Abstract
Cyclic di-AMP (cdiA) is a second messenger predicted to be widespread in Gram-positive bacteria, some Gram-negative bacteria, and Archaea. In the human pathogen Listeria monocytogenes, cdiA is an essential molecule that regulates metabolic function and cell wall homeostasis, and decreased levels of cdiA result in increased antibiotic susceptibility. We have generated fluorescent biosensors for cdiA through fusion of the Spinach2 aptamer to ligand-binding domains of cdiA riboswitches. The biosensor was used to visualize intracellular cdiA levels in live L. monocytogenes strains and to determine the catalytic domain of the phosphodiesterase PdeA. Furthermore, a flow cytometry assay based on this biosensor was used to screen for diadenylate cyclase activity and confirmed the enzymatic activity of DisA-like proteins from Clostridium difficile and Methanocaldococcus jannaschii. Thus, we have expanded the development of RNA-based biosensors for in vivo metabolite imaging in Gram-positive bacteria and have validated the first dinucleotide cyclase from Archaea.
Graphical abstract
Cyclic dinucleotides are an expanding class of second messengers with key roles in bacterial and eukaryotic signaling.1 A recent addition to this class, cyclic di-AMP (cdiA), regulates processes including bacterial sporulation, ion transport, cell wall homeostasis, and central metabolism in many Gram-positive and pathogenic bacteria, and is the only cyclic dinucleotide predicted in Archaea.2,3 In the bacterial pathogens Staphylococcus aureus and Listeria monocytogenes, decreased levels of cdiA result in increased susceptibility to peptidoglycan-targeting antibiotics,4,5 which makes this pathway interesting from a drug targeting perspective. Additionally, secretion of cdiA by L. monocytogenes into the cytosol of infected murine macrophage cells elicits a type I interferon response,6 and further studies have shown the efficacy of cyclic dinucleotides as small molecule adjuvants.7 However, many aspects of cdiA signaling still need to be elucidated, including its homeostasis, metabolic regulatory activity, secretory mechanism, and its function in other microorganisms.
To target this pathway or to further elucidate the role of cdiA in pathogenesis, a robust method for in vivo detection of cdiA is required. Direct methods for detection of cdiA include HPLC-MS, dye intercalation assays, and a competitive ELISA assay,8–10 each of which are limited to in vitro detection. A cell line harboring an IFNβ-luciferase reporter has been used to indirectly detect secreted cdiA.5,6 However, to our knowledge, no sensor for live cell imaging of cdiA has yet been reported.
Recently, we and others have generated novel fluorescent biosensors by combining the ligand-sensing domain of different riboswitches or in vitro selected aptamers with the profluorescent dye-binding aptamer Spinach (Figure 1a).11,12 Here we report the development of two RNA-based biosensors for cdiA that exhibit ligand-induced fluorescence activation in vitro and in vivo. These fluorescent biosensors were used to examine the levels of cdiA in live L. monocytogenes strains by fluorescence microscopy and flow cytometry. To our knowledge, we have generated the first in vivo biosensor for cdiA and demonstrated the first application of RNA-based biosensors in a Gram-positive bacterium. Finally, we have applied the biosensor to detect the activity of putative diadenylatecyclases from Clostridium difficile, a nosocomial human pathogen, and from Methanocaldococcus jannaschii, a thermophilic methanogenic archaea. The latter result provides the first experimental evidence that cyclic dinucleotide signaling extends to Archaea, in addition to bacteria and eukaryotes.1,13
Figure 1.
