Abstract
Despite advances in contemporary chemotherapeutic strategies, long term survival still remains elusive for patients with metastatic colorectal cancer. A better understanding of the molecular markers of drug sensitivity to match therapy with patient is needed to improve clinical outcomes. In this study, we used in vitro drug sensitivity data from the NCI-60 cell lines together with their Affymetrix microarray data to develop a gene expression signature to predict sensitivity to oxaliplatin. In order to validate our oxaliplatin sensitivity signature, Patient-Derived Colorectal Cancer Explants (PDCCEs) were developed in NOD-SCID mice from resected human colorectal tumors. Analysis of gene expression profiles found similarities between the PDCCEs and their parental human tumors, suggesting their utility to study drug sensitivity in vivo. The oxaliplatin sensitivity signature was then validated in vivo with response data from 14 PDCCEs treated with oxaliplatin and was found to have an accuracy of 92.9% (Sensitivity=87.5%; Specificity=100%). Our findings suggest that PDCCEs can be a novel source to study drug sensitivity in colorectal cancer. Furthermore, genomic-based analysis has the potential to be incorporated into future strategies to optimize individual therapy for patients with metastatic colorectal cancer.
Keywords: Oxaliplatin, Murine, Xenograft, Colorectal, Cancer
Introduction
Colorectal cancer is the third most common cancer in the world with approximately 150,000 new cases in the United States each year and ranks second only behind lung cancer as the leading cause of cancer-related deaths (1). At initial diagnosis approximately 20% of patients will have distant metastasis, and another 25-30% of patients with early stage disease will develop metastasis (2, 3). Currently the use of chemotherapy in the metastatic setting is predominantly for disease control and palliation of symptoms. If left untreated, patients with metastatic colorectal cancer have an overall survival of 6 to 9 months, but with combination therapy, survival can be improved to greater than 20 months (4-6). Although the prolonged survival with current combination therapy represents a significant achievement, metastatic colorectal cancer still remains an incurable disease, and new therapeutic approaches are required to improve clinical outcomes.
Therapy based upon the biology of an individual's tumor rather than established histopathological and anatomical classification is an approach which promises to optimize the use of existing therapies and identify novel targets for future therapies. Currently, either a single gene or a small collection of genes is used to determine response to therapeutic agents. Gene expression analysis offers the potential to measure genome-wide gene activity which can be used to complement currently available clinical and biochemical markers in order to identify discrete clinically and biologically relevant phenotypes to better characterize a disease (7, 8). As a result, clinical medicine becomes a data-intensive, quantitative genomic science, and such data can be used to uncover patterns and trends that can distinguish between biological phenotypes in order to help guide existing therapies and discover new therapeutic targets (9-11).
The ability to create a predictive model that can determine which patient may derive the most benefit from a particular agent is the first step in guiding therapy. Previous studies have shown that the NCI-60 cell line panel can be used to create predictive therapeutic models (12-14); however, it remains unclear whether or not responses to therapeutic agents in vitro are predictive of clinical response. Therefore, similarly to the incorporation of new therapeutic agents in the clinical setting, predictive biomarkers must be assessed for their therapeutic potential in preclinical models.
In the past, mouse xenografts have been developed to screen new cancer drugs (15). Initially, athymic mice (nu/nu) and SCID mice were used to establish xenografts from human tumor cell lines in order to test their response to cancer drugs (16). More recently, the direct transplantation of resected human tumors into mice to study sensitivities to therapeutic agents in gastrointestinal cancers has been performed (17, 18). However, it remains unclear whether or not responses to therapeutic agents in vivo are predictive of clinical responses; thus, the need for a clinically relevant preclinical model arises.
In this study, we have developed a predictor of sensitivity to oxaliplatin in order to identify patients who would derive the most benefit from oxaliplatin-based therapy along with a preclinical murine model of patient-derived colorectal cancer explants (PDCCEs) to validate our predictive signature. Together, these approaches describe a widely applicable system that facilitates the preclinical development and characterization of therapeutic agents alone and in combination in order to maximize response to chemotherapeutic drugs and change the current paradigm of clinical cancer therapy evaluation in colorectal cancers.
Materials and Methods
Development of in vitro oxaliplatin sensitivity predictor
An oxaliplatin sensitivity signature was generated as follows. Briefly, the GI50, TGI, and LC50 data for oxaliplatin on the NCI-60 cell line panel obtained from the NCI Developmental Therapeutics Program (19) were compared to determine relative oxaliplatin sensitivity. We subsequently chose cell lines within the NCI-60 panel that would represent the extremes of sensitivity in order to develop an in vitro gene-expression-based predictor of oxaliplatin sensitivity from the pharmacologic data used in the NCI-60 drug screen studies. RMA-normalized expression data from the NCI-60 cell lines were estimated from CEL files downloaded from CellMiner (20, 21) and were used in a supervised analysis using Bayesian regression methodologies to develop a signature for sensitivity to oxaliplatin. Specifically, a Bayesian probit regression model was fit to the most differentially expressed genes, as summarized by the top components of a singular value decomposition. The predictive probability of chemosensitivity was computed as the average of the posterior distribution of the Bayesian model. Complete details are in the supplemental material and methods.
