Abstract
Toxicogenomics is proposed to be a useful tool in human health risk assessment. However, a systematic comparison of traditional risk assessment approaches with those applying toxicogenomics has never been done. We conducted a case study to evaluate the utility of toxicogenomics in the risk assessment of benzo[a]pyrene (BaP), a well-studied carcinogen, for drinking water exposures. Our study was intended to compare methodologies, not to evaluate drinking water safety. We compared traditional (RA1), genomics-informed (RA2) and genomics-only (RA3) approaches. RA2 and RA3 applied toxicogenomics data from human cell cultures and mice exposed to BaP to determine if these data could provide insight into BaP's mode of action (MOA) and derive tissue-specific points of departure (POD). Our global gene expression analysis supported that BaP is genotoxic in mice and allowed the development of a detailed MOA. Toxicogenomics analysis in human lymphoblastoid TK6 cells demonstrated a high degree of consistency in perturbed pathways with animal tissues. Quantitatively, the PODs for traditional and transcriptional approaches were similar (liver 1.2 vs. 1.0 mg/kg-bw/day; lung 0.8 vs. 3.7 mg/kg-bw/day; forestomach 0.5 vs. 7.4 mg/kg-bw/day). RA3, which applied toxicogenomics in the absence of apical toxicology data, demonstrates that this approach provides useful information in data-poor situations. Overall, our study supports the use of toxicogenomics as a relatively fast and cost-effective tool for hazard identification, preliminary evaluation of potential carcinogens, and carcinogenic potency, in addition to identifying current limitations and practical questions for future work.
Keywords: Benchmark dose, carcinogens, dose–response, environmental pollutant, genomics, human health risk assessment, mode of action, point of departure, polycyclic aromatic hydrocarbon, transcriptomics
1. Introduction
Regulatory agencies worldwide suffer from a backlog of chemicals in need of human health risk assessment. Some of this backlog is due to chemicals that have few or no data on which to base evaluations. To address this problem, a paradigm shift has been proposed that moves away from examining every apical endpoint towards broadly examining perturbations of pathways using new approaches, such as genomics and high-throughput assays, followed by testing for specific predicted toxicities (National Research Council [NRC], 2007a, 2007b).
Whole-genome microarrays examine the response of all genes within an organism's genome following a chemical exposure (toxicogenomics). The application of bioinformatics tools allows the categorization of these responses into specific biological functions, thus providing insight into the potential hazards of a toxicant. While the use of toxicogenomics at this time cannot fully replace traditional tests, its use in the prediction of a chemical's effects can offer considerable advantages over standard practices, including increased throughput, reduced animal use, details on mode of action (MOA; see Box1 for risk assessment definitions), as well as cost and time savings. Traditional tests are generally both costly and time-consuming: for example, a 2-year rodent cancer assay can cost approximately $2–4 million and take about 3 years per compound (reviewed in Thomas et al., 2007b); the battery of genotoxicity assays costs approximately $60 000 and requires several months. In contrast, a short-term in vivo toxicogenomics experiment could be done much more cost-effectively in a 1-month period. Thus, the application of toxicogenomics has been proposed to alleviate some of the problems associated with data development in support of risk assessment.
Recently, techniques that allow quantitative dose–response analysis from genomics data have been developed that are expected to facilitate the integration of genomics in human health risk assessment. Specifically, Thomas and colleagues (Thomas et al., 2007a, 2011, 2012) developed an approach to derive transcriptional benchmark dose (BMD) values for quantitative risk assessment. Using this approach, they found that the BMDs for the most sensitive toxicogenomics responses (i.e. the pathways, biological functions or biological processes with the lowest median gene expression BMD values) correlate well with BMDs for apical cancer and noncancer endpoints. The rationale for this approach is that these BMD values estimate the doses at which the system begins to be perturbed in response to the toxicant. These approaches are especially useful for chemicals that act via nonselective mechanisms, perturbing multiple different biological pathways (Thomas et al., 2013). In contrast, other chemicals operate through “selective” means (i.e. interacting with specific receptors or signaling pathways), having only one or a few biological processes that they target directly. In these cases, toxicogenomics data can be used to develop detailed MOAs and derive BMDs for key events or molecular initiating events for these toxicants.
In the present study, we incorporate both MOA and BMD approaches to derive points of departure for risk assessment of benzo[a]pyrene (BaP), a by-product of the incomplete combustion of organic materials. BaP is an extensively studied polycyclic aromatic hydrocarbon (PAH) and a well-characterized carcinogen with a genotoxic MOA (summarized in detail below). Following metabolism of BaP by cytochrome P450s (CYPs), metabolites of BaP that escape detoxication can bind deoxyribonucleic acid (DNA) to form adducts that cause mutations, which increase the risk of several forms of cancer. Epidemiological studies and experimental animal studies associate exposure to BaP with an increased risk of several forms of cancer, including tumors in forestomach, oral cavity, liver and lung (International Agency for Research on Cancer [IARC], 2012). Current guideline values for oral exposure to BaP through drinking water range from 0.007 to 0.7 μg/L (World Health Organization [WHO], California Environmental Protection Agency [Cal/EPA], Australia, Netherlands, New Zealand, United States Environmental Protection Agency [US EPA] and Health Canada) (Supplementary Table 1). Because of the extensive research that has been conducted on BaP, it provides an excellent model to compare how toxicogenomics may be useful under both data-rich and data-poor circumstances for hazard and risk assessment.
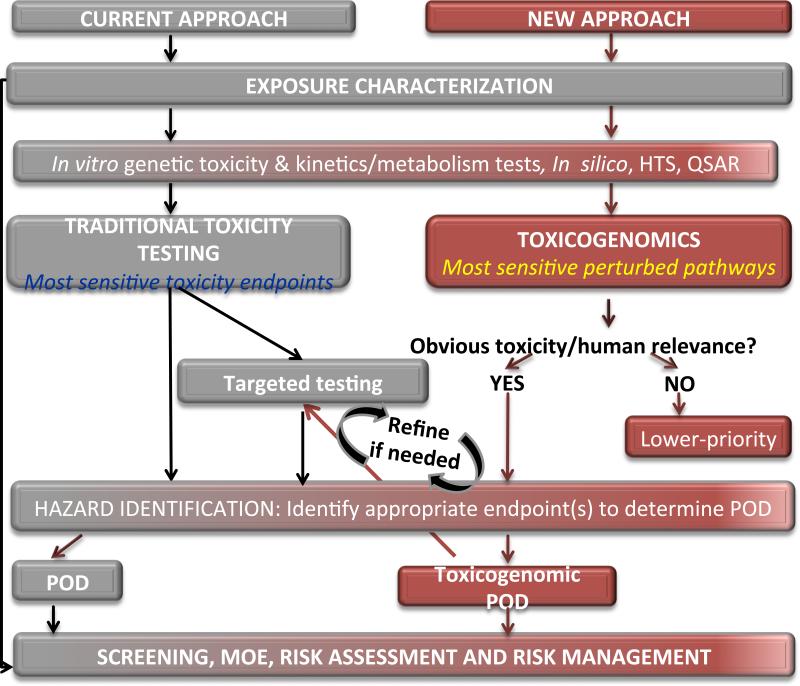
The objective of this case study is to evaluate approaches for utilizing toxicogenomics data in risk assessment. Our study was intended to compare methodologies, not to evaluate drinking water safety. The case study focuses specifically on exposure to BaP in drinking water. To accomplish this goal, we compared three risk assessment approaches (summarized in Figure 1) to derive three separate points of departure (PODs; the lower confidence bound on the lowest dose at which an endpoint significantly deviates from normal levels for studies considered critical in exposure risk) in order to compare current standard approaches with toxicogenomics-informed approaches:
-
1)
Risk assessment 1 (RA1): Traditional approach. A comprehensive literature review was performed, and toxicity data on all apical endpoints (observable outcomes in a tissue, organ system, or entire organism resulting from a toxicant exposure (Krewski et al., 2010)) were collected. A POD was derived based on the most sensitive apical adverse effect reported that was of relevance to humans.
-
2)
Risk assessment 2 (RA2): Genomics-informed approach. Toxicogenomics information from human cell culture and rodent BaP exposures was included in the traditional risk assessment, along with other MOA data, and used to inform the POD selection.
-
3)
Risk assessment 3 (RA3): Genomics-only approach. This approach assumed that no data from standard toxicity testing were available for BaP. Genomics data were used exclusively to provide insight into the MOA and for the selection of a POD.
Figure 1.
Comparison of traditional and genomics risk assessment approaches for determination of a point of departure (POD) for BaP in drinking water. RA1: Traditional risk assessment approach. A comprehensive literature review was performed and a POD was selected based on the most sensitive apical adverse effect reported (Section 2). RA2: Genomics-informed approach. Genomics information was included in the traditional assessment and used to inform the MOA and POD selection (Section 3.1 for general methods and Section 3.2 for RA2-specific details). RA3: Genomics-only approach. This approach assumed a data-poor chemical with little or no information other than genomics information. The definitions of “data-rich” and “data-poor” compounds relate to the amount of toxicity information available for a given compound and may be agency-specific. Only genomics information was used to select a POD for BaP (Section 3.1 for general methods and Section 3.3 for RA3-specific details).
We have focused our risk assessment comparisons on BaP in drinking water, although it should be noted that this work does not represent an evaluation of the safety of BaP in drinking water. The purpose of our work is to compare risk assessment approaches and to provide an example of the potential use of toxicogenomics in risk assessment.
2. Traditional risk assessment approach (RA1)
The information presented below is consistent with the type of information found in Health Canada's technical documents for its Canadian drinking water guidelines; some examples can be found at: http://www.hc-sc.gc.ca/ewh-semt/water-eau/drink-potab/guide/index-eng.php. A literature review was performed to examine sources of BaP exposure, treatment technologies, kinetics and metabolism, human health effects and effects on experimental animals. For potential health effects, Scopus and PubMed databases were searched (1 January 1966 to November 2013) using the following search string:
((CASREGNUMBER(50-32-8) AND LANGUAGE(english)) AND PUBYEAR AFT 1965) AND (TITLE-ABS-KEY(“acceptable daily intake” OR noel OR noael OR “reproductive effect” OR teratogen* OR “developmental effect” OR “neurological effect” OR “endocrine effect” OR “immunological effect” OR “mode of action” OR “mechanism of action” OR pbpk OR toxic OR toxicity OR toxicol* OR neurotox* OR embryotox* OR cytotox* OR hepatotox* OR fetotox* OR genotox* OR maternotox* OR immunotox* OR dermatotox* OR cardiotoxic* OR nephrotoxic* OR ototoxic* OR cancer* OR mutagen* OR carcin* OR tumour* OR tumor* OR malign*))
All of the studies that we evaluated are summarized in Supplementary Table 2 (human health effects) and Supplementary Table 3 (animal health effects). The relevant studies used to determine a POD for human health effects for exposure to BaP in drinking water are discussed below. Relevance was based on potential association with an adverse outcome in humans for animal studies and adequate monitoring of BaP exposure concentrations for epidemiological studies, as well as various other parameters indicating general study quality (e.g. control for confounders, sufficient population size and appropriate data collection protocols).
2.1 Use, identity and sources of BaP exposure
BaP is a by-product of the incomplete combustion of organic substances. Thus, the vast majority of BaP released in the atmosphere results from anthropogenic activity (e.g. burning of fossil fuels for industrial applications, transportation, waste incineration) and, to a lesser extent, natural processes (e.g. forest fires, volcanic eruptions). Additional sources of BaP affecting water and soil include oil spills, municipal effluents and urban and agricultural runoff. BaP is not manufactured in Canada. Physicochemical properties of BaP (Table 1), such as low water solubility, low vapor pressure and elevated octanol–water partition coefficient (log Kow), strongly favor the adsorption of BaP onto particles, soil and sediment (Mackay and Paterson, 1991). As such, approximately 82% of BaP is estimated to partition to soil, 17% to sediment, 1% to water and < 1% to air, according to a multimedia transport model (Hattemer-Frey and Travis, 1991). However, exposures through a variety of the above-mentioned environmental vehicles have been identified as potentially important sources of exposure of humans to BaP (refer to Supplementary File A for information on exposures through soil, air, as well as, food and comsumer products).