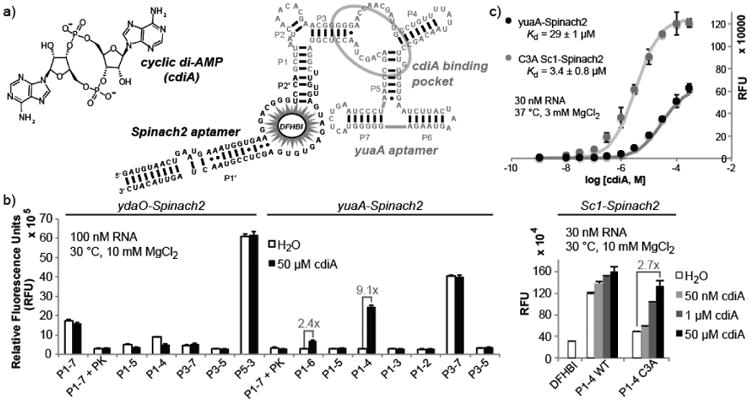
Development of cyclic di-AMP biosensors based on riboswitch-Spinach2 aptamer fusions. (a) Sequence and secondary structure model of yuaA-Spinach2 P1-4 construct. The yuaA riboswitch aptamer (grey) binds cyclic di-AMP and the Spinach2 aptamer (black) binds the profluorescent dye DFHBI. (b) Fluorescence activation screen for the site of Spinach2 attachment to different riboswitch aptamers. Labels indicate the riboswitch stem number followed by the number of base pairs included in the stem. Error bars represent standard deviation between two (Sc1) or three (ydaO, yuaA) independent replicates. (c) In vitro binding affinity measurements for two biosensors for cyclic di-AMP. The average from three independent replicates and best-fit curve are shown. Hill coefficients were 0.94 ± 0.05 for yuaA-Spinach2 and 0.97 ± 0.03 for C3A Sc1-Spinach2. Background fluorescence (without ligand) was subtracted from all data points.
The ydaO riboswitch class recently was identified to bind cdiA with high affinity and selectivity.14 Structural probing experiments did not pinpoint the ligand binding site, although it was shown that the pseudoknot outside of the first pairing stem (P1) was not required for ligand binding.14 Thus, several truncations of the riboswitch aptamer from the ydaO gene in Bacillus subtilis were fused to the Spinach2 aptamer15 and tested for cdiA-dependent fluorescence activation (Figures 1b and S1). However, none demonstrated fluorescence activation. In contrast, two constructs in which the related riboswitch aptamer from the yuaA gene in B. subtilis was fused to Spinach2 showed some fluorescence activation (2.4-fold for P1-6 and 9.1-fold for P1-4). Screening P1-4 variants of other phylogenetic representatives of this riboswitch class did not identify any with improved fluorescence activation (Figure S2). We instead observed several that exhibit consistent fluorescence deactivation, similar to ydaO P1-4 and P1-5. While turn-off biosensors may be useful to pursue in the future, we initially focused on two candidate biosensors with P1-4 stems, yuaA-Spinach2 and Sc1-Spinach2, because they demonstrated high fold turn-on and ligand sensitivity, respectively.
In contrast to yuaA-Spinach2, the Sc1-Spinach2 biosensor displays some fluorescence activation with 50 nMcdiA, but also exhibits much higher background. Several types of destabilizing mutations to the P1-4 stem of Sc1-Spinach2 were made in order to reduce fluorescence background (Figure S3). Replacement of a C-G base pair with an A-G mismatch via the C3A mutation led to a biosensor with 2.7-fold fluorescence activation and stronger binding affinity relative to yuaA-Spinach2 (Figure 1c). Both biosensors responded selectively to cdiA versus other cyclic dinucleotides and adenosine containing compounds at ligand concentrations up to 100 μM. (Figure S4). The lower dissociation constant for C3A Sc1-Spinach2 appears to be due primarily to faster association kinetics (Figure S5).
The x-ray crystal structures of ydaO class riboswitch aptamers recently revealed that two molecules of cdiA are bound in an RNA fold that exhibits pseudo two-fold symmetry.16–18 This finding was unexpected, and in retrospect, attachment to the P1-4 stem removes the 3′ peripheral end of the pseudoknot that forms part of one cdiA binding site. Fortunately, mutation of this binding site reduced ligand affinity only five-fold,16 and our results also corroborate that this binding site is non-essential. However, the importance of this region to the global RNA fold may explain why biosensor constructs are so sensitive to minor changes in P1 stem length, and why most phylogenetic variants are non-functional.
To our advantage, both biosensors harbor a single ligand binding site and display 1:1 stoichiometry of binding to cdiA that gives a good dynamic range of detection (Figure 1c). The in vitro fluorescence signal changed from 10% to 90% between 3.2 to 260 μM cdiA for yuaA-Spinach2 and between 0.37 to 30 μM for Sc1-Spinach2. Together, these biosensors can detect cdiA concentrations spanning four orders of magnitude. However, it should be noted that the fold change in fluorescence is more modest and not directly proportional to the fold change in ligand concentration.