Development of Patient-Derived Colorectal Cancer Explants (PDCCEs)
Colon tumor tissue specimens were obtained from patients (n = 14) with histologically confirmed colorectal cancer who had undergone complete surgical resections at the Duke University Medical Center (Durham, NC) between November 15, 2007, and August 27, 2009. This investigation was approved by the Institutional Review Board of the Duke University Medical Center, and all patients provided informed consent. All specimens were sectioned, stained with H&E, and examined by microscopy by a board certified pathologist. Colorectal tumors resected at the time of surgery were washed with PBS and then minced into 2~3 mm cubes. The samples were then placed in an enzyme medium [RPMI media containing collagenase IV (6 mg/ml), hyaluronidase (1 mg/ml), and deoxyribonuclease (0.25 mg/ml) (Sigma, Hamburg, Germany)] and agitated at room temperature for 18-24 hours. After agitation, the cells were centrifuged at 2000 RPM for 15 minutes at room temperature, washed with PBS, and passed through a 70μM cell strainer (BD Biosciences, Bedford, MA). After washing with PBS, the cells were again centrifuged at 2000 RPM for 15 minutes at room temperature, resuspended in serum-free RPMI/Matrigel mixture (1:1 volume), and then injected into the flanks of 4-week-old female JAX NOD.CB17-PrkdcSCID-J mice.
All mouse experiments were performed in accordance with the animal guidelines and with the approval of the Institutional Animal Care and Use Committee (IACUC) at the Duke University Medical Center.
In vivo oxaliplatin sensitivity assay of PDCCEs
To test the sensitivity of oxaliplatin in the PDCCEs, colorectal cancer cells extracted from previously generated, earlier passaged explants (passages 4-8) were injected subcutaneously into the flanks of five JAX NOD.CB17-PrkdcSCID-J mice (four-week-old female) and measured every 2-3 days with a vernier caliper until the volume of the tumor [V = L×2W×0.52 (L = longest diameter, W = shortest diameter)] reached approximately 500 mm3. The mice were then randomized and treated either with oxaliplatin at a standard dose of 10 mg/kg weekly via intraperitoneal injection or with saline for 2.5 weeks with each group containing five mice each. Tumors were then measured at least 2X/week with a vernier caliper, and both tumor volume and tumor growth inhibition ratio [TGI % = 1 - (average tumor volume of oxaliplatin group)/(average volume tumor of control group) × 100%] were calculated at each time point. At the end of three weeks, the tumors from both groups were harvested and placed immediately in Optimal Cutting Temperature (OCT) compound (Sakura Finetek, Torrance, CA) and frozen on dry ice or placed in formalin overnight and paraffin embedded the next day.
Sample Processing. Fresh Frozen Samples
Frozen PDCCE samples were sectioned at 8μm and placed onto histological slides. An initial section was stained with hematoxylin and eosin (Sigma) for histological characterization of the tissue, and the sample was subsequently macrodissected to ensure > 80% tumor. Approximately 100 μg of tissue was macrodissected, and total RNA was isolated from the homogenized tissue using the RNAase Isolation Kit (Qiagen, Valencia, CA). RNA were quantified using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE), and quality was assessed by spectrophotomeric analysis on an Agilent 2100 Bioanalyzer conductor using the RNA 6000 nano assay Kit (Agilent Technologies, Santa Clara, CA).
3-4 μg of total RNA was used to generate gene expression data. Briefly, first strand cDNA synthesis is generated using a T7-linked oligo-dT primer followed by second strand synthesis. An in vitro transcription reaction is performed to generate the cRNA-containing biotinylated UTP and CTP, which is subsequently chemically fragmented at 95°C for 35 minutes. The fragmented, biotinylated cRNA is hybridized in MES buffer (2-[N]-morpholino-ethansulfonic acid) containing 0.5 mg/ml acetylated bovine serum albumin to Affymetrix GeneChip Human U133A 2.0 arrays at 45°C for 16 hours, according to the Affymetrix protocol (Affymetrix, Santa Clara, CA). Generated CEL files are available at GEO (GSE28691).
Formalin Fixed, Paraffin Embedded Samples (FFPE)
Tumors from the PDCCEs were fixed in formalin overnight and paraffin embedded the following day. FFPE PDCCE samples were sectioned at 10 μm and placed onto histological slides. An initial section was stained with hematoxylin and eosin for histological characterization of the tissue to ensure > 80% tumor. RNA was then isolated from 8-10 μm FFPE sections using the RecoverAll-MagMAX Custom Kit and protocol (Applied Biosystems, Foster City, CA) with the following modifications: RNA isolation digestions were incubated at 50°C for 15 minutes and then at 80°C for 15 minutes; Lysis Binding Solution was reconstituted using 22 ml of 100% isopropanol (Mallinckrodt Chemicals, Phillipsburg, NJ); Wash Solution 1H was reconstituted using 12 ml of 100% isopropanol; and Wash Solution 2 was reconstituted using 44 ml of 100% ethanol (Pharmco-Aaper, Brookfield, CT).
RNA was amplified according to the MessageAmp Premier protocol (Ambion, Austin, TX). Affymetrix DNA microarray analysis was prepared according to the manufacturer's instructions, and targets were hybridized to the Human U133A 2.0 GeneChip (Affymetrix, Santa Clara, CA). Generated CEL files are available at GEO (GSE28691).
Validation of Oxaliplatin Sensitivity Signature
In order to validate the accuracy of the Bayesian probit regression model, first, the RMA-normalized gene expression data of the training data set (NCI-60 Oxaliplatin Sensitivity Signature) and validation data set (PDCCE fresh frozen or PDCCE FFPE samples) were merged together utilizing an in-house program, File Merger (22). Next, the model was used to estimate the relative probabilities and associated measures of uncertainty for each sample in the validation set as described previously (23). Samples scoring below 0.5 were considered belonging to the oxaliplatin-resistant class, while samples scoring above 0.5 were considered belonging to the oxaliplatin-sensitive class. The associations between the oxaliplatin sensitivity predictor and PDCCE TGIs are evaluated using pearson correlation coefficients and two-sided p-values. Complete details are in the supplemental material and methods.