Table 1.
Physicochemical properties of benzo(a)pyrene.
| Property | Value |
|---|---|
| Chemical Abstracts Service registry number | 50-32-8*†‡±¥ |
| Chemical formula | C20H12*†‡±¥ |
| Molecular weight (g/mol) | 252.3*†‡±¥ |
| State at ambient temperature | Solid, plates or needles*†± |
| Vapor pressure (mmHg) | 5.6 × 10–9 (20°C)†; 7.47 × 10–7 (25°C)†; 5.49 × 10–9 (25°C)‡±¥ |
| Half-life (days) | 84.6 (river, calculated)¥, 931.3 (lake, calculated)¥ |
| Photolysis half-life in water (hours) | 0.54 (irradiation in upper layer of clear water, partitioning to sediment will significantly increase half-life)‡ |
| Henry's Law constant | 0.034 Pa·m3/mol (20°C*); 4.9 × 10–7 atm·m3/mol†; 1.13 × 10–6 atm·m3/mol‡; 4 5710–7 atm·m3/mol (25°C)±¥ |
| Boiling point | 310–312°C (10 mmHg)*†±; 495°C (760 mmHg)*†‡¥ |
| Melting point | 176.5–179.3°C*†‡±¥ |
| Specific gravity | 1.351†± |
| n-Octanol–water partition coefficient (log Kow) | 6.35*; 6.06 (25°C)†; 5.97‡; 6.13 (25°C )±¥ |
| Water solubility | Slightly, 0.001 62 mg/L (25°C)*‡±¥; 3.8 × 10–6 g/L (25°C), 2.3 × 10–3 mg/L† |
| Volatility | Poor† |
| Taste and odor threshold | No data† |
CHEMFATE (2013): http://esc.syrres.com/scripts/CASCFcgi.exe?CASNUM=50328
HSDB Hazardous Substances Data Bank (HSDB): http://toxnet.nlm.nih.gov (search term 50-32-8).
US EPA. (2013). Estimation Programs Interface Suite™ for Microsoft® Windows, v4.11. United States Environmental Protection Agency, Washington, DC, USA.
BaP primarily enters source waters through atmospheric deposition. Mean concentrations of BaP in the Great Lakes range from 0.03 to 0.7 ng/L (Environment Canada, 1991). Given BaP's strong association with soil, it is not expected to leach into groundwater. Nevertheless, concentrations as high as 0.32 μg/L have been reported in groundwater in proximity to a New Brunswick wood preserving plant (Intera Technologies Ltd et al., 1989).
BaP primarily enters drinking water by leaching from asphaltic or bituminous (coal tar) lining of water storage tanks and distribution system pipes. Pipes and tanks installed prior to the 1960s may contain this type of lining and may still be a source of BaP in drinking water. Factors such as disinfectant dose and type, hydraulic disturbances and increased residence time of the water in the distribution system can contribute to the release of BaP into the water (Maier et al., 2000). Canadian drinking water is estimated to contain BaP at concentrations of < 0.005–3 ng/L (Canadian Council of Ministers of the Environment [CCME], 2010), whereas the concentration of BaP in US drinking water is typically 0.55 ng/L (Santodonato et al., 1981). Global concentrations of BaP in drinking water range from < 0.04 to 914 ng/L, as reported by WHO (2003). Thus, based on a daily water intake of 1.5 L, a body weight (bw) of 70 kg for the average adult and the global concentrations of BaP in drinking water reported by WHO (2003), daily BaP ingestion is calculated to be 0.000857–19.6 ng/kg bw.
2.2 Analytic methods and treatment technology
A number of analytical methods are available for the routine analysis of BaP in drinking water. These methods include liquid-based extraction followed by gas chromatography/mass spectrometry or high-performance liquid chromatography/ultraviolet/fluorescence detection, with method detection limits as low as 16 ng/L in the methods, approved by the US EPA for routine monitoring (US EPA, 1990,1995, 2003, 2009).
Conventional treatment (i.e. coagulation, sedimentation and filtration) can reduce the concentration of BaP to < 0.001 mg/L in the drinking water at the treatment plant. Adsorption technologies such as granular activated carbon and powdered activated carbon are capable of reducing BaP concentrations in drinking water to below 0.2 mg/L and 10 mg/L, respectively. Point-of-use treatment devices such as activated carbon filters are also capable of removing BaP at the tap (US EPA, 2007). The leaching of BaP from coal tar–lined pipes in the distribution system can also be minimized by managing distribution system operations and water quality and minimizing water age (Maier et al., 2000).
2.3 Absorption, distribution, metabolism,excretion, and pharmacokinetics
Each section is presented in relation to the route of exposure, which plays an important role in the potential effects induced by BaP exposure. Overall, it is clear that BaP is rapidly metabolized during the first few hours after oral exposure. Uno et al. (2004) analyzed the clearance of BaP in the blood of mice following gavage exposure, and showed that the levels of BaP in the blood increased until 45 minutes post-exposure and were undetectable by four hours post-exposure. Oral BaP is known to be detoxified in the small intestine very efficiently (Uno et al. 2006 and Nebert et al., 2013a), where Cyp1a1 gene expression persists for over 30 days post-exposure (Shi et al., 2010a), implying that the major contributor to BaP detoxication occurs in the small intestine. While the small intestine plays a major role in detoxifying BaP, BaP distribution to distal organs following oral exposure also occurs. The literature shows that BaP is absorbed in the stomach lining upon ingestion and is absorbed into the circulatory system or into the hepatic portal vein.
2.3.1 Absorption
BaP is rapidly absorbed following administration by oral, inhalation and dermal routes. The delivery vehicle (the substance in which the BaP is administered, such as the oil/fat content of the gastrointestinal tract) significantly affects BaP absorption by the oral, inhalation and dermal routes. Based on the broad absorption profile of BaP, it is clear that major tissues and organs involved in its absorption come into direct contact with the compound. Therefore, the widespread distribution of BaP contributes to its toxicity across numerous tissues. Absorption of BaP is described in greater detail in Supplementary File A.
2.3.2 Distribution
Available reports unequivocally confirm the distribution of BaP and its metabolites to major organs and tissues following intravenous, oral, inhalation and dermal exposure (reviewed in Agency for Toxic Substances and Disease Registry [ATSDR], 1995). BaP is rapidly distributed to the stomach in mice exposed to BaP via oral or topical administration (Carlson et al., 1986). Oral BaP exposure in rats leads to more rapid distribution of BaP to blood plasma compared with inhalation exposure; for example, BaP concentrations in the serum peaked 1 and 8 hours following oral and inhalation exposures to BaP, respectively (Ramesh et al., 2002). Lung and liver were the major organs in which BaP metabolites were detected following oral, inhalation and intravenous exposures of rats to BaP (Ramesh et al., 2002). The distribution of BaP to various organs following exposure, and its subsequent elimination within 24–72 hours, suggests rapid metabolism and excretion of BaP and its metabolites in rodents.
The lipophilicity of absorbed BaP favors its association with lipoproteins in the blood (Busbee et al., 1984), enabling BaP to rapidly access distant organs via the systemic circulation. For example, 5 minutes following intravenous administration of 15 mg of 14C-labelled BaP per kilogram body weight in rats, the liver contained the highest amount of radioactivity, followed by lung, heart, kidney, blood, brain, spleen, testes and adipose tissue (Moir et al., 1998). Another study (Marie et al., 2010) confirmed the distribution of BaP to various organs and tissues in rats following intravenous administration; 2 hours after administration, the largest proportion of the administered BaP was found in the lungs (17.0%), followed by skin (7.6%), adipose tissue (2.5%), liver (2.3%) and kidney (0.5%). These results suggest that BaP is translocated readily to other organs via circulation, potentially leading to systemic effects.
Overall, multiple studies demonstrate that BaP is widely distributed across various organs and tissues following oral, topical, inhalation and intravenous exposures, such that a considerable fraction of the administered dose is bioavailable. Vehicle is an important factor for BaP distribution. The broad distribution profile of BaP aids in the interpretation of the diverse health effects induced by BaP, described in detail below. Distribution of BaP is described in greater detail in Supplementary File A.
2.3.3 Metabolism
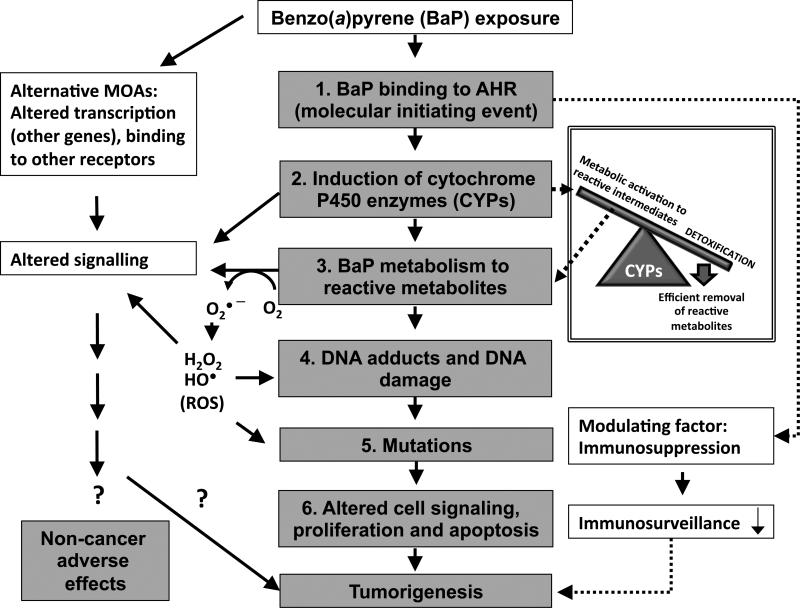
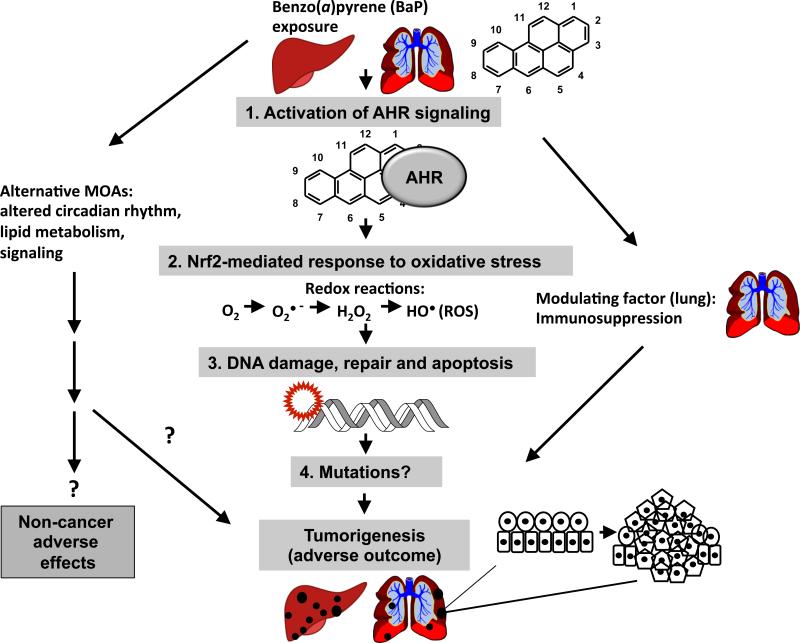
The metabolism of BaP was systematically reviewed recently (IARC, 2010) and is briefly summarized here. BaP is subject to phase I (activation) and phase II (conjugation/detoxication) xenobiotic metabolism reactions, driven by its interaction with the aryl hydrocarbon receptor (AHR).