For intracellular detection of cdiA, yuaA-Spinach2 was preferred due to its low background fluorescence. While no in vivo measurements of cdiA levels have previously been made, HPLC analysis of cell extracts of Bacillus subtilis and S. aureus cultures indicate low micromolar concentrations of cdiA in cells.4,8 Thus, this biosensor should be able to detect cdiA at physiologically relevant concentrations.
We were interested in testing the utility of cdiA biosensors in L. monocytogenes, a Gram-positive bacterium and human pathogen that has served as an important model for cdiA signaling. Fluorescence microscopy confirmed that DFHBI was able to permeate through the thicker peptidoglycan layer, and expression of Spinach2 in a tRNA scaffold in turn leads to observable cellular fluorescence in L. monocytogenes (Figure S6). This result also demonstrates that a strong, constitutive promoter provides sufficient expression of Spinach2-based constructs for visualization in bacteria, avoiding the need to engineer an induction system (Figure S7).
To test the ability of the yuaA-Spinach2 fluorescent biosensor to detect cdiA in live cells, plasmid encoding the biosensor in a tRNA scaffold was transformed into strains of L. monocytogenes. For quantitation by flow cytometry, the recently described DFHBI-1T dye19 dramatically improved the resolution (Figure S8). In comparison to wild type, a ΔpdeA mutant carrying a deletion of the pdeA gene that encodes a cdiA phosphodiesterase5 exhibited increased cellular fluorescence (Figure 2a, b). Conversely, decreased cellular fluorescence is observed for the ΔdacAΔrelAPQ deletion strain, which rescues lethality of knocking out dacA, the only diadenylate cyclase gene.20 In contrast, expression and fluorescent visualization of Spinach2 showed no significant change in these strains (Figure S6). Thus, the data are consistent with the yuaA-Spinach2 biosensor giving a fluorescent read-out of differences in endogenous cdiA concentrations for mutant versus wild-type strains of Listeria.
Figure 2.
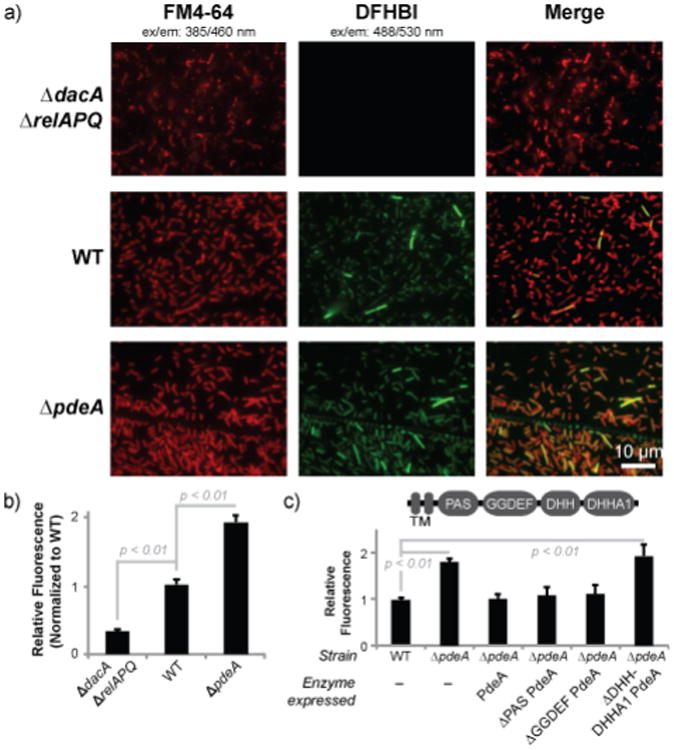
The yuaA-Spinach2 biosensor detects altered cyclic di-AMP levels in Listeria monocytogenes.(a) Fluorescence microscopy images of L. monocytogenes 10403s strains expressing yuaA-Spinach2 tRNAs after incubation with the membrane dye FM4-64 or DFHBI (left and middle panels, respectively; right panels show merge). (b) Flow cytometry analysis of L. monocytogenes strains expressing yuaA-Spinach2 tRNAs. Error bars represent standard deviation between four independent biological replicates. (c) Flow cytometry analysis of WT or ∆pdeA strains with no complementation or complemented with different pdeA enzyme constructs. The domain arrangement for PdeA is shown; the transmembrane (TM) domain was not included in the enzyme constructs. Error bars represent standard deviation between three independent biological replicates.