Statistical Analysis
Expression estimates were obtained from the Affymetrix CEL files using MAS5 and RMA (24). To check for sample outliers and batch effects, 3D principal components analysis of the global gene expression was performed. Batch effects were normalized using the ComBat algorithm (25). Unsupervised hierarchical clustering of the human tumors and matching PDCCEs was performed on the 20% of genes with the greatest coefficient of variation. Agglomerative clusters were generated using the pearson correlation coefficient and complete linkage. To determine whether clusters were statistically robust, the AU (Approximate Unbiased) and BP (Bootstrap Probability) values were calculated by 10,000 resamples using the R package: pvclust. The associations between cell-line phenotypes and genomic predictors are evaluated using spearman correlation coefficients and two-sided p-values.
Results
Development of Oxaliplatin Sensitivity Signature
For patients with metastatic colorectal cancer, standard of care first line treatment options are either oxaliplatin or irinotecan-based therapies. However, response rates for either drug regimen range between 40-45% (26). As a first step in the goal to optimize therapy for colon cancer and to determine which patients would benefit from oxaliplatin-based therapy, we employed expression data from NCI-60 cell lines (20) with known sensitivities to oxaliplatin to develop a binary Bayesian model to predict oxaliplatin response. Genes whose expression was most highly correlated with sensitivity to oxaliplatin were identified, and these genes were then used to develop a predictive model that could differentiate between oxaliplatin sensitivity and resistance.
First, we identified NCI-60 cell lines that were most resistant or sensitive to oxaliplatin as defined by their oxaliplatin GI50 (Growth Inhibition of 50%) values while also taking into consideration their TGI (Total Growth Inhibition) and LC50 (concentration of drug resulting in a 50% reduction in the measured protein at the end of the drug treatment as compared to that at the beginning) values. Cell lines with a GI50 < 0.5 μM were considered sensitive, and cell lines with a GI50 > 20 μM were considered resistant (Suppl Table 1). From these cell lines, corresponding RMA-estimated gene expression array data were used for subsequent analysis. However, one sensitive cell line, MCF-7, was observed to be a single-outlier by 3D principal components analysis of global expression values of all of the NCI-60 cell lines (Suppl Figure 1). Because of this, MCF-7 was omitted from the training set prior to developing the predictor (Table 1).
Table 1.
List of NCI-60 Cell Lines in Oxaliplatin Sensitivity Predictor with Corresponding Oxaliplatin Sensitivity Values
| Cell Line | Source | GI50 (μM) | TGI (μM) | LC50 (μM) | Classificationa |
|---|---|---|---|---|---|
| ADR-RES | Ovarian | 0.0317 | 30.4 | 100 | Sensitive |
| SW-620 | Colon | 0.0469 | 83.75 | 100 | Sensitive |
| RPMI-8226 | Leukemia | 0.0527 | 91.2 | 100 | Sensitive |
| MOLT-4 | Leukemia | 0.1365 | 15.63 | 100 | Sensitive |
| HT29 | Colon | 0.1754 | 8.24 | 72.44 | Sensitive |
| CCRF-CEM | Leukemia | 0.3499 | 100 | 100 | Sensitive |
| HCT-116 | Colon | 0.3784 | 100 | 100 | Sensitive |
| NCI-H460 | Lung | 0.4355 | 38.28 | 100 | Sensitive |
| TK-10 | Renal | 21.18 | 73.96 | 100 | Resistant |
| HOP-92 | Lung | 25.47 | 73.96 | 100 | Resistant |
| NCI-H322M | Lung | 35.32 | 100 | 100 | Resistant |
| HOP-62 | Lung | 46.13 | 100 | 100 | Resistant |
| SK-OV-3 | Ovarian | 66.22 | 100 | 100 | Resistant |
| EKVX | Lung | 88.72 | 100 | 100 | Resistant |
| MDA-MB-231 | Breast | 100 | 100 | 100 | Resistant |
| HS 578T | Breast | 100 | 100 | 100 | Resistant |
Cell lines with a GI50 < 0.5 μM were classified as sensitive; cell lines with a GI50 > 20 μM were classified as resistant.
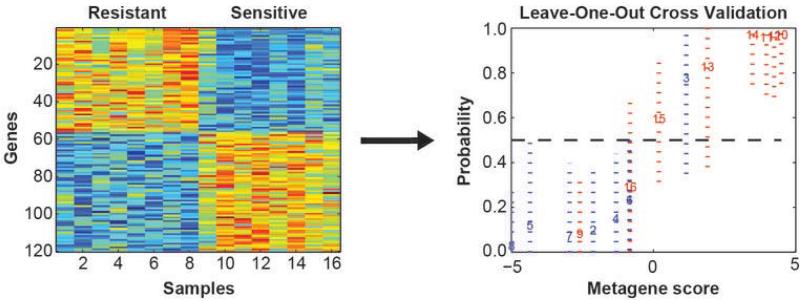
In order to tune parameters in the model to give the largest separation between binary phenotypes, a data-driven empirical approach was taken to select the optimal number of genes to include in the predictor. The discriminatory power was evaluated using the misclassification rate under leave-one-out performance (LOOP) with an a priori defined threshold of Pr ≥ 0.5. Gene set sizes of 50 to 200 were considered. Note: because the LOOP is used to optimize the multi-gene models, no statistical inferences are drawn from the cross-validation, and an independent validation is required to fully assess their predictive value. After consideration, the final developed model consisted of 120 probes (Suppl Table 2) based on prediction of oxaliplatin sensitivity and LOOP (Figure 1).