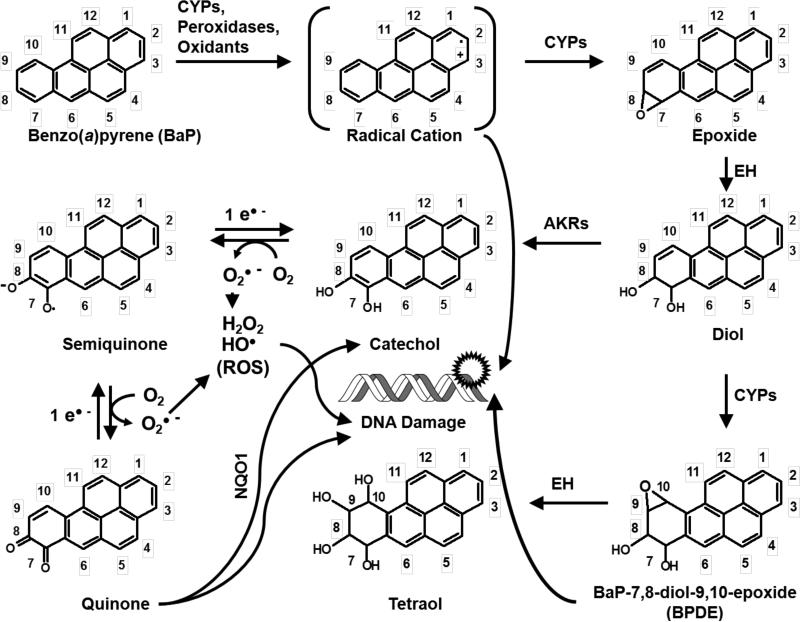
In phase I activation, BaP is converted to an epoxide by CYP enzymes (primarily CYP1A1 and CYP1B1) and other enzymes (e.g. prostaglandin synthase) (Trush et al., 1982). BaP epoxides undergo hydration by epoxide hydrolase (EH) to form diols (Figure 2). Aldo-keto reductases (AKRs) convert BaP diols to catechols. Most notably, the BaP-7,8-dihydrodiol-AKR pathway generates BaP 7,8-catechol that autooxidizes to produce DNA-reactive BaP-7,8-quinone (Lan et al., 2004). Secondary epoxidation by CYPs yields diol epoxides. BaP-7,8-diol-9,10-epoxide (BPDE) is the most mutagenic and well-studied BaP diol epoxide, forming covalent adducts with DNA at the N2 position of deoxyguanosine (Fang et al., 2001). Alternatively, CYPs, as well as some chemical oxidants and peroxidases, can also catalyze one-electron oxidation of BaP to form a BaP radical cation (Cavalieri and Rogan, 1992) with an electrophilic C-3 position (Figure 2). Oxygen transfer to C-3 by CYPs can give rise to 3-hydroxybenzo[a]pyrene (3-OH BaP) (Cavalieri and Rogan, 1995), which is proposed as a biomarker of dermal exposure to BaP in humans (Payan et al., 2009). The primary metabolites of BaP in vivo as well as the main products of BaP detoxication are thought to be glucuronide conjugates of BaP (Recio and Hsie, 1984) and 3-OH BaP (Saunders et al., 2006. In addition to 3-OH BaP, other BaP phenol metabolites have been reported, such as 6-hydroxybenzo[a]pyrene (6-OH BaP) (Cavalieri and Rogan, 1995) and 9-hydroxybenzo[a]pyrene (9-OH BaP) (Saunders et al., 2006). Phenols such 6-OH BaP can yield autooxidation products BaP-1,6-, -3,6-, -6,12-diones (Cavalieri and Rogan, 1995). BaP radical cations are genotoxic (DNA-reactive) species and form unstable adducts with purinic bases, resulting in apurinic sites (IARC, 2010). Genotoxic BaP metabolites can result in mutations in proto-oncogenes, eventually leading to tumorigenesis (IARC, 2010). In addition, CYPs can undergo “uncoupling” of their catalytic reactions, during which an electron “leaks out” of the substrate-bound catalytic site of an enzyme into the subcellular milieu and is accepted by molecular oxygen (O2), creating superoxide (O2•−) and other reactive oxygen species (ROS), including hydrogen peroxide (H2O2) and hydroxyl radicals (HO•) (Shertzer et al., 2004). ROS can also be generated during BaP catechol–quinone redox cycling (Figure 2). ROS are genotoxic and can produce mutations (primarily G to T transversions), contributing to the overall mutagenicity and carcinogenicity of BaP (Lan et al., 2004). Thus, both metabolites of BaP and ROS can interact with DNA and cause mutations following BaP exposure.
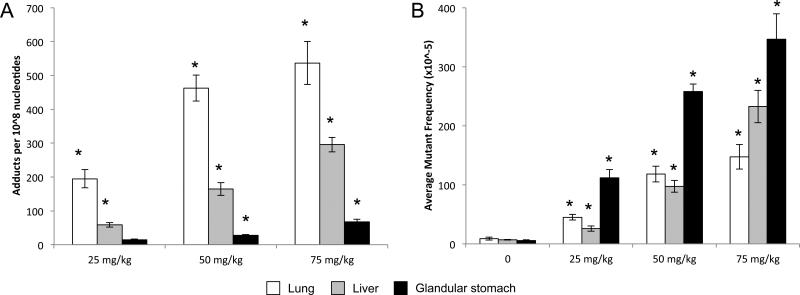
Figure 2.
DNA adduct formation (a) and lacZ mutant frequency (b) in the lungs, livers, and glandular stomach from Muta™Mouse exposed to 25, 50, and 75 mg/kg-bw per day BaP for 28 consecutive days and excised 3 days post-exposure. Levels of dG-N2-BPDE adducts were determined using the nuclease P1 enrichment version of the 32P-postlabeling method. Data are represented as average ± SEM (n = 5 mice/group). Average lacZ mutant frequency was determined using the P-Gal positive selection assay. Values shown are average frequencies × 10−5 ± SEM. Asterisk (*) indicates significance (p < 0.05) compared with controls. Please note, no adducts were detected in mice dosed with vehicle control. All data were previously published in Lemieux et al., 2011; Malik et al., 2012; and Labib et al., 2012.
During phase II xenobiotic metabolism reactions (or detoxication), BaP metabolites are conjugated with hydrophilic moieties (glutathione, glucuronic acid or sulfate) to enhance their solubility in water, rendering them suitable for urinary and fecal excretion (Bock and Bock-Hennig, 2010; Garg et al., 2008; Meinl et al., 2008). Phase II metabolism enzymes include glutathione S-transferases, (GSTs), uridine diphosphate–glucuronosyl transferases (UGTs) and sulfotransferases (SULTs). Detoxication of BaP is efficiently conducted in the small intestine by CYP1A1 upon oral exposure (Uno et al., 2006 and Nebert et al., 2013a). It appears that tight coupling between phase I and phase II metabolism is required to reduce the detrimental effects of BaP exposure, as shown by knockout studies in mice (reviewed in Nebert and Dalton, 2006). This coupling is achieved through activation of the AHR and nuclear factor (erythroid-derived 2)-like 2 (Nrf2 or Nfe2l2). These transcription factors regulate the expression of phase I and phase II metabolism enzymes, respectively, with some apparent functional redundancy (i.e. several enzymes are regulated by both factors).
The AHR is the principal transcription factor governing the activation of most phase I and some phase II enzymes and has been studied extensively (Michaelson et al., 2011). Upon binding of BaP to the AHR, the BaP–AHR complex translocates to the nucleus of the cell, where it dimerizes with the AHR nuclear translocator protein. Once in the nucleus, the complex can then bind to the promoters of genes containing AHR response elements to regulate their expression, including phase I and phase II xenobiotic metabolizing enzymes. Thus, BaP binding to the AHR drives its own metabolism. Genetic polymorphisms in AHR that affect the receptor's ability to bind ligands impact the sensitivity of animals to AHR ligands, including the extent of CYP1A1 induction (reviewed in Okey et al., 2005). However, the significance of AHR polymorphisms in humans and their influence on AHR ligand binding is still unknown (Okey et al., 2005). In general, humans are typically less affected than mice and rats following exposure to AHR ligands, perhaps due to a lower binding affinity of human AHR to its ligands compared with rodents (Okey et al., 2005). Sequencing of the AHR gene of 108 people from six ethnic backgrounds revealed six single nucleotide polymorphisms primarily outside the ligand binding domain (Rowlands et al., 2010). However, it is possible that differential ligand binding affinity resulting from polymorphisms in the AHR gene in humans may contribute to differences in susceptibility to AHR-mediated effects of BaP among individuals, but more research is needed to clarify this hypothesis. In addition, it is conceivable that polymorphisms in other genes that belong to the AHR signaling pathway (e.g. AHR repressor [AHRR]) may also contribute to differential sensitivity to BaP across species and individuals.
Nrf2 is the master regulator of inducible gene expression of phase II enzymes (reviewed by Kaspar et al., 2009). Nrf2 is activated by various pro-oxidants (electrophiles and ROS) and is regulated by subcellular localization, such that it is kept inactive in the cytoplasm under normal homeostatic conditions by interaction with the protein Kelch-like ECH-associated protein 1 (reviewed by Mitsuishi et al., 2012). Nrf2 activation triggers its nuclear translocation and subsequent activation of Nrf2-driven gene expression. Nrf2 deficiency leads to increased formation of DNA adducts and probability of carcinogenesis, whereas increased Nrf2 activity decreases the probability of chemically induced carcinogenesis in mice (reviewed by Mitsuishi et al., 2012). Mutations in Nrf2 and Keap1 that affect Keap1–Nrf2 interaction have been identified in various human cancers (reviewed by Mitsuishi et al., 2012).
Finally, male and female rats appear to metabolize BaP differently. For example, significantly lower amounts of metabolites were detected in the feces and urine of females relative to males following oral exposure (Van de Wiel et al., 1993). This sex-specific difference in BaP metabolism (albeit reported only in rats, to the best of our knowledge) may contribute to the greater incidence of BaP-induced liver (Lavoie et al., 1987; Wislocki et al., 1986) and lung (Lavoie et al., 1987) tumors in male rodents. In general, the very short half-life of BaP in rats (about 1, 4 and 6 hours for oral, dermal and inhalation exposures, respectively [Carlson et al., 1986; Ramesh et al., 2002], and approximately 2–5 hours in mouse blood after oral administration [Uno et al., 2006]) suggests that BaP is rapidly metabolized and excreted. However, following oral administration a small amount of BaP escapes detoxication and induces mutations (refer to section 2.5.7.2 Mutations).
2.3.4 Excretion
In general, BaP is eliminated rapidly from the body through urinary and fecal excretion, as evidenced by the detection of its metabolites in urine and feces. BaP clearance from tissues with relatively high lipid content is prolonged (from hours to several days). Only minor traces of BaP were detectable in most tissues in intravenously exposed rats 32 hours postinjection, whereas there was some lag time in the elimination of BaP from adipose tissue, brain and testes (Moir et al., 1998). Inefficient elimination of BaP from adipose tissue could be due to poor circulation to the tissue in combination with the lipophilicity of BaP, which may result in its prolonged residence in the adipose tissue. In addition, BaP metabolites are highly retained in the brain (Saunders et al., 2003), potentially as a result of the high lipophilicity of this tissue. This may provide insight into the manifestation of BaP toxicity in distal organs and tissues, such as brain and testes. Excretion of BaP is described in greater detail in Supplementary File A.
2.3.5 Physiologically based pharmacokinetic (PBPK) modeling
The currently available PBPK models are not adequately developed to describe the series of animal experiments studying the pharmacokinetics of BaP. Only one PBPK model has been developed specifically for BaP (Crowell et al., 2011). This study presents a PBPK model to describe available time series data from the scientific literature on BaP in rats and mice. However, the model fails to use appropriate physiological values. In particular, the cardiac output does not match the reference values cited by Brown et al. (1997), and the blood flow values applied were incorrect. Even after correcting for errors in blood flow values, the PBPK model predictions are not consistent with the data presented and validated in Crowell et al. (2011). Extrapolation of a PBPK model for the pharmacokinetics of pyrene (Haddad et al., 1998) to BaP yielded inconsistent results. Heredia-Ortiz et al. (2011) presented an alternative toxicokinetic compartmental model to describe the pharmacokinetics of BaP in rats. While the model is consistent with experimental data on rats, the model uses rate constants instead of physiological parameters. These rate constants are consistent with the fitted data only and cannot be extrapolated to other species, such as humans and mice, or to dose conditions beyond those observed in the study. Thus, the available PBPK models are not adequately developed to describe the pharmacokinetics of BaP.