Furthermore, we showed that the yuaA-Spinach2 biosensor could be employed in a complementation assay in Listeria. The in vivo catalytic activity of pdeA gene constructs harboring different domain deletions was assessed by co-expression with the biosensor in the ΔpdeA mutant background. Whereas the full-length enzyme or truncated versions lacking either the PAS or GGDEF domains rescued phosphodiesterase activity, constructs without the C-terminal DHH and DHHA1 domains showed no rescue of activity (Figure 2c). This result is consistent with the DHH domain being the catalytic domain of the phosphodiesterase, as shown for GdpP phosphodiesterase, a homologue in B. subtilis.21
Finally, we applied the biosensor towards assaying the in vivo catalytic activity of putative diadenylatecyclases (DACs) by heterologous expression in E. coli (Figure 3a). Co-expression of biosensors specific for each bacterial cyclic dinucleotide11,22 with each of three confirmed dinucleotide cyclases demonstrated that each biosensor distinguishes production of specific cyclic dinucleotides (Figure 3b, Table S1). For instance, the yuaA-Spinach2 biosensor exhibited a ∼12-fold fluorescence activation in response to co-expression of a known DAC, DisA from Bacillus subtilis, relative to no enzyme. In contrast, co-expression with WspR led to little change and co-expression with DncV led to a modest change (∼2-fold) in fluorescence that may be due to promiscuous enzyme activity23 or slight cross-sensitivity of the biosensor.
Figure 3.
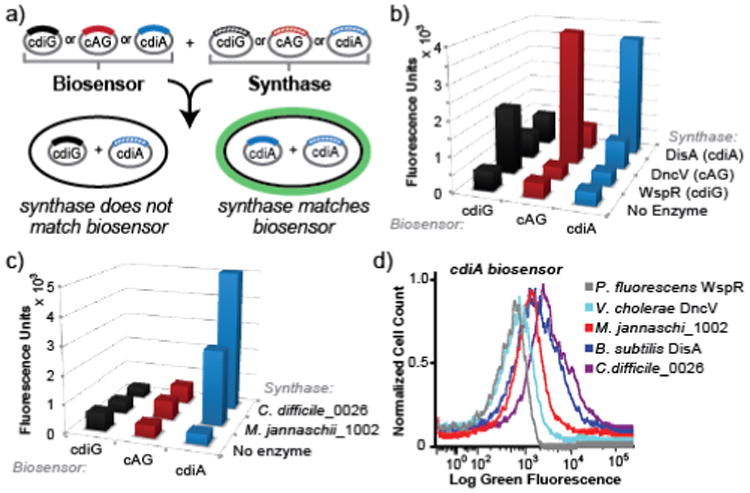
An in vivo activity assay for novel diadenylatecyclases using cyclic dinucleotide-selective biosensors. (a) Schematic of the two plasmid screening system. E. coli is co-transformed with a biosensor-encoding plasmid and an enzyme-encoding plasmid. After induction of expression, cells are incubated with DFHBI-1T and fluorescence is analyzed. (b) Flow cytometry data for biosensors upon coexpression with no enzyme or a known diguanylate cyclase (WspR), cyclic AMP-GMP synthase (DncV), or diadenylate cyclase (DisA). (c) Flow cytometry data for biosensors upon coexpression with predicted diadenylatecyclases from M. jannaschii and C. difficile. Fluorescence values from (b) and (c) are the average of three independent biological replicates and are shown with standard deviations in Table S1. (d) Representative flow cytometry graph for the yuaA-Spinach2 biosensor using logicle scaling on x-axis.
While more than 2,000 DAC domains have been predicted across bacteria and select archaea,24 only a handful of enzymes have been confirmed in bacteria. Furthermore, cyclic dinucleotide signaling has yet to be demonstrated in Archaea. Thus, we analyzed single DAC genes predicted in the genomes of Clostridium difficile (gene id: 0026), a ubiquitous human pathogen, and Methanocaldococcus jannaschii (gene id: 1002), an important methane-producing archaea. Co-expression of either putative DAC genes with the cdiG or cAG-specific biosensor produced no significant fluorescence above background. With the cdiA biosensor, however, ∼13- and ∼7-fold fluorescence activation over background was observed in response to the C. difficile and M. jannaschii enzymes, respectively (Figure 3c, d). These results demonstrate that these enzymes are active DACs, which was confirmed through cell extract analysis (Figure S9), and thus expand cyclic dinucleotide signaling to all three domains of life.