Figure 1. NCI-60 Oxaliplatin Signature Bayesian Regression Heatmap and Leave-One-Out Performance.
A subset of cell lines that represent the extremes of sensitivity to oxaliplatin were identified from the NCI-60 panel. The left panel is the expression plot for genes selected for discriminating the oxaliplatin sensitive and resistant cell lines from the NCI-60 set based on the expression levels of the 120 genes selected during the Bayesian binary regression analysis. Blue represents lowest expression and red highest. Each column represents an individual cell line and each row one of the discriminating genes in the oxaliplatin predictor. From these 120 genes, phenotype discriminatory power from sample misclassification was optimized using leave-one-out performance (right panel). The acuracy of the predictor on each samples in the leave-one-out performance are displayed with 95% CI.
Development of Patient-Derived Colorectal Cancer Explants
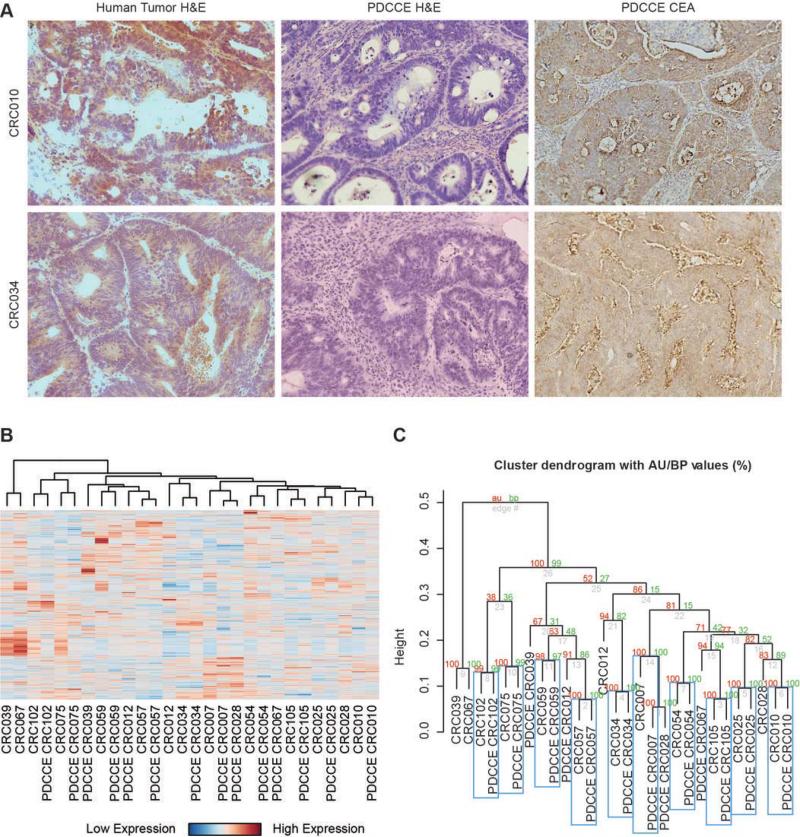
The true value of a predictor lies in its ability to predict sensitivity in an independent in vivo setting. To generate a validation set to test our oxaliplatin sensitivity signature, a murine model was developed by generating human metastasis-derived colorectal cancer explants (PDCCEs) in NOD-SCID mice. Following surgical resection and pathologic assessment, excess tissue to be discarded was immediately processed to generate PDCCEs as described above. A total of 20 resected tumors were injected into SCID mice, and 14 PDCCEs (Table 2) have been established for an uptake rate of 70% (14/20). Of these patients, 9/14 did not receive any chemotherapy prior to surgical resection (neoadjuvant) and 9/14 received chemotherapy after surgical resection (adjuvant). Figure 2a revealed that sections stained with hematoxylin and eosin (H&E) were consistent with adenocarcinoma and that immunohistochemistry (IHC) stains with carcinoembryonic antigen (CEA) were consistent with a colorectal cancer.
Table 2.
Patient-Derived Colorectal Cáncer Explants (PDCCEs) with Human Origin and Chemotherapy History
| Tumor IDa | Primary Site | Metastatic Siteb | Neoadjuvant Chemo. | Adjuvant Chemo. |
|---|---|---|---|---|
| CRC007 | Colon | Liver | none | FOLFIRI |
| CRC010 | Colon | Liver | none | none |
| CRC012 | Colon | Liver | none | N/A |
| CRC025 | Rectal | Lung | none | XELOX |
| CRC028 | Colon | Liver | none | FOLFOX + Bevacizumab |
| CRC034 | Colon | Colon | N/A | XELOX |
| CRC039 | Colon | Liver | FOLFOX + Bevacizumab | none |
| CRC054 | Colon | Lung | Xeloda | FOLFOX + Bevacizumab |
| CRC057 | Colon | Liver | none | FOLFIRI + Bevacizumab |
| CRC059 | Colon | Colon | N/A | none |
| CRC067 | Colon | Liver | FOLFOX + Bevacizumab | Xeloda + Bevacizumab |
| CRC075 | Colon | Liver | none | FOLFOX + Bevacizumab |
| CRC102 | Colon | Liver | XELOX + Bevacizumab | XELIRI + Bevacizumab |
| CRC105 | Colon | Liver | FOLFOX | none |
Human tumors were extracted from their respective metastatic sites and implanted into NOD-SCID mice.
Tumors labeled as “Colon” in the Metastatic Site column are primary colon tumors.
