3 weeks, colonies larger than 2 mm in diameter were counted.
Supplementary Material
Acknowledgments
We thank Mien-Chie Hung (University of Texas M.D. Anderson Cancer Center, Houston, TX) for GLI1 HA-tagged and Flag-tagged constructs; Hui-Kuan Lin (University of Texas M.D. Anderson Cancer Center, Houston, TX) for LKB1 shRNA constructs; Keith R. Laderoute for the AMPK MEF cells (SRI International, Menlo Park, CA); Xiaoqi Liu and W. Andy Tao (Purdue University, West Lafayette, IN) for technical support. We also thank Sherri Huang and Ljiljana Milenkovic (Stanford University, Stanford, CA) for commenting on the manuscript. This work was supported by a Showalter Research Scholar grant (207655 to J.-Y. Y.) ; P30 CA023168 to the Purdue University Center for Cancer Research in support of the use of facilities; American Cancer Society Institutional Research Grant #58-006-53 to the Purdue University Center for Cancer Research and Purdue Start-up Fund (J. -Y.Y.). J. -Y.Y. was partly supported by NIH Tumor Biology Training Fellowship (NIH T32CA09151) and Lucile Packard Foundation, Stanford CTSA (UL1RR025744) when the project was initiated in Matthew Scott's Lab. Y.-H.L. was supported by Postdoctoral Research Abroad Program #103-2917-I-564-036, Taiwan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
J.Y.Y designed, performed and coordinated research; J.Y.Y., Y.H.L., J.L., Y.Y.C., V.E.H., J.C., C.C.L. and M.B. performed research; J.Y.Y. analyzed the data; all authors contributed to discussions of results and interpretations, and J.L. and M.P.S wrote part of the manuscript and J.Y.Y. wrote the paper.
References
- Abi KS, Haddad-Zebouni S, Roukoz S, Smayra T, Kamal H, Menassa-Moussa L, Aoun NJ, Ghossain MA. Ultrasound as an adjunct to radiography in minor musculoskeletal pediatric trauma. J Med Liban. 2011;59:70–74. [PubMed] [Google Scholar]
- Alaimo PJ, Shogren-Knaak MA, Shokat KM. Chemical genetic approaches for the elucidation of signaling pathways. Curr Opin Chem Biol. 2001;5:360–367. doi: 10.1016/s1367-5931(00)00215-5. [DOI] [PubMed] [Google Scholar]
- Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal affinity purification of direct kinase substrates. J Am Chem Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nature medicine. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- Banko MR, Allen JJ, Schaffer BE, Wilker EW, Tsou P, White JL, Villen J, Wang B, Kim SR, Sakamoto K, et al. Chemical genetic screen for AMPKalpha2 substrates uncovers a network of proteins involved in mitosis. Molecular cell. 2011;44:878–892. doi: 10.1016/j.molcel.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Evans AM, Peers C, Wyatt CN, Kumar P, Hardie DG. Ion channel regulation by the LKB1-AMPK signalling pathway: the key to carotid body activation by hypoxia and metabolic homeostasis at the whole body level. Advances in experimental medicine and biology. 2012;758:81–90. doi: 10.1007/978-94-007-4584-1_11. [DOI] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell metabolism. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. The Journal of biological chemistry. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature reviews. Molecular cell biology. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henin N, Vincent MF, Gruber HE, Van den Berghe G. Inhibition of fatty acid and cholesterol synthesis by stimulation of AMP-activated protein kinase. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1995;9:541–546. doi: 10.1096/fasebj.9.7.7737463. [DOI] [PubMed] [Google Scholar]
- Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- Jacob LS, Wu X, Dodge ME, Fan CW, Kulak O, Chen B, Tang W, Wang B, Amatruda JF, Lum L. Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling. Science signaling. 2011;4:ra4. doi: 10.1126/scisignal.2001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen N, Sundelin EI, Moller AB. AMP kinase in exercise adaptation of skeletal muscle. Drug discovery today. 2014 doi: 10.1016/j.drudis.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Jishage K, Nezu J, Kawase Y, Iwata T, Watanabe M, Miyoshi A, Ose A, Habu K, Kake T, Kamada N, et al. Role of Lkb1, the causative gene of Peutz-Jegher's syndrome, in embryogenesis and polyposis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8903–8908. doi: 10.1073/pnas.122254599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell metabolism. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Kim JS, Romero R, Tarca AL, LaJeunesse C, Han YM, Kim MJ, Suh YL, Draghici S, Mittal P, Gotsch F, et al. Gene expression profiling demonstrates a novel role for foetal fibrocytes and the umbilical vessels in human fetoplacental development. J Cell Mol Med. 2008;12:1317–1330. doi: 10.1111/j.1582-4934.2008.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Foller M. Regulation of ion channels and transporters by AMP-activated kinase (AMPK). Channels (Austin) 2013;8 doi: 10.4161/chan.27423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Chen HY, Li YW, Wu SY, Wangta L, Lin GH, Hu SY, Chang ZK, Gong HY, Liao CH, et al. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Scientific reports. 2013;3:1176. doi: 10.1038/srep01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell reports. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Chereji RV, McClean MN, Morozov AV, Broach JR. Noise and interlocking signaling pathways promote distinct transcription factor dynamics in response to different stresses. Molecular biology of the cell. 2013;24:2045–2057. doi: 10.1091/mbc.E12-12-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix KN, Devarakonda CV, Fox MM, Stevens LE, Claffey KP. AMPKalpha2 Suppresses Murine Embryonic Fibroblast Transformation and Tumorigenesis. Genes & cancer. 2012;3:51–62. doi: 10.1177/1947601912452883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J. 2007;403:139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Scott JW, Oakhill JS, van Denderen BJ. AMPK/SNF1 structure: a menage a trois of energy-sensing. Front Biosci. 2009;14:596–610. doi: 10.2741/3266. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- Suter M, Riek U, Tuerk R, Schlattner U, Wallimann T, Neumann D. Dissecting the role of 5'-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. The Journal of biological chemistry. 2006;281:32207–32216. doi: 10.1074/jbc.M606357200. [DOI] [PubMed] [Google Scholar]
- Teperino R, Amann S, Bayer M, McGee SL, Loipetzberger A, Connor T, Jaeger C, Kammerer B, Winter L, Wiche G, et al. Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell. 2012;151:414–426. doi: 10.1016/j.cell.2012.09.021. [DOI] [PubMed] [Google Scholar]
- van der Velden YU, Wang L, Zevenhoven J, van Rooijen E, van Lohuizen M, Giles RH, Clevers H, Haramis AP. The serine-threonine kinase LKB1 is essential for survival under energetic stress in zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4358–4363. doi: 10.1073/pnas.1010210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet B, Athea Y, Mounier R, Guigas B, Zarrinpashneh E, Horman S, Lantier L, Hebrard S, Devin-Leclerc J, Beauloye C, et al. AMPK: Lessons from transgenic and knockout animals. Front Biosci (Landmark Ed) 2009;14:19–44. doi: 10.2741/3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer cell. 2012;21:374–387. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick AN, Drury DR, Nakada HI, Wolfe JB. Localization of the primary metabolic block produced by 2-deoxyglucose. The Journal of biological chemistry. 1957;224:963–969. [PubMed] [Google Scholar]
- Wilding M, Coppola G, Dale B, Di Matteo L. Mitochondria and human preimplantation embryo development. Reproduction. 2009;137:619–624. doi: 10.1530/REP-08-0444. [DOI] [PubMed] [Google Scholar]
- Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Liu X, Zheng X, Yao Y, Wang M, Liu Q. The transcriptional activity of Gli1 is negatively regulated by AMPK through Hedgehog partial agonism in hepatocellular carcinoma. International journal of molecular medicine. 2014;34:733–741. doi: 10.3892/ijmm.2014.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nature cell biology. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Park IJ, Yun H, Im DU, Ock S, Kim J, Seo SM, Shin HY, Viollet B, Kang I, et al. AMP-activated protein kinase alpha2 and E2F1 transcription factor mediate doxorubicin-induced cytotoxicity by forming a positive signal loop in mouse embryonic fibroblasts and non-carcinoma cells. The Journal of biological chemistry. 2014;289:4839–4852. doi: 10.1074/jbc.M113.496315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.