2.4 Human health effects
Overall, 74 epidemiological studies of chronic BaP-containing complex mixtures were reviewed in detail (Supplementary Table 2). These include primarily studies on inhalation of BaP-containing complex mixtures in occupational settings and ingestion of BaP-containing complex mixtures from charred and burnt meats. Twenty studies (summarized in Table 2) were selected on the basis of having reported specific exposure concentrations as well as various other parameters that indicated general study quality (e.g. control for other risk factors and/or confounders, sufficient population size and appropriate data collection protocols). Many potentially adverse health effects of BaP exposure at doses that varied considerably between studies were found. Cancer was the most common outcome and was associated with BaP doses as low as 850 ng/m3 -year (i.e. for lung and stomach cancer) and 10.4 ng/day (i.e. for pancreatic cancer) for chronic inhalation and oral exposure, respectively. The most sensitive of the endpoints was neurological function; exposure to BaP via inhalation at 19.5 ng/m3 led to a general decrease in learning, memory and neurotransmitter levels. The most sensitive effect that was correlated with oral exposures was pancreatic cancer (i.e. dose of 10.4 ng/day), as measured by daily meat intake and preparation methods. Thus, there is sufficient evidence that exposure to complex mixtures containing BaP is associated with excess cancer risk in humans. However, due to co-exposure to chemicals other than BaP, as well as the variability in air concentrations and oral doses of BaP at which these effects occur, route of administration (exposure) and subsequent target organ affected, it is not possible to conduct dose-response analysis to determine acceptable exposure levels for risk assessment in humans. In addition, genetic cancer risk prediction in individuals exposed to an environmental toxicant is extremely difficult and defined as a multifactorial trait (Nebert et al., 2013b). For these reasons, our risk assessment of BaP relied more heavily on animal data at the population level.
Table 2.
Human health effects potentially associated with benzo(a)pyrene as a component of a complex mixture exposure.
| Exposure | LOAEL* | Duration | Response | Reference |
|---|---|---|---|---|
| Air | 136.6 mg/m3·year | Chronic | Bladder cancer | Armstrong et al. (1986) |
| 100 μg/m3·year | Chronic | Lung cancer | Armstrong and Thériault (1996) | |
| 35.60 μg/100 m3 | Chronic | Lung cancer mortality | He et al. (1991) | |
| 20 μg/m3·year | Chronic | Lung and bladder cancer | Gibbs and Sevigny (2007) | |
| 0.85 μg/m3·year | Chronic | Lung and stomach cancer | Xu et al. (1996) | |
| 80 μg/m3·year | Chronic | Bladder and stomach cancer | Spinelli et al. (2006) | |
| 273 ng/m3 | Chronic | Ischemic heart disease mortality | Burstyn et al. (2005) | |
| 7.79 μg/m3·year | Chronic | Ischemic heart disease mortality | Friesen et al. (2010) | |
| 78.21 ng/m3 | Chronic | Immunological alterations | Jeng et al. (2011) | |
| 19.5 ng/m3 | Chronic | Neurological function | Niu et al. (2010) | |
| 0.1 μg/m3 | Chronic | Serum p53 | Pan et al. (1998) | |
| 0.05 μg/m3 | Chronic | DNA adducts in white blood cells | Perera et al. (1988) | |
| Oral | 42.2 μg/100 g | Chronic | Lung cancer | De Stefani et al. (2009) |
| 10.4 ng/d | Chronic | Pancreatic cancer | Anderson et al. (2005) | |
| 33.7 ng/d | Chronic | Colon cancer | Butler et al. (2003) | |
| 99.0 ng/d | Chronic | Esophageal cancer | Hakami et al. (2008) | |
| In vitro | 0.005 μM | Acute | Cell cycle, growth and apoptosis pathways (correlated with S & G2/M phase cell cycle arrest and cytotoxicity) in human amniotic epithelial cells | Lu et al. (2009) |
| 2.5 and 3.0 μM | Acute | Cellular response to xenobiotic exposure and metabolism in human hepatic cell line | Hockley et al. (2006) | |
| 0.017 μM | Acute | Stress response and DNA repair response in human lymphoblastoid cell line | Luo et al. (2005) | |
| Genomics | 4 μM | Acute | Cyp1b1 expression in normal mammary epithelial cells | Gwinn et al. (2005) |
Estimated values.
2.5 Effects on experimental animals, mechanisms and their human relevance
Effects observed in experimental animals were compared to epidemiological data of BaP containing complex mixtures to establish potential relevance to human adverse health outcomes. All studies that were evaluated are presented in Supplementary Table 3, and a very brief overview is given below for each toxicity endpoint. Table 3 provides a summary of the effects of BaP on experimental animals that occurred at the lowest doses. A detailed description of each toxicity endpoint was not possible within this document, but we refer the reader to the supplementary materials (Supplementary Table 3 and Supplementary File B) for further toxicity and mechanistic information. Given the extensive and well-documented evidence demonstrating the link between BaP exposure and genotoxicity/carcinogenicity, we have developed these sections in more detail. Lowest-observed-adverse-effect levels (LOAELs) and no-observed-adverse-effect levels (NOAELs) are presented when available.
Table 3.
Summary of the animal studies used to derive PODs for each of the toxicities reviewed in RA1.
| Toxic effect | Oral exposure duration (vehicle) | Species | NOAEL/LOAEL (mg/kg bw per day) | Calculated BMD/BMDL# (mg/kg bw per day) | Reference |
|---|---|---|---|---|---|
| Reproductive toxicity | Subchronic (corn oil) | Male rat | NA/1 | No dose–response | Zheng et al. (2010) |
| Neuro-developmental toxicity | Acute (peanut oil) | Male & female rat | 0.02/0.2 | 0.09/0.05 (0.02†) | Chen et al. (2012) |
| Hepatotoxicity | Subchronic (soybean oil) | Male & female rat | NA/3 | No dose–response | Wester et al. (2012) |
| Renal toxicity | Subchronic (peanut oil) | Male rat | 5/50 | Data not available | Knuckles et al. (2001) |
| Cardiovascular toxicity | Chronic (olive oil) | Male mouse | NA/2.5 | No dose–response | Yang et al. (2009) |
| Immunotoxicity | Subchronic (soybean oil) | Male rat | 3/10 | 14/8.9 (4.8†) | De Jong et al. (1999) |
| Liver mutations | Subchronic (olive oil) | Male mouse | NA/25 | 2.2/1.4 | Lemieux et al. (2011) |
| Lung mutations | Subchronic (olive oil) | Male mouse | NA/25 | 7.2/4.8 | Lemieux et al. (2011) |
| Forestomach mutations | Subchronic (olive oil) | Male mouse | NA/25 | 0.5/0.3 | Lemieux et al. (2011) |
| Liver tumors | Chronic (soybean oil) | Male & female rat | *2.1/7.1 | *3.3/2.4 (1.2†) | Wester et al. (2012) |
| Forestomach tumors | Chronic (soybean oil) | Male & female rat | *NA/2.1 | *1.5/0.8 (1.1†) | Wester et al. (2012) |
| Forestomach tumors | Chronic (diet) | Female mouse | 0/0.65 | 0.8/0.5 | Culp et al. (1998) |
Dose adjustment for time (dose × 5/7 dosing days) before modeling.
For comparison of rat and mouse BMDLs, scaled from rat to mouse by multiplying rat values by (0.03/0.35)¼ based on the assumption that the physiological processes scale with body weight to the ¾ power (allometric scaling).
Benchmark response: BMD10/BMDL10 for quantal data (tumor) and BMD1SD/BMDL1SD continuous data (Neurotoxicity, immunotoxicity and genotoxicity). Refer to supplemental table 4 for BMD model fit data.
NA, not available
2.5.1 Reproductive toxicity
Twenty-seven studies of reproductive toxicity were found (Supplementary Table 3). Those pertaining to carbon black–bound BaP (i.e. inhalation studies) were excluded due to the confounding effects of particle inhalation. These papers demonstrate that adverse reproductive effects of BaP, including alterations in gonadal tissues and steroid hormone levels and reduced fertility, are observed in both male and female rodents exposed to BaP as adults or in utero. Doses as low as 1 mg/kg bw per day for 90-day exposures (Zheng et al., 2010) were effective in perturbing reproductive parameters. Major effects include impaired fertility in exposed adults (Arafa et al., 2009; Chen et al., 2011; Xu et al., 2010; Zheng et al., 2010) and developing rodents (Kristensen et al., 1995; Mackenzie and Angevine, 1981; Nakamura et al., 2012), which appears to be mediated by an AHR-dependent MOA (Neal et al., 2010; Sadeu and Foster, 2011). Overall, it is apparent that BaP affects the reproductive fitness of exposed organisms by impacting germ cell quality and quantity, hormone balance and gonadal tissue growth and development at exposure doses as low as 1 mg/kg bw per day in rodents (LOAEL).
There has been nly one study that has identified an effect on human reproductive toxicity (Tang et al., 2006); thus, human relevance cannot be clearly established at this time.
2.5.2 Developmental toxicity
Twenty studies pertaining to developmental toxicity effects of BaP were found (Supplementary Table 3), all of whichreported developmental effects. The studies indicate that there are several detrimental effects of gestational exposure to BaP on the developing fetus and newborns, including decreased fetal viability (Bui et al., 1986; Shum et al., 1979; Wu et al., 2003), reduced birth weight (Mackenzie and Angevine, 1981; Perera et al., 2004), increased predisposition to cancer (Holladay, 1999; Turusov et al., 1990; Urso and Gengozian, 1982, 1984; reviewed by US EPA, 2007) and neurodevelopmental effects (Chen et al., 2012). The mechanisms leading to developmental toxicity appear to be governed by both the AHR and genotoxicity (Bolognesi et al., 1985; Lu et al., 1986; Shugart and Matsunami, 1985). The most sensitive developmental endpoints are neurodevelopmental, with effects occurring at BaP doses as low as 0.2 mg/kg bw per day (LOAEL) and no effects occurring at 0.02 mg/kg bw per day (NOAEL). The use of high doses or single doses in many of these studies impaired our ability to evaluate developmental toxicity.
Human relevance for developmental toxicity is limited. There is some evidence that co-exposure to BaP causes developmental neurotoxicity in children (reduced IQ and cognitive development; Perera et al., 2006 and 2008).
2.5.3 Renal and hepatic toxicity
Three studies pertaining to noncancer effects in the liver and kidney following BaP exposure were found that document increased liver to body weight ratios (De Jong et al., 1999; Knuckleset al., 2001; Wester et al., 2012) and increased tubular casts (Knuckles et al., 2001) in rodents exposed to BaP. The most sensitive endpoint is increased kidney tubular casts, occurring at doses as low as 50 mg/kg bw per day (NOAEL 5 mg/kg bw per day).
No studies were identified that could offer information on the mechanisms underlying these effects, nor were there any human data to suggest that these findings are relevant to humans.
2.5.4 Cardiovascular toxicity
Several studies have demonstrated that BaP exposure can lead to cardiovascular toxicity. Of the 23 studies on cardiovascular toxicity reviewed (Supplementary Table 3), those that are relevant for dose–response evaluation demonstrate an increase in atherosclerosis (Knaapen et al., 2007; Yang et al., 2009), an increase in heart to body weight ratios and cardiotoxicity biomarkers (Aboutabl et al., 2009, 2011) and impaired cardiovascular function later in life (Jules et al., 2012) following BaP exposure. Mechanisms in cardiovascular toxicity may be related to AHR-mediated responses (Aboutabl et al., 2009, 2011; Kerley-Hamilton et al., 2012; N'Diaye et al., 2006, 2009; Oesterling et al., 2008; Owens et al., 2009; Podechard et al., 2009). The most sensitive cardiovascular endpoint (atherosclerosis) occurred at doses as low as 2.5 mg/kg bw per week (LOAEL; 0.36 mg/kg per day, if scaled from once per week to daily exposure).