FOLFOX = Oxaliplatin + 5-FU/Leucovorin
XELOX = Oxaliplatin + Xeloda
FOLFIRI = Irinotecan + 5-FU/Leucovorin
XELIRI = Irinotecan + Xeloda
Figure 2. Global Comparison of Human Tumors and Matching PDCCEs.
A. PDCCEs and matching human tumors were sectioned on histology slides and stained with hemotoxalin and eosin (H&E) to confirm presence of tumor tissue. PDCCE slides were also stained for carcinoembryonic antigen (CEA) by immunohistochemistry.
B. Global gene expression from 14 human tumors and matching PDCCEs were estimated using RMA and normalized for batch effects using ComBat. After filtering, samples were subjected to unsupervised hierarchical clustering using pearson correlation and complete linkage. Patient-Derived Colorectal Cancer Explants are prefixed with “PDCCE”.
C. The R library pvclust was used to determine statistical significance of clusters, with Approximate Unbiased (AU) values in red and Bootstrap Probability (BP) values in green. Cluster pairs with statistically significant expression correlations (AU, BP > 95%) are boxed in blue. From this it was found that 71% (10/14) of samples cluster together with statistical significance.
To determine the extent to which the underlying biology of a resected colorectal cancer metastatic tumor is maintained when explanted into a murine model, global gene expression analysis between the matched resected patient colorectal tumor and PDCCE was performed. Initial 3D principal components analysis between the patient tumors and the corresponding explants revealed batch effects; therefore, to minimize these effects, the two sets were subsequently normalized using ComBat. An unsupervised hierarchical clustering was then performed on CV-filtered expression data to generate a heatmap of clustered gene expression (Figure 2b) which revealed that 10/14 (71%) of the matched patient tumor samples and corresponding PDCCEs clustered together with greater than 95% probability under resampling (Figure 2c), suggesting that the global biology between the matched samples are similar.
In vivo validation of the oxaliplatin predictor
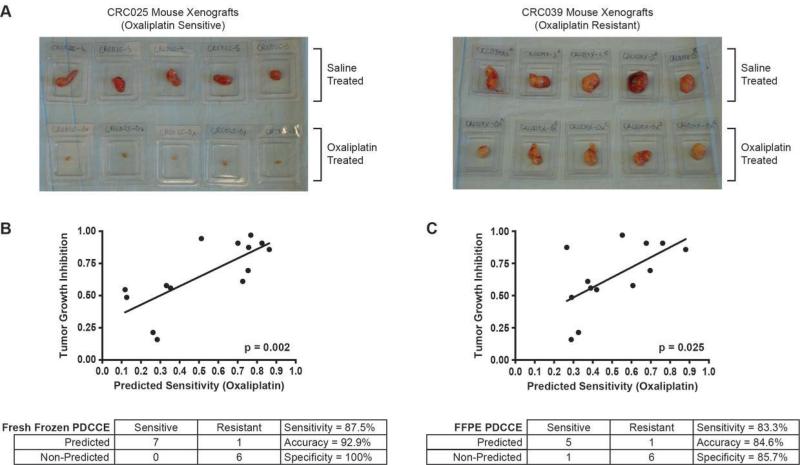
To identify oxaliplatin-sensitive tumors, the 14 PDCCEs were treated with oxaliplatin as described above, and tumor growth inhibition (TGI) was monitored and recorded for each PDCCE during treatment (Suppl Table 3). Figure 3a shows pictures taken at time of PDCCE extraction to illustrate the difference between oxaliplatin-sensitive (e.g. CRC025) and oxaliplatin-resistant (e.g. CRC039) PDCCEs. The cut-off for sensitivity was defined as the arithmetic mean of the TGI values (mean = 0.665). From these studies, 7 PDCCEs were identified as sensitive (TGI % < mean), and 7 PDCCEs were identified as resistant (TGI % > mean).
Figure 3. Metastasis-Derived Colorectal Cancer Explants and Tumor Growth Inhibition Versus Predicted Oxaliplatin Sensitivity.
A. Displayed is a representative sample from each phenotype, with both saline-treated and oxaliplatin-treated PDCCEs, to illustrate differences in oxaliplatin response.
B. In fresh frozen PDCCEs, a Pearson correlation between tumor growth inhibition and predicted oxaliplatin sensitivity was found to be statistically significant (p = 0.002). The signature was found to have an accuracy of 92.9%.
C. In FFPE PDCCEs, a Pearson correlation between tumor growth inhibition and predicted oxaliplatin sensitivity was found to be statistically significant (p = 0.025). The signature was found to have an accuracy of 84.6%.
The accuracy of the oxaliplatin sensitivity predictor was then determined using drug sensitivity data derived from the PDCCEs treated with oxaliplatin. Using the defined cut-off for sensitivity as described above, the oxaliplatin sensitivity predictor was then applied to each PDCCE (Table 3) and was found to have an accuracy of 92.9% (Sensitivity = 87.5%, Specificity = 100%; Figure 3b). Figure 3b shows that there is a statistically significant correlation between predicted probability of oxaliplatin sensitivity and TGI % to oxaliplatin treatment (p=0.002).
Table 3.