In summary, we have developed RNA-based biosensors for cdiA that enable live cell imaging of this second messenger in Listeria and E. coli. The application of these biosensors has been demonstrated for screening bacterial strains, mapping enzymatic domains, and testing candidate enzymes from heterologous organisms. Most notably, we validated the activity of an archaeal DAC and the only predicted DAC in C. difficile. However, to monitor subtle fluctuations in cdiA levels that may occur in response to cellular and environmental conditions, the sensitivity and fluorescence turn-on properties of these biosensors would need to be improved. Also, robust methods to normalize the fluorescence signal under different conditions will need to be developed.
It is interesting that despite the structural similarity of the three bacterial cyclic dinucleotides, distinct enzyme classes have been found to produce each one. Whether any organisms have evolved other mechanisms to synthesize cyclic dinucleotides remains an open question. We envision that the selective biosensors reported here will enable validation of other predicted dinucleotide cyclases and will potentiate the discovery of previously unidentified signaling domains.
Supplementary Material
Acknowledgments
We thank Zachary Hallberg for assistance in cell extraction assays. This work is supported in part by NIH 1DP2 OD008677 (M.C.H.), NIH 1P01 AI63302 (D.A.P.), 1R01 AI27655 (D.A.P.), DoD NDSEG fellowship (C.A.K.), and NSF graduate fellowship DGE 1106400 (A.T.W.). M.C.H. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Funding Sources: Daniel A. Portnoy has a consulting relationship with and a financial interest in Aduro BioTech, and both he and the company stand to benefit from the commercialization of the results of this research.
Footnotes
Supporting Information: Methods and supplementary data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Danilchanka O, Mekalanos JJ. Cell. 2013;154:962. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corrigan RM, Gründling A. Nat Rev Microbiol. 2013;11:513. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 3.Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, Jurado AR, Goo YA, Sadilek M, Iavarone AT, Sauer JD, Tong L, Woodward JJ. Cell. 2014;158:1389. doi: 10.1016/j.cell.2014.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. MBio. 2013;4 doi: 10.1128/mBio.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodward JJ, Iavarone AT, Portnoy DA. Science. 2010;328:1703. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libanova R, Becker PD, Guzmán CA. Microb Biotechnol. 2012;5:168. doi: 10.1111/j.1751-7915.2011.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. EMBO Rep. 2011;12:594. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai Y, Yang J, Eisele LE, Underwood AJ, Koestler BJ, Waters CM, Metzger DW, Bai G. J Bacteriol. 2013;195:5123. doi: 10.1128/JB.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Sayre DA, Zheng Y, Szmacinski H, Sintim HO. Anal Chem. 2014;86:2412. doi: 10.1021/ac403203x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. J Am Chem Soc. 2013;135:4906. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaap P. IUBMB Life. 2013;65:897. doi: 10.1002/iub.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Nat Chem Biol. 2013;9:834. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strack RL, Disney MD, Jaffrey SR. Nat Methods. 2013;10:1219. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao A, Serganov A. Nat Chem Biol. 2014;10:787. doi: 10.1038/nchembio.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren A, Patel DJ. Nat Chem Biol. 2014;10:780. doi: 10.1038/nchembio.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones CP, Ferré-D'Amaré AR. 2014;33:2692. doi: 10.15252/embj.201489209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song W, Strack RL, Svensen N, Jaffrey SR. J Am Chem Soc. 2014;136:1198. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteley AT, Pollock AJ, Portnoy DA. Cell Host & Microbe in press. doi: 10.1016/j.chom.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. J Biol Chem. 2010;285:473. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellenberger CA, Wilson SC, Hickey SF, Gonzalez TL, Hallberg ZF, Brewer TF, Iavarone AT, Carlson HK, Hsieh YF, Hammond MC. Proc Natl Acad Sci USA. 2015;112:5383. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Cell. 2012;149:358. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romling U. Sci Signal. 2008;1:pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