We found two epidemiological studies to suggest some human relevance of this toxicity: (1) asphalt workers showed a significant positive correlation between BaP exposure and ischemic heart disease (Burstyn et al., 2005); and (2) aluminum smelter workers showed a modest association between BaP exposure and myocardial infarction (Friesen et al., 2010).
2.5.5 Neurotoxicity
We identified a total of 12 reports on BaP-induced neurotoxicity in adult animals. These studies demonstrate that BaP exposure can cause very specific behavioral alterations and span various adversities, including impaired spatial learning and memory (Chen et al., 2012; Grova et al., 2007; Qiu et al., 2011; Xia et al., 2011) and other types of behavioral change (Bouayed et al., 2012; Grova et al., 2008; Saunders et al., 2006). In addition, studies indicate that BaP exposure may also cause neurodevelopmental toxicity (Chen et al., 2012) (Section B5, Supplementary File B). The studies demonstrate that BaP exposure can cause alterations in neurobehavior at doses as low as 0.02 mg/kg bw per day (LOAEL). The mechanism behind these alterations appears to be associated with changes in N-methyl-D-aspartate glutamate receptor (NMDAR) subunit levels.
Human relevance is supported by several studies showing various neurobehavioral effects in humans exposed to BaP mixtures occupationally (Niu et al., 2010) or in utero from maternal exposure (e.g. Perera et al., 2004).
2.5.6 Immunotoxicity
The immunosuppressive properties of BaP are well established (reviewed in Holladay, 1999). In total, 35 studies of BaP-induced immunotoxicity were found in our literature survey (Supplementary Table 3). Effects of BaP exposure include reduced antibody production (Dean et al., 1983; Urso and Gengozian, 1984; White and Holsapple, 1984), diminished lymphocyte response (Urso and Gengozian, 1984; Wojdani and Alfred, 1984) and various other immunotoxicities (De Jong et al., 1999; Fischer et al., 2011). The study by De Jong et al. (1999) was considered to be the most relevant for a detailed evaluation and inclusion in risk assessment because it (1) applied methods from an Organisation for Economic Co-operation and Development (OECD) guideline protocol (No. 407), (2) assayed several well-established markers of immunotoxicity and (3) employed a wide range of BaP concentrations. These authors demonstrated that BaP-induced immunotoxicity occurs at doses as low as 10 mg/kg bw per day (LOAEL). BaP-induced immunosuppression is hypothesized to provide a favorable environment for BaP-induced tumors (Urso and Gengozian, 1984). Current clinical immunotherapy aimed at overcoming the immunosuppression that is associated with tumors (Mellman et al., 2011) suggests that weakening the immune system (decreased immunosurveillance) by chemical exposure to AHR agonists favors tumor formation and survival (Ridolfi et al., 2010).
In support of BaP-induced immunotoxicity occurring in humans, Davila et al. (1996) showed that BaP co-exposures affects human peripheral blood T cell mitogenesis, and Allan et al. (2006) showed that BaP exposure inhibits B cell growth.
2.5.7 Genotoxicity
It is well established that BaP is metabolized to reactive metabolites (Section 2.3.3) that are capable of binding to DNA. These DNA adducts can lead to the formation of mutations if they are not properly repaired, and these mutations may subsequently initiate carcinogenesis if they occur in genes that are involved in cancer pathways. A previous detailed review by IARC summarizes the evidence demonstrating that BaP induces DNA adducts, leading to genotoxicity and mutagenicity, as measured in various bacterial and eukaryotic bioassays (including human) in vivo and in vitro (IARC, 2010).
2.5.7.1 DNA adducts
The 2010 IARC monograph reviews all of the in vitro and in vivo cases of BaP–DNA adducts in cultured cells exposed to BaP, mouse skin after topical treatment and internal organs and blood lymphocytes following oral, inhalation and intraperitoneal administration of BaP (IARC, 2010). Of all the in vivo studies reviewed, a study by Garner et al. (1985) had the lowest dose administered that caused adducts. In this study, male Wistar rats were given a single dose of BaP (2 mg/kg bw), and DNA adducts were measured in lung and liver (Garner et al., 1985) (LOAEL 2 mg/kg bw).
2.5.7.2 Mutations
Various studies have demonstrated that BaP induces mutations in proto-oncogenes and tumor suppressor genes. For example, BaP induced three types of Ki-ras codon 12 mutations in lung adenomas in A/J mice: GGT→TGT (56.3%), GGT→GTT (25%) and GGT→GAT (19%) (Masset al., 1993). This mutation spectrum confirms that deoxyguanosine is a primary target for active metabolites of BaP. BaP also induced codon 13 (DiGiovanni et al., 1993) and codon 61 (Chakravarti et al., 1995) mutations in c-Ha-ras in Sencar mouse skin papillomas. BaP caused mutations in murine embryonic fibroblasts from human TP53 knock-in (Hupki) mice that were similar to those found in smoking-related lung cancers in humans (Liu et al., 2005). In our own experiments, male Muta™Mouse mice exposed to BaP at 25, 50 or 75 mg/kg bw per day for 28 days and sacrificed 3 days after the final exposure showed increased lacZ transgene mutant frequencies in the lungs (Labib et al., 2012), liver, glandular stomach, small intestine and bone marrow (Lemieux et al., 2011) at all doses (Figure 2; LOAEL 25 mg/kg bw per day). A dose-dependent increase in mutant Pig-a phenotypes was also observed in these mice. A meta-analysis of published in vivo dose–response genotoxicity studies used the BMD approach to calculate the dose representing a specified 10% change in effect in exposed animals versus controls (BMD10) (Hernandez et al., 2011). BMDs represent a more appropriate and advanced measure than NOAELs (described in Section 2.6). The meta-analysis did not provide starting values; as such, no NOAEL or LOAEL could be determined. However, a BMD10 of 1.52 mg/kg bw per day was observed for mutations (Hernandez et al., 2011).
2.5.7.3 Micronuclei
Shimada et al. (1990) treated male CD-1 mice orally once, twice or three times with BaP at 0, 250, 500, 1000 or 2000 mg/kg bw per day at 24-hour intervals with a posttreatment sampling time of 24 hours. These mice had a significant increase in micronucleus frequency at all doses. In our own work, male Muta™Mouse mice exposed to BaP at 25, 50 or 75 mg/kg bw per day for 28 days by oral gavage and sacrificed 3 days after the final exposure also had significant increases in the frequency of micronucleated reticulocytes and normochromatic erythrocytes at all doses (Lemieux et al., 2011) (LOAEL 25 mg/kg bw per day). A meta-analysis of historical in vivo genotoxicity studies using the BMD approach found a BMD1SD of 1.28 mg/kg bw per day for the dose–response relationship between BaP dose and micronucleus frequency in blood (Hernandez et al., 2011). The meta-analysis did not provide starting values; as such, no NOAEL or LOAEL could be determined.
2.5.7.4 Other genotoxicity endpoints
In addition to the genotoxicity endpoints described above, BAP also induces single- and double-strand DNA breaks, sister chromatid exchanges and chromosomal aberrations (ATSDR, 1995). For example, male DBA/2 mice given two BaP doses of either 10 or 100 mg/kg bw per day intragastrically experienced significant increases in the frequency of sister chromatid exchanges in bone marrow cells at both doses, whereas male C57BL/6J mice exhibited only a small effect at 100 mg/kg bw per day (Wielgosz et al., 1991). A single exposure of adult Brown Norway rats to BaP at 62.5 mg/kg bw intragastrically did not cause the induction of unscheduled DNA synthesis (Mullaart et al., 1989). Intestinal cells from these adult Brown Norway rats had a significant increase in DNA single-strand breaks relative to controls at the 62.5 mg/kg bw (LOAEL) dose (Mullaart et al., 1989). Thus, the lowest LOAEL established for these genotoxicity endpoints is 10 mg/kg bw per day (Wielgosz et al., 1991), and a NOAEL was not achieved.
2.5.7.5 Mechanisms
BaP is biologically inert and requires metabolic activation to exert its genotoxicity. The potential reactive metabolites of BaP and the enzymes involved in producing them are described in detail in Section 2.3.3. Diol epoxides have the ability to form a carbonium ion, which has a high affinity for reacting with DNA and proteins (nucleophiles) in the form of covalent adducts. Owing to their stereoselectivity, the BaP diol epoxides can react with DNA to form cis or trans adducts, primarily with deoxyguanosine and, to a lesser extent, deoxyadenosine. BaP diol epoxides have the ability to intercalate into DNA and thus perturb the structure of the DNA at the site of binding (Volk et al., 2003). Improper DNA repair can lead to mutations. While double-strand breaks are not generated directly by BaP diol epoxides, they may be produced as by-products of DNA repair (Zhou and Shephard, 2006); bulky DNA adducts are repaired by nucleotide excision repair (Geacintov et al., 2002). Finally, error-prone polymerases such as DNA polymerase kappa (Polκ) can bypass bulky DNA adducts, which can cause mutations during DNA replication.
2.5.7.6 Human relevance
A large amount of data demonstrates that exposure to BaP in various media can result in DNA adducts in humans. Furthermore, evaluation of genotoxicity in human cells in vitro and in tissue slices demonstrates that genotoxicity is relevant to human health. BPDE–DNA adducts have been detected in human lung samples from smokers and nonsmokers (Lodovici et al., 1998), in lymphocytes from coke oven workers (Rojas et al., 1995) and cigarette smokers (Rojas et al., 1995) and in human mammary epithelial cells exposed to BaP (Moore et al., 1987). BPDE–DNA adducts in human cells in culture are well documented to cause genotoxicity and mutagenicity (ATSDR, 1995). A recent study using human skin ex vivo and in vitro models showed that DNA-reactive BaP metabolites were generated in these models following exposure to BaP at 50 nmol/cm2 (Brinkmann et al., 2013). Furthermore, these metabolites caused DNA strand breaks in human cultured keratinocytes (NHEK cells) and in human dermal fibroblasts exposed to 3.5 μg BaP. At concentrations of 0.5–50 μM, BaP induced dose-dependent increases in DNA strand breaks (measured using the comet assay) and micronucleus formation in human intestinal Caco-2 cells (Le Hégarat et al., 2012) as well as in human hepatoma HepaRG cells (Le Hégarat et al., 2012). Human liver slices (ex vivo, in vitro) exposed to 10–100 μM BaP for 24 hours showed a concentration-dependent increase in DNA strand breaks (comet assay) at all of the concentrations tested (Plazar et al., 2007).
2.5.7.7 Summary
In summary, a large amount of evidence supports the induction of genotoxicity in human cells exposed to BaP. A review of all of the literature on the genotoxicity of BaP in vivo in rodent models revealed that genotoxicity (specifically DNA adducts and mutations) can occur at doses as low as 2 mg/kg bw.
2.5.8 Carcinogenicity
BaP is classified as carcinogenic to humans (Group 1 carcinogen; IARC, 2012). This classification is based on strong and extensive experimental evidence for the carcinogenicity of BaP in multiple mammalian species, supported by consistent and coherent mechanistic evidence from experimental and human studies that provide biological plausibility. Several routes of exposure, including dermal, oral and inhalation, lead to tumorigenesis. Below, we describe the data for specific tumor sites.
2.5.8.1 Skin tumors
Dermal application of BaP to different strains of mice results in benign and malignant skin tumors (mainly squamous cell carcinomas) (reviewed in IARC, 2012) at doses as low as 2 μg BaP (LOAEL) per animal twice a week for 63–109 weeks (Habs et al., 1984). The lowest dose at which dermal tumours were observed was in a study of C3H/HeJ mice. Shaved dorsal skin of mice exposed to 0.5 and 5.0 μg in cyclohexane/acetone twice per week developed skin tumours at rates of 20% (5/30) and 90% (27/30), respectively (Sivak et al., 1997).