List of Fresh Frozen PDCCEs with Corresponding Class and Predicted Response
| Mouse Explant | Tumor Growth Inhibition | Identified Classificationa | Oxaliplatin Predicted Probability | Predicted Responseb |
|---|---|---|---|---|
| CRC007 | 0.907 | Sensitive | 0.826 | Respond |
| CRC010 | 0.875 | Sensitive | 0.757 | Respond |
| CRC012 | 0.486 | Resistant | 0.127 | Non-respond |
| CRC025 | 0.969 | Sensitive | 0.768 | Respond |
| CRC028 | 0.907 | Sensitive | 0.701 | Respond |
| CRC034 | 0.559 | Resistant | 0.353 | Non-respond |
| CRC039 | 0.159 | Resistant | 0.284 | Non-respond |
| CRC054 | 0.546 | Resistant | 0.118 | Non-respond |
| CRC057 | 0.943 | Sensitive | 0.512 | Respond |
| CRC059 | 0.214 | Resistant | 0.262 | Non-respond |
| CRC067 | 0.578 | Resistant | 0.332 | Non-respond |
| CRC075 | 0.694 | Sensitive | 0.754 | Respond |
| CRC102 | 0.858 | Sensitive | 0.863 | Respond |
| CRC105 | 0.611 | Resistant | 0.726 | Respond |
Each sample was identified as either resistant or sensitive to oxaliplatin based on the TGI cutoff of 0.665.
Predicted response to oxaliplatin was determined by the oxaliplatin predicted probability cutoff of 0.5.
In vivo validation of the oxaliplatin predictor in formalin-fixed paraffin-embedded (FFPE) samples
Finally, although microarray analyses are best performed using minimally degraded RNA from freshly collected cell lines or tumor tissue, the challenge of incorporating a genomic signature into the clinical setting is that fresh tissue samples can be limited and therefore constrains our ability to take these studies forward to broad validations of the initially identified predictive profiles. As a result, the ultimate use of these profiles in a clinical diagnostic setting may best be done on standard pathological samples, including formalin-fixed, paraffin-embedded samples (FFPE), and an ability to assay gene expression patterns making use of FFPE samples would clearly represent a major advance and an opportunity to validate the initial signatures.
The oxaliplatin sensitivity predictor was then applied to the FFPE-derived PDCCE tumors as described above to predict their sensitivity to oxaliplatin and was found to have an accuracy of 84.6% (Sensitivity = 83.3%, Specificity = 85.7%; Figure 3c). One FFPE PDCCE sample, CRC057, was omitted from analysis due to not meeting quality control standards (Suppl Figure 2). The mean TGI of the 13 remaining samples was 0.643, and this number was used as the cutoff for classification of sensitivity (Suppl Table 4). Figure 3c shows that there is a statistically significant correlation between predicted probability of oxaliplatin sensitivity and TGI to oxaliplatin (p=0.025).
Discussion
Recent advances in molecular profiling technologies such as gene expression profiling, proteomic profiling, and genetic analysis are currently being utilized to tailor medical care to an individual's needs. In medical oncology, the challenges are to first, develop a method to predict which patient would derive the most benefit from these specific therapies and second, develop a preclinical model to test these therapies. This form of personalized medicine requires the ability to assay tumors for molecular features associated with responsiveness to the proposed therapy along with the development of a reliable preclinical model to test drug sensitivities.
Recent advances in the use of microarrays to assess the entire complement of the expressed genome have documented the power of this method to identify characteristics unique to an individual patient's tumors (8, 10, 11). This information has the potential to best match existing therapeutic agents to individual patient tumors as well as to identify novel therapeutic agents that could be used to treat individual patients. In metastatic colorectal cancer, cytotoxic chemotherapy with an oxaliplatin or irinotecan-based regimen remains the backbone of therapy (26, 27). However, the use of oxaliplatin-based therapy as a standard of care first line therapy results in only a 50-60% response rate (4, 6). Thus, the challenge is develop a predictive marker of oxaliplatin therapy.
Several mechanisms have been proposed to mediate oxaliplatin resistance, including increasing drug efflux and decreased cellular uptake, drug detoxification, apoptosis, and DNA repair (28, 29). In colorectal cancer, in vitro models have implicated genes involved in apoptosis, drug transport, and DNA repair as potential predictive biomarkers of oxaliplatin sensitivity (30-32). ERCC1 mRNA level and polymorphic variants within the ERCC1 gene have been shown to be prognostic markers of colorectal patients treated with oxaliplatin (33, 34). However, it remains unclear whether proteins involved in nucleotide excision repair such as ERCC1 can serve as predictive markers for oxaliplatin sensitivity in patients with colorectal cancer.
In this study, we have used a genomic-based assay to develop a gene signature of oxaliplatin sensitivity. However, genes identified in the predictor did not include previously identified genes known to be involved in oxaliplatin resistance such as ERCC1 and other repair proteins, but instead consisted of genes known to be involved in oncogenesis [epidermal growth factor (EGFR), insulin-like growth factor binding protein (IGFBP7), vascular endothelial growth factor C (VEGFC), platelet-derived growth factor C (PDGFC), and CD44]. These genes were found to have lower expression in oxaliplatin sensitive cell lines and are consistent with studies that show low levels of VEGFA and VEGFC in oxaliplatin-sensitive cell lines (35), and that cetuximab, which targets the EGFR pathway, can overcome oxaliplatin resistance (36).
A frequently used strategy for identifying and testing potentially effective drugs is to administer them to immunodeficient mice that have been implanted with well-characterized cancer cell lines that mimic human malignancies. However, these cell lines, having been derived and repeatedly cultured over many years, may have little resemblance to tumors growing in patients. Thus, a more reliable model is needed. Recently, the direct transplantation of resected gastrointestinal human tumors in mice has been developed as a more comparable model to study drug sensitivities (17, 18), but it remains unclear whether or not responses seen in these models are predictive of clinical response. In our current study, we rapidly engrafted metastatic colorectal cancers into NOD-SCID mice and observed that the biology of the metastasis-derived colorectal cancer explants were similar to their corresponding patient tumors based on gene expression profiling. It was, however, observed that two of the PDCCEs (CRC067 and CRC039) did not cluster with the rest of the samples, suggesting that these tumors may have different biology, but this may be a result of these 2 patients receiving more chemotherapy [FOLFOX (Oxaliplatin/5-FU) + Bevacizumab] than the other 12 patients prior to resection of their cancer (Table 2). As a whole, we feel that these findings provide greater reassurance that the observed antitumor effects in our murine model would be similar to those observed in patients and thus provide a preclinical model to study drug sensitivities and mechanisms.