2.5.8.2 Respiratory tract tumors
Respiratory tract tumors are formed following both inhalation and oral administration of BaP. For example, the incidence of respiratory tract tumors was measured in male hamsters administered BaP via inhalation at 0, 2.2, 9.5 or 46.5 mg/m3 for 4.5 hours per day for 10 weeks and then for 3 hours per day for the rest of their lifetimes (total average doses of 0, 29, 127 and 383 mg BaP per animal) (Thyssen et al., 1981). Hamsters treated with the two highest doses showed an increased incidence of papillomas and squamous cell carcinomas in the upper respiratory tract when exposed to as little as 9.5 mg/m3. Female A/J mice fed a diet containing 0, 16 or 98 parts per million (ppm) BaP (total dose 0, 11 and 67 mg) for 260 days developed lung tumors at the highest dose (Weyand et al., 1995). In another study, female B6C3F1 mice fed a diet containing coal tar had an estimated NOAEL/LOAEL for BaP exposure of 0.3 and 0.8 mg/kg bw per day, respectively (Culp et al., 1998).
2.5.8.3 Digestive tract tumors
The most sensitive site for tumor formation following oral exposure to BaP is the digestive tract. In a 2 year study, female B6C3F1 mice fed a diet containing 0, 5, 25 or 100 ppm BaP (calculated intake of 0, 0.65, 3.5 or 15.2 mg/kg bw per day) developed forestomach tumors at the middle and high doses and esophageal tumors at the high dose (Culp et al., 1998) (Table 4). In another study, male and female Wistar rats administered BaP at doses of 0, 3, 10 or 30 mg/kg bw per day by gavage for 5 days per week for 104 weeks developed forestomach tumors at all doses in males and at the top two doses in females (Wester et al., 2012) (Table 4). Additional studies exist that support BaP-induced forestomach tumors, however, their design limit their use for POD determination. Firstly, male and female CFW mice fed a diet containing BaP at doses up to 32.5 mg/kg bw per day developed papillomas and squamous cell carcinomas in the forestomach at 5.85 mg/kg bw per day and higher exposures (Neal and Rigdon, 1967). Further, female A/J mice fed a diet containing 0, 16 or 98 ppm BaP (total dose 0, 11 and 67 mg) for 260 days developed forestomach tumors at all exposure doses (Weyand et al., 1995). However, mice were exposed for less than 2 years (not optimal for extrapolating lifetime risk) and the A/J strain is not common for cancer assessment and is prone to spontaneous lung adenomas. In addition, male hamsters exposed to total average BaP doses of 0, 29, 127 or 383 mg per animal via inhalation (exposure to 0, 2.2, 9.5 or 46.5 mg/m3 for 4.5 hours per day for 10 weeks, and then 3 hours per day for the rest of their lifetimes) (Thyssen et al., 1981) showed an increased incidence of papillomas and squamous cell carcinomas in the upper digestive tract at the two highest doses.
Table 4.
Organs in which benzo(a)pyrene significantly (at least two doses) and dose-dependently increased tumor incidence (number of tumor-bearing animals/number of total animals is presented).
| Study | Species, sex | BaP dose | Daily intake (mg/kg bw per day) | Forestomach | Liver | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diet (ppm) | Gavage (mg/kg bw per day) | Papillomas and/or carcinomas | Papillomas | Carcinomas | Adenomas and/or carcinomas | Adenomas | Carcinomas | |||
| Culp et al. (1998) | Female mice | 0 | 0* | 1/48 | ||||||
| 5 | 0.65* | 3/47 | ||||||||
| 25 | 3.5* | 36/46‡ | ||||||||
| 100 | 15.2* | 46/47‡ | ||||||||
| Wester et al. (2012) | Male rats | 0 | 0§ | 0/52 | 0/52 | 0/52 | 0/52 | 0/52 | 0/52 | |
| 3 | 2.14§ | 8/52‡ | 7/52‡ | 1/52 | 4/52 | 3/52 | 1/52 | |||
| 10 | 7.14§ | 43/52‡ | 18/52‡ | 25/52‡ | 38/52‡ | 15/52‡ | 23/52‡ | |||
| 30 | 21.43§ | 52/52‡ | 17/52‡ | 35/52‡ | 49/52‡ | 4/52 | 45/52‡ | |||
| Female rats | 0 | 0§ | 1/52 | 1/52 | 0/52 | 0/52 | 0/52 | 0/52 | ||
| 3 | 2.14§ | 6/51 | 3/51 | 3/51 | 2/52 | 2/52 | 0/52 | |||
| 10 | 7.14§ | 30/51‡ | 20/51‡ | 10/51‡ | 39/52‡ | 7/52‡ | 32/52‡ | |||
| 30 | 21.43§ | 50/52‡ | 25/52‡ | 25/52‡ | 51/52‡ | 1/52 | 50/52‡ | |||
Calculated daily intake, corrected for food consumption (Cal/EPA (2010) calculation based on food consumption and body weight).
† Reported by Schneider et al. (2002).
Significantly different from control (p < 0.05) as reported in Benford et al. (2010) or in Cal/EPA (2010).
Administered dose of 5 days per week was adjusted to 7 days per week daily dose (dose × 5/7).
2.5.8.4 Liver tumors
Exposure of male and female Wistar rats to BaP by gavage at a dose of 0, 3, 10 or 30 mg/kg bw per day for 5 days per week for 104 weeks caused tumors in the livers of both males and females at the middle and high doses (Wester et al., 2012) (Table 4).
2.5.8.5 Other tumors
Rats gavaged with 50 μmol BaP once per week for 8 weeks exhibited increased numbers of mammary gland adenocarcinomas (El-Bayoumy et al., 1995). Female B6C3F1 mice fed a diet providing BaP (0, 0.65, 3.25 or 13 mg/kg bw per day) for 2 years developed tumors in the tongue and larynx at the high dose (Culp et al., 1998).
2.5.8.6 Mechanistic information
Well-established mode of action for BaP-induced carcinogenicity is genotoxicity, as described in detail in Section 2.5.7. Unrepaired adducts can lead to mutations, increasing a cell's susceptibility to carcinogenic transformation.
BaP causes tumorigenesis in tissues directly at the site of contact, as well as in those removed from the site of contact. The most sensitive reported site of tumorigenesis in rodents is the forestomach, with a LOAEL of 3.5 mg/kg bw per day for chronic exposure (Culp et al., 1998). In vitro human cell culture models indicate that human cells are capable of metabolically transforming BaP to reactive metabolites causing genotoxicity (Section 2.5.7). Further, epidemiological work supports the ability for BaP to cause cancer in humans (Section 2.4). The carcinogenicity of BaP may be further modulated by AHR-driven immunosuppression (Section 2.5.6), which may contribute to tissue specificity.
2.6 Selection of the point of departure for risk assessment 1 (RA1)
Only toxicological outcomes with sufficient information to derive probable adverse health outcomes that are applicable to humans (i.e. biologically relevant for humans) were considered. It is advantageous to use BMD modeling in place of the NOAEL and LOAEL approach, as the entire dataset is employed in deriving the POD. By using this approach, our results do not solely rely on the specific doses selected within each study, and confidence limits can also be established. Moreover, BMDs are necessary in deriving cancer slope factors for genotoxic substances such as BaP and are thus critical to the risk assessment of such chemicals. The NOAEL/LOAEL values, on the other hand, are used for non-cancer effects or non-genotoxic carcinogens and thus cannot be applied to BaP. The US EPA (2012) recommends an extra risk of 10% in the benchmark response for standard reporting of quantal data, to serve as a basis for comparisons across chemicals and endpoints, since the 10% response is near the limit of sensitivity in most cancer and some noncancer bioassays. For continuous data, the US EPA (2012) recommends that the BMD (and BMDL) corresponding to a change in the mean response equal to one standard deviation from the control mean always be presented for comparison purposes. This value would serve as a standardized basis for comparison, akin to the BMD corresponding to 10% extra risk for quantal data. Thus, BMD modeling was employed when possible (i.e. when a sufficient number of doses were tested, when significance of the effect was established at least at the higher doses and when at least one of the BMD models was an appropriate representation of the data trend) (Table 3). The toxicological outcomes for lung, liver, and forestomach with the lowest BMDL, and thus the most sensitive endpoint, were selected as the PODs for comparison to RA2 and RA3.
2.6.1 Calculation of BMD/BMDL
Benchmark responses were BMD/BMDL10 for quantal data and BMD/BMDL1SD for continuous data. BMDs and BMDLs (the estimated 95% lower-bound confidence limit on the BMD) were calculated for apical endpoint data using the US EPA's Benchmark Dose Software (BMDS) version 2.2 (Davis et al., 2011). Only datasets with at least two doses that were statistically significant compared with control levels (p < 0.05) were modeled. Prior to modeling, the data were screened for homogeneity of variance. Continuous endpoints (neurotoxicity, immunotoxicity and genotoxicity) were fit against five dose–response models (Hill, polynomial, linear, power and exponential), whereas tumor endpoints were fit against nine dichotomous dose–response models (gamma, logistic, log-logistic, log-probit, multistage, multistage-cancer, probit, Weibull, quantal-linear). For continuous transcriptomics data, the Hill model was considered only when the k parameter was more than one third of the lowest positive dose, in order not to artificially minimize BMDs and BMDLs (Black et al., 2012). Curves were visually inspected for a good fit to the data, a goodness of fit > 0.05 and scaled residuals within ± 2.0. The lowest Akaike's Information Criterion (AIC) was applied as a cut-off for selection of the appropriate model when more than one model was suitable and the BMDs for these were within 3-fold of each other. In all other instances, the lowest BMD was selected. BMDLs that could be calculated for key effects in experimental animals are presented in Table 3 (refer to Supplementary Table 4 for BMD model fit parameters).
2.6.2 Selection of the key effect with the lowest biologically relevant BMD/BMDL
The estimated human ingestion of BaP through drinking water was calculated to be 0.000857–19.6 ng/kg bw per day (Section 2.1). BaP doses at the high end of this range and above could possibly cause carcinogenicity at multiple tissue sites in humans (Table 2 and Supplementary Table 2). However, the variability in the estimation of the oral doses of BaP at which these effects can occur makes it difficult to draw firm conclusions regarding acceptable exposure levels for risk assessment. More reliable dose–response data come from studies of laboratory animals.
Tumors in rodent liver and forestomach occur at BMDLs as low as 0.5–1.2 mg/kg bw per day (Table 3; Section 2.5). Even lower doses of BaP are reported to induce neurotoxic effects in developing and adult rodents, with a BMDL as low as 0.05 mg/kg bw per day, based on diminished learning and memory in BaP-treated rats (Table 3). If the rodent data are scaled to humans using allometric scaling, one would get a human equivalent of 0.076 and 0.013mg/kg bw per day for cancer and neurotoxic endpoints, respectively. These two values are approximately 4000 and 700 times greater than the maximal human exposure of 19.6 ng/kg bw per day. Despite some emerging insights into the neurotoxic MOA of BaP (Chepelev et al., manuscript in preparation), the literature database of the neurotoxic effects of BaP is not as comprehensive as that for carcinogenicity. Therefore, BaP-induced carcinogenicity was selected as the endpoint of concern for POD selection, based on:
evidence of tumors in humans (Section 2.4);
strong and extensive evidence of tumor induction in rodents (Section 2.5.8);
consistent and coherent mechanistic evidence from rodent and human studies of a genotoxic MOA in cancer (Sections 2.3.3 and 2.5.7); and
the classification of BaP as “Group 1: Carcinogenic to humans” (IARC, 2012).