Our study has now shown both the ability to develop a genomic-based predictor for oxaliplatin sensitivity and also the capability to validate the accuracy of the predictor in a preclinical model. Furthermore, we have also shown that our genomic-based predictors can also be applied to formalin-fixed, paraffin-embedded (FFPE) samples which are much more prevalent in the clinical setting. However, we do realize the limitations of our study. First, in utilizing the NCI-60 cell lines as our training set, there were only two colon cancer cell lines that were classified as sensitive to oxaliplatin and no colon cancer cell lines that were classified as resistant to oxaliplatin. This raises the question as to whether or not a more robust colorectal cancer oxaliplatin sensitivity predictor could be generated if only colorectal cancer cell lines are used. To attempt this, we treated 25 colorectal cancer cell lines with oxaliplatin but found that there was only a 10 fold difference in oxaliplatin IC50s between the most sensitive and most resistant cell lines (data not shown), which is consistent with another published report (31). Furthermore, when attempting to develop an oxaliplatin sensitivity signature from these colorectal cancer cell lines, this signature was not able to predict response to oxaliplatin in the PDCCEs (data not shown). The discrepancy between the performance of the NCI-60 oxaliplatin signature and the colorectal cancer cell line signature in predicting oxaliplatin response in the PDCCE model is most likely due to the greater than 100 fold difference in oxaliplatin IC50 values in the NCI-60 cell lines as opposed to the 10 fold difference in the purely colorectal cancer cell lines, suggesting that in order to develop a reliable and accurate chemotherapy sensitivity predictor, the difference in sensitivity to a drug must be greater than 2 logs.
Second, we must also be very careful in extrapolating results from a preclinical model to potential patient outcome. In our preclinical model, the PDCCEs were only treated for 2.5 weeks, and outcome was measured by response to drug. Although, there are studies in colorectal cancer suggesting that response rate can be a surrogate for survival (37), response rate is still not an accepted end-point in clinical trials. Additionally, although single agent oxaliplatin is not typically used in the initial treatment of colorectal cancer due to poor response rates (2-10%), it must be noted that these trials were mainly small phase II trials (38-40). However, given these limitations, we must be careful with interpreting these results within a clinical setting.
Nevertheless, while this only serves as a proof-of-concept study, it is still a crucial first step in bringing a predictive biomarker to the clinic. However, before a predictive biomarker can become clinically relevant, it must undergo rigorous preclinical testing to gauge its accuracy, reliability, and reproducibility. The next crucial step is the retrospective validation of our oxaliplatin sensitivity predictor in patient samples, and this must be performed on multiple patient samples to further validate the signature's predictive capabilities before it can finally be prospectively tested in a clinical trial. Thus, the strength of our study lies in the power of our preclinical murine model coupled with gene expression technology to identify and test novel combinations of therapeutic agents and also to develop both predictive and prognostic biomarkers which can then be systematically brought forth into the clinical setting. More importantly, this now lays down the foundation for the development and validation of future genomic-based biomarkers in a preclinical model prior to clinical assessment.
Finally, the capacity of a genomic-based signature to predict response in preclinical models begins to define a strategy for personalized medicine and also presents the ability to identify cytotoxic agents that best match individual patients with advanced colorectal cancer. Although these strategies will need to be eventually validated in clinical trials, this model is the first step in evaluating the performance of genomic-signature-based selection in the individualized treatment strategy for patients with metastatic colorectal cancer.
Supplementary Material
Acknowledgements
The authors wish to thank the Duke microarray core facility for collecting the microarray data.