Rodent forestomach tumors were the carcinogenic endpoint with the lowest BMDL (0.5 mg/kg bw per day) (Culp et al., 1998). However, as humans do not have forestomachs, the application of rodent forestomach tumor data for predicting cancer risk in humans for a particular chemical should meet specific criteria, including a clear carcinogenic MOA and induction of tumors at multiple sites and in various species (Proctor et al., 2007). BaP meets these criteria, because: (1) BaP acts through a genotoxic MOA and causes tumors at multiple sites by oral exposures in various species and in both sexes; and (2) the MOA of BaP in carcinogenesis is generally understood and considered applicable to humans (Proctor et al., 2007). Thus, the specific criteria outlined by Proctor et al. (2007) are satisfied, and it is appropriate to use BaP-induced forestomach tumor data to predict cancer risk in humans.
Therefore, a BMDL of 0.5 mg/kg bw per day for rodent forestomach tumors was selected as the POD most relevant for human health risk assessment. Tissue-specific PODs of 1.2 and 0.8 mg/kg bw per day were also calculated for liver (Wester et al., 2012) and lung (Culp et al., 1998) tumors, respectively.
3. Genomics approaches (RA2 and RA3)
The aim of this project is to determine if genomics can be helpful in a quantitative risk assessment framework. In RA2 and RA3, we investigate the utility of toxicogenomics for various components of risk assessment, including: (1) hazard identification; (2) dose–response characterization; (3) development of an MOA; (4) supporting human relevance; and (5) deriving potential PODs. In order to accomplish these goals, we employed toxicogenomics data produced in-house, but we note that data available in public repositories could easily serve the same purpose. The methods and data for the toxicogenomics experiments used in the present study are described below.
3.1 Overview of toxicogenomics data and analyses used in RA2 and RA3
3.1.2 Functional annotation
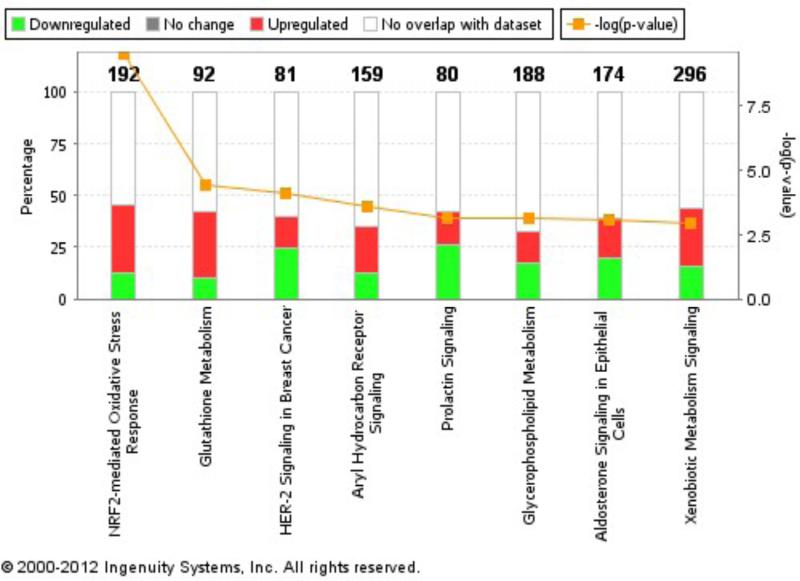
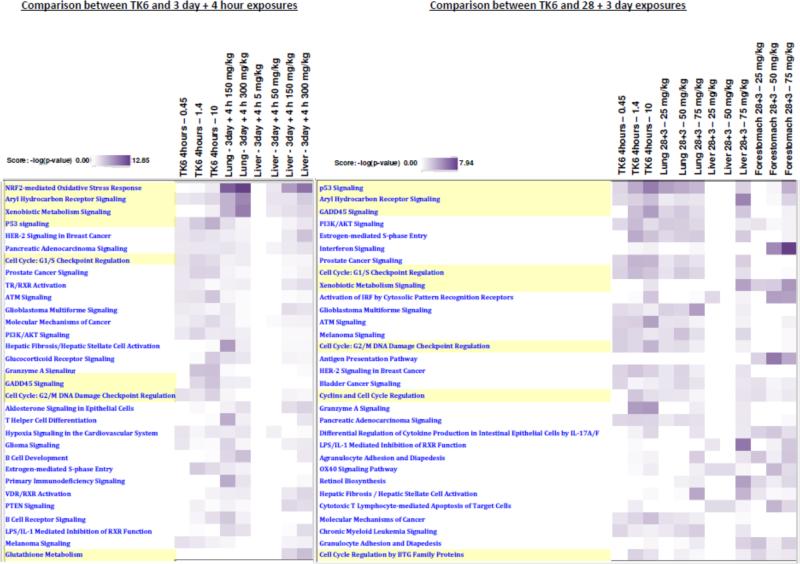
Expression profiles were analyzed using Ingenuity® Pathway Analysis (IPA) to determine which functional pathways and processes were enriched within the observed transcriptional changes. Transcriptional BMDs were calculated (as described below in Section 3.1.4) and consolidated with IPA pathways.
3.1.1 Microarray analysis
We analyzed global transcriptional changes in response to BaP exposures over time and across doses in a variety of mouse strains and tissues and in a human cell culture model. All of the data for these studies are publicly available and comply with the minimal information requirements for a microarray experiment (Brazma et al., 2001). All gene expression datasets have been uploaded to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: GSE4363, GSE24751, GSE18789, GSE24910, GSE35718 and GSE43438.
3.1.1.1 Animal data
We relied primarily on data that have recently been produced by coauthors of this manuscript. The experiments were conducted on either whole tissue homogenates or random slices from tissues, and were not selected for any particular cell type. Experimental details from three experiments conducted at different times are described briefly below:
Experiment 1—In the first experiment, 5-week-old male B6C3F1 mice received BaP in corn oil by oral gavage at a concentration of 0 (corn oil alone), 5, 50, 150 or 300 mg/kg bw per day for 3 consecutive days (n = 5 per dose group). Samples were collected 4 or 24 hours after the last exposure. Full experimental details were published in Yauk et al. (2011) and Halappanavaret al. (2011). Samples were analyzed using 4×44K Agilent gene expression microarrays as per the manufacturer's protocol (Agilent Technologies). Lung microarray data have been published in Halappanavar et al. (2011), and partial results for liver microarrays are found in Yauk et al. (2011); all information regarding array quality control and normalization can be found in these manuscripts. The full liver gene expression dataset (unpublished) has been uploaded to the Gene Expression Omnibus under accession number GSE4363.
Hepatic gene expression analysis in mice exposed to 5, 50, 150 or 300 mg BaP/kg bw per day for 3 days revealed 4, 36, 373, and 900 genes, respectively, that were altered relative to solvent-treated mice (fold change [FC] ≥ 1.5 and false discovery rate [FDR] adjusted p-value ≤ 0.05) 4 hours after exposure and 1, 11, 410, and 579 genes, respectively, 24 hours after exposure. In the lungs of these mice, 558 and 1267 genes were differentially expressed (FC ≥ 1.5 and FDR p-value ≤ 0.05) following 150 and 300 mg/kg bw per day BaP exposure compared with controls, respectively, 4 hours after exposure. The overall gene expression response in the liver was consistent with the known physiological function of the organ, including its primary role in the detoxication of xenobiotic compounds. For example, we measured significantly increased transcription of the Cyp1a1 metabolism gene in liver and lung (Table 5). The pulmonary gene expression response to BaP included changes in biological pathways involved in B cell receptor signaling, inflammation and DNA damage response. Additional real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) array analyses were also performed on the lung tissue across all doses and time points. Ribonucleic acid (RNA) samples were processed, and data were analyzed as described in Labib et al. (2012) on custom PCR arrays that included 162 genes (Supplementary Table 5 includes the 38 genes referred to in the present study).
Table 5.
Cyp1a1 and Cyp1b1 gene expression in mouse lung and liver exposed to 5, 50, 150, and 300 mg/kg-bw/day BaP for three days and excised 4 or 24 hours post-exposure. Those not statistically significant are indicated by ns.
| Dose (mg/kg-bw/day) | LUNG (fold change) | LIVER (fold change) | |||
|---|---|---|---|---|---|
| 4 hours | 24 hours | 4 hours | 24 hours | ||
| Cyp1a1 | 5 | ↑46.9 | ↓7.5 | ↑25.5 | ns |
| 50 | ↑103.3 | ↓1.5 | ↑109.9 | ns | |
| 150 | ↑179.3 | ↑14.7 | ↑146.7 | ns | |
| 300 | ↑210.1 | ↑42.9 | ↑165.4 | ↑9.0 | |
| Cyp1b1 | 5 | ↑10.4 | ↑1.5 | ns | ns |
| 50 | ↑23.5 | ↑1.7 | ns | ns | |
| 150 | ↑161.2 | ↑21.8 | ns | ns | |
| 300 | ↑208.6 | ↑28.6 | ns | ns | |
Experiment 2—Adult (25-week-old) Muta™Mouse (transgenic mouse strain 40.6) males were exposed to BaP (Sigma Aldrich, Canada) in olive oil by oral gavage at a concentration of 0 (olive oil alone), 25, 50 or 75 mg/kg bw per day for 28 consecutive days (n = 5 per group). Mice were sacrificed 72 hours after the final exposure. The right lobe of the lung, the median lobe of the liver and the forestomach were excised, flash frozen in liquid nitrogen and stored at −80°C until use. All samples were analyzed using Agilent 4×44K or 8×60K gene expression microarrays. The full DNA microarray and real-time quantitative PCR array analyses are published for liver (Malik et al., 2012), lung (Labib et al., 2012) and forestomach (Labib et al., 2013); the reader should refer to these publications for details relating to microarray data processing and normalization. Data for liver, lung and forestomach are available in the Gene Expression Omnibus (accession numbers GSE24910, GSE35718 and GSE43438, respectively). Microarray analysis of variance (MAANOVA) revealed significant changes in the transcript levels (FDR p-value ≤ 0.05 and FC ≥ 1.5) of 6, 7 and 121 genes in livers, 20, 145 and 373 in lungs and 9, 135 and 408 in forestomachs of mice in the 25, 50 and 75 mg/kg bw per day exposure groups, respectively. In the liver, the affected genes were primarily associated with biological processes such as xenobiotic metabolism and p53 signaling. In the lung, there was significant enrichment of p53 signaling and cancer pathways. Although the p53 signaling pathway was also perturbed in the forestomach, the most significantly enriched processes were associated with antigen processing and presentation, immune response, chemotaxis and keratinocyte differentiation. The results in the lung and liver are consistent with expected changes in gene expression in response to a genotoxic carcinogen, whereas the forestomach data point to additional modulating factors at work (a pro-immune, inflammatory response) (Hochstenbach et al., 2012). Please note that BPDE-DNA adducts and lacZ transgene mutant frequency were also assessed in the lung, liver, and glandular stomach tissues from the same mice (Lemieux et al., 2011, Malik et al., 2012, Labib et al., 2012) (Figure 2). Adducts were detected via the 32P-postlabeling method and were found in all three dose groups in all tissues, with the highest relative adduct labeling in the lung tissue at three days post-exposure. LacZ mutant frequency was assessed using the P-Gal positive selection assay and was highest in the glandular stomach at this time point.