This work was supported by grants from the Charles Scott Research Fund and the Mentored Research Scholars Grant (119824-MRSG-10-195-01-TBG) from the American Cancer Society
Abbreviation List
- FFPE
Formalin Fixed, Paraffin Embedded
- LOOP
Leave one out performance
- PDCCEs
Patient Derived Colorectal Cancer Explants
- TGI
Total Growth Inhibition
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Andre T, Bensmaine MA, Louvet C, Francois E, Lucas V, Desseigne F, et al. Multicenter phase II study of bimonthly high-dose leucovorin, fluorouracil infusion, and oxaliplatin for metastatic colorectal cancer resistant to the same leucovorin and fluorouracil regimen. J Clin Oncol. 1999;17:3560–8. doi: 10.1200/JCO.1999.17.11.3560. [DOI] [PubMed] [Google Scholar]
- 3.August DA, Sugarbaker PH, Ottow RT, Gianola FJ, Schneider PD. Hepatic resection of colorectal metastases. Influence of clinical factors and adjuvant intraperitoneal 5-fluorouracil via Tenckhoff catheter on survival. Ann Surg. 1985;201:210–8. doi: 10.1097/00000658-198502000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:2013–9. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- 5.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–42. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 6.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:4697–705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 7.Golub TR. Genome-wide views of cancer. N Engl J Med. 2001;344:601–2. doi: 10.1056/NEJM200102223440809. [DOI] [PubMed] [Google Scholar]
- 8.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 9.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 10.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 11.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, et al. Diffuse large B- cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 12.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13086–91. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riddick G, Song H, Ahn S, Walling J, Borges-Rivera D, Zhang W, et al. Predicting in vitro drug sensitivity using Random Forests. Bioinformatics. 2011;27:220–4. doi: 10.1093/bioinformatics/btq628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, et al. Chemosensitivity prediction by transcriptional profiling. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10787–92. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suggitt M, Bibby MC. 50 years of preclinical anticancer drug screening: empirical to target-driven approaches. Clin Cancer Res. 2005;11:971–81. [PubMed] [Google Scholar]
- 16.Alley M, Hollingshead MG, Dykes DJ, et al. Human Tumor Xenograft Models in NCI drug Development. In: TBaA PA, editor. Anticancer Drug Development Guide. Humana Press; Totowa: 2004. pp. 125–52. [Google Scholar]
- 17.Pitts TM, Tan AC, Kulikowski GN, Tentler JJ, Brown AM, Flanigan SA, et al. Development of an integrated genomic classifier for a novel agent in colorectal cancer: approach to individualized therapy in early development. Clin Cancer Res. 2010;16:3193–204. doi: 10.1158/1078-0432.CCR-09-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res. 2006;12:4652–61. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 19.dtp.nci.nih.gov. [internet] National Cancer Institute (Developmental Therapeutics Program); Washington DC.: [updated Nov 2, 2011].available from dtp.nci.nih.gov/docs/cancer/cancer_data.html. [Google Scholar]
- 20.Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Molecular cancer therapeutics. 2007;6:820–32. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 21.discover.nci.nih.gov [internet] National Cancer Institute (Genomics and Bioinformatics Group); Washington DC.: [updated Jan 12, 2012]. available at discoverncinihgov/cellminer/. Washington DC. NCI. [Google Scholar]
- 22.filemerger.genome.duke.edu [internet] Duke University; Durham: [updated July 1, 2011]. available at filemerger.genome.duke.edu. [Google Scholar]
- 23.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Tournigand C, Andre T, Achille E, Lledo G, Flesh M, Mery-Mignard D, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22:229–37. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 27.Grothey A, Sargent D, Goldberg RM, Schmoll HJ. Survival of patients with advanced colorectal cancer improves with the availability of fluorouracil-leucovorin, irinotecan, and oxaliplatin in the course of treatment. J Clin Oncol. 2004;22:1209–14. doi: 10.1200/JCO.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nature reviews Drug discovery. 2005;4:307–20. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 29.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–79. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 30.Plasencia C, Martinez-Balibrea E, Martinez-Cardus A, Quinn DI, Abad A, Neamati N. Expression analysis of genes involved in oxaliplatin response and development of oxaliplatin-resistant HT29 colon cancer cells. International journal of oncology. 2006;29:225–35. doi: 10.3892/ijo.29.1.225. [DOI] [PubMed] [Google Scholar]
- 31.Arango D, Wilson AJ, Shi Q, Corner GA, Aranes MJ, Nicholas C, et al. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. British journal of cancer. 2004;91:1931–46. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnould S, Hennebelle I, Canal P, Bugat R, Guichard S. Cellular determinants of oxaliplatin sensitivity in colon cancer cell lines. European journal of cancer. 2003;39:112–9. doi: 10.1016/s0959-8049(02)00411-2. [DOI] [PubMed] [Google Scholar]
- 33.Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H, Danenberg KD, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 34.Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J, et al. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol. 2003;1:162–6. [PubMed] [Google Scholar]
- 35.Dallas NA, Xia L, Fan F, et al. Resistance of colon cancer cells to 5-FU or oxaliplatin enriches for tumor stem cells. Gastrointestinal Cancer Symposium. 2008 Abstract 305. [Google Scholar]
- 36.Prewett M, Deevi DS, Bassi R, Fan F, Ellis LM, Hicklin DJ, et al. Tumors established with cell lines selected for oxaliplatin resistance respond to oxaliplatin if combined with cetuximab. Clin Cancer Res. 2007;13:7432–40. doi: 10.1158/1078-0432.CCR-07-1768. [DOI] [PubMed] [Google Scholar]
- 37.Buyse M, Thirion P, Carlson RW, Burzykowski T, Molenberghs G, Piedbois P. Relation between tumour response to first-line chemotherapy and survival in advanced colorectal cancer: a meta-analysis. Meta-Analysis Group in Cancer. Lancet. 2000;356:373–8. doi: 10.1016/s0140-6736(00)02528-9. [DOI] [PubMed] [Google Scholar]
- 38.Becouarn Y, Ychou M, Ducreux M, Borel C, Bertheault-Cvitkovic F, Seitz JF, et al. Phase II trial of oxaliplatin as first-line chemotherapy in metastatic colorectal cancer patients. Digestive Group of French Federation of Cancer Centers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:2739–44. doi: 10.1200/JCO.1998.16.8.2739. [DOI] [PubMed] [Google Scholar]
- 39.Diaz-Rubio E, Sastre J, Zaniboni A, Labianca R, Cortes-Funes H, de Braud F, et al. Oxaliplatin as single agent in previously untreated colorectal carcinoma patients: a phase II multicentric study. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 1998;9:105–8. doi: 10.1023/a:1008200825886. [DOI] [PubMed] [Google Scholar]
- 40.Machover D, Diaz-Rubio E, de Gramont A, Schilf A, Gastiaburu JJ, Brienza S, et al. Two consecutive phase II studies of oxaliplatin (L-OHP) for treatment of patients with advanced colorectal carcinoma who were resistant to previous treatment with fluoropyrimidines. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 1996;7:95–8. doi: 10.1093/oxfordjournals.annonc.a010489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.