Experiment 3— Eight-week-old male C57BL/6J mice were treated four times a week with BaP in corn oil at a dose of 13 mg/kg bw per day or corn oil vehicle (7.4 mg/kg bw per day, adjusted for daily exposure). Mice were dosed by oral gavage on days 0, 2, 4 and 6 (autopsy on day 7) and euthanized 24 hours after the final exposure. Each treatment group consisted of four mice. Liver, spleen, kidney, bone marrow and urinary bladder were isolated. Tissues were stored in RNAlater using the manufacturer's protocol (Qiagen, Valencia, CA, USA). Total RNA was isolated using the miRNeasy kit (Qiagen, Valencia, CA, USA) and the QIAcube (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. All samples passed RNA quality control using capillary gel electrophoresis (RNA integrity number [RIN] > 7.6) (Bioanalyzer 2100; Agilent Technologies, Amstelveen, Netherlands). Messenger RNA (mRNA) was amplified and labeled with the GeneChip Expression 3′-Amplification One-Cycle cDNA Synthesis Kit and GeneChip Expression 3′-Amplification Reagents for IVT Labeling according to the manufacturer's instructions (Eukaryotic Sample and Array Processing 701025 Rev.5; Affymetrix Inc., Santa Clara, CA, USA). Amplified materials were hybridized to Mouse Genome 430 2.0 Array for 16 hours at 45°C, subsequently washed and stained with the EukGE-WS2v5_450 protocol and finally scanned using the GeneChip Scanner 3000-7G (Affymetrix Inc., Santa Clara, CA, USA). Image generation and feature extraction were performed using Affymetrix GCOS Software version 1.4.0.036. Quality control and correction of significant hybridization and experimental blocking effects, annotation, RNA normalization and subsequent data analysis were performed as previously described (Schaap et al., 2012). Gene expression data for the liver are available in the Gene Expression Omnibus (accession number GSE43977). Data for the other tissues will become publicly available (M. Luijten et al., unpublished data). Gene expression profiles for the spleen, kidney, and liver were similar to what we observed at the lowest dose (5 mg/kg bw per day) in our 3-day experiments described above for the lung and liver (i.e. only a few genes were significantly affected by the treatment). Unlike the aforementioned tissues, there was no significant change in gene expression profiles in the bone marrow or bladder samples of BaP-treated animals compared with control animals. This suggests that toxicogenomics is useful in identifying the most relevant tissues and organs affected by a chemical treatment.
3.1.1.2 Human cell culture data
To gain insight into BaP-induced genotoxic hazard and pathway perturbations in human cells, we compared gene expression profiles of BaP-treated human cells in culture to those derived from our rodent studies. Gene expression profiles were generated following in vitro BaP exposure of TK6 cells, a human lymphoblastoid cell line (ATCC No. CRL-8015; Manassas, VA, USA). Detailed methodology and data can be found in Supplementary File C (J. Buick et al., submitted). TK6 cells were exposed to BaP in dimethyl sulfoxide (DMSO) (0, 0.45, 1.4 or 10 μg/mL) in the presence of 1% pentobarbital/benzoflavone-induced rat liver S9 with reduced nicotinamide adenine dinucleotide phosphate (NADPH) generating system cofactors (for metabolism of BaP) for 4 hours. Cells were collected immediately post-exposure (4-hour time point) or were placed in fresh media and collected 20 hours later (24-hour time point). Three technical replicates were produced per concentration for both time points. Cisplatin was used as a positive control for genotoxicity. Because cisplatin is direct acting, no metabolic activation system was required. In addition, negative controls (media only) and vehicle controls were included, in both the presence and absence of the metabolic activation system. The in vitro chemical exposures were performed by Integrated Laboratory Systems Inc. (ILS; Research Triangle Park, NC, USA), in addition to the associated cytotoxicity and genotoxicity testing. The CellTiter96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) was used to measure cell viability and proliferation in treated cells by means of a colorimetric assay. In addition, flow cytometry–based cytotoxicity and micronucleus assays were conducted using the In Vitro MicroFlow kit (Litron Laboratories, Rochester, NY, USA) to assess relative survival, percentage of apoptotic/necrotic cells and micronucleus frequency, as described in Avlasevich et al. (2006). RNeasy Mini Kits were used to extract total RNA from treated and control cells following the Purification of Total RNA from Animal Cells Using Spin Technology Protocol (Qiagen, Valencia, CA, USA). RNA samples were labeled with Cyanine 3 and Cyanine 5 using Low Input Quick Amp Labeling kits (Agilent, Santa Clara, CA, USA) according to the manufacturer's instructions. Labeled complementary RNA (cRNA) samples were hybridized to Agilent SurePrint G3 Human 8×60K microarrays (n = 3 per dose and time point), following the manufacturer's directions, using a two-color dye-swap design.
The toxicogenomics analysis was performed as described in Goodsaid et al. (2010) and H. Liet al. (unpublished data). The gene expression profiles of BaP-treated cells were measured using Agilent 60K oligo microarrays using a dye-swap model. The presence of a genotoxic signature in profiles of BaP-treated TK6 cells was evaluated utilizing a genotoxic signature/classifier that is capable of differentiating genotoxic and non-genotoxic agents (Goodsaid et al., 2010; Li et al., submitted). The classifier is based on a reference database comprising 28 model agents run on Agilent DNA microarrays. Thus, at this time the approach can only be applied using Agilent whole genome microarray platform. In addition, the classifier is currently limited by the spectrum of chemicals used to train and test the signature. However, the classifier is currently under a formal validation by the Health and Environmental Sciences Institute of the International Life Sciences Institute (ILSI HESI), which has markedly increased the number of agents tested. Finally, the classifier was built using direct acting chemicals and only a few chemicals requiring metabolic activation have been tested and analyzed thus far. In our experience determining the most effective S9 in terms of concentration and type of induction can also be a challenge.
Exposure of TK6 cells to BaP caused a concentration-dependent increase in cytotoxicity and micronucleus formation and upregulation of known BaP-induced stress response genes (Figure 4; Supplementary Table 6). Expression profiles of BaP-treated TK6 cells clustered with those of DNA-reactive genotoxic toxicants (Figure 5).
Figure 4.
Cytotoxicity and genotoxicity measurements in human TK6 cells following exposure to BaP using a flow cytometry–based assay (In Vitro Microflow kit; Litron Laboratories). Relative survival (shown in blue), percentage of apoptotic/necrotic cells (shown in green) and percentage of micronuclei (MN; shown in red) are depicted following 24 hours of exposure (4-hour exposure + 20-hour recovery). * denotes p < 0.05 compared with vehicle control (VC; + S9), and error bars are standard error.
Figure 5.
Hazard identification of BaP as a genotoxic compound by comparison of its transcriptome profile with the genotoxicity biomarker following BaP exposure in human TK6 cells in the presence of metabolic activation (1% rat liver S9) at 4, 8 and 24 hours. (a) Hierarchical clustering of the expression levels of genes in a genotoxicity signature derived from a training set of twenty-eight genotoxic (pink) and non-genotoxic (blue) agents indicates that BaP clusters with genotoxic agents at the mid- and high concentrations at all three time points. The GenBank accession numbers for the 65 classifier genes contained within the predictive gene signature are indicated on the right hand-side of the heatmap. Gene expression fold-changes relative to control are shown by means of the colour scale: upregulated genes are shown in red, downregulated genes are shown in green and genes that are not regulated are shown in black. (b) BaP transcriptome profiles were analyzed using the 65-gene classifier to predict genotoxicity. Nearest shrunken centroids classification probabilities for BaP treatments are shown using the 65-gene classifier. The NSC method was employed to classify BaP transcriptome profiles by examining them for similarities with the transcriptome profiles of the reference chemicals in the database using statistical and bioinformatics tools. Briefly, the standardized centroid (SC) was computed by applying the NSC method for each class of training chemicals, in which SC is the mean expression level for each gene in a class divided by its within-class standard deviation. For each class, the SC is shrunken in the direction of the overall centroid to create the NSC. BaP was then classified through comparison of its gene expression profiles to the class of NSCs for each concentration and time point. Sample classification was achieved by assigning it to a class that is closest to it in squared distance [Tibshirani et al., 2002].
3.1.3 Identification of key events and proposal of MOA
To derive a MOA based on the International Programme on Chemical Safety (IPCS)/ (ILSI) MOA framework (Meek et al. 2014; Boobis et al., 2006; Meek et al., 2003; Sonich-Mullin et al., 2001), key events were identified by interrogating all genomics data based on functional annotation. Key events are empirically observable, necessary precursor events in the MOA or biologically based markers for such events (US EPA, 2005). We interrogated our putative key events, comprising our proposed MOA, based on their temporal and dose–response patterns of gene expression, as well as based on modified Bradford Hill criteria (Hill, 1965), in accordance with the MOA/human relevance framework analysis. Once the key events were identified, IPA pathways were assigned to each key event using knowledge acquired from the literature.
3.1.4 Calculation of transcriptional PODs
Dose-related changes in gene expression were used to calculate transcriptional BMDs using BMDExpress version 1.4.1 (Yang et al., 2007). Briefly, analyses were performed on all genes using four models: Hill, power, linear and polynomial 2°. The best fit selected was the model that: (1) described the data with the least complexity; (2) had a nested chi-squared test cut-off of 0.05; (3) had the lowest AIC (measure of the relative goodness of fit); and (4) the BMD was lower than the highest dose. Other settings included maximum iterations of 250, confidence level of 0.95, BMR factor of 1.349 (the number of standard deviations defining the BMD, which corresponds to a 10% change compared with controls) (Yang et al., 2007) and power restricted to ≥ 1. The use of the Hill model was restricted such that it was flagged if the k parameter of the model was less than one third of the lowest BaP dose used in an experiment (Black et al., 2012). A flagged Hill model was excluded if at least one of the other models had a p-value > 0.05. In such a case, the next model with least complexity and with a p-value > 0.05 was selected. In the case when no model had a p-value > 0.05, the Hill model BMD was modified to 0.5 of the lowest BMD value. The resulting BMDL datasets were mapped to IPA pathways (probes that mapped to multiple annotated genes were removed). Supplemental Table 7 displays model fit criteria for key PODs determined in RA2 and RA3. We then calculated the average BMD/BMDLs at the 10th percentile of all genes affected by BaP treatment in the assigned IPA pathway. Given that a standard benchmark response applied to apical endpoints is 10 percent (akin to 1 standard deviation; U.S. EPA, 2005), the 10th percentile is an appropriate threshold for comparative purposes. As pathways can contain many genes that can be expressed at a given time and the gene expression analysis represents a snapshot in time, only a few genes within an enriched pathway may be expected to be differentially expressed at a single time point. Multiple IPA pathways can be representative of a single key event. Thus, the IPA pathway with (1) at least 5 genes passing the modeling criteria above; (2) BMD/BMDL < 10; and (3) the lowest BMDL aligned with each key event in our MOA was selected as representative of that key event.
The transcriptional data were used to inform the genomics-informed risk assessment (RA2; Section 3.2) and the genomics-only risk assessment (RA3; Section 3.3).
3.2 Genomics-informed approach (RA2)
The objective of the genomics-informed approach (RA2) was to evaluate the ability of toxicogenomics to derive relevant PODs and to provide mechanistic information to support the human relevance of BaP-induced effects for human health risk assessment. As carcinogenicity was identified in RA1 as the endpoint of concern for POD selection, RA2 focused on this endpoint as well. Although MOAs are described in detail for many of the toxicity endpoints described in RA1, the availability of genomics data for RA2 provides a means to expand upon the cancer MOA and derive quantitative measures of response for key events for this specific adverse outcome. Thus, we relied on toxicogenomics information (primarily our own data) and all available published BaP information (i.e. RA1) to propose an MOA for BaP-mediated carcinogenicity and to guide the selection of a POD for risk assessment.
We would like to emphasize that our primary goal was to integrate biologically relevant toxicogenomics information into our risk assessment (i.e. focus on pathways and genes that are relevant to existing toxicological knowledge, rather than including all and any gene that responded). Toxicogenomics data were used first to propose an MOA for genotoxicity leading to cancer, as the most notable effect associated with BaP exposure in mouse liver, lung and forestomach in our study. We then calculated transcriptional BMDs for genes and pathways comprising the key events in our proposed MOA in order to derive PODs for risk assessment. Our approach contrasts with that of Thomas et al. (2007a, 2011, 2012), who showed that the lowest BMDs for transcription (i.e. the pathways or GO terms with the lowest median BMD values regardless of biological function) correlate well with BMDs for apical cancer and noncancer endpoints and suggested that these values should be used as PODs. This approach is valuable for chemicals operating as nonspecific toxicants that perturb multiple biological pathways, as such perturbations may be adverse to normal physiological functions. In our example, we aim to demonstrate how toxicogenomics can be used to obtain information relating to key events to develop more specific MOAs, which has been noted as an important potential application of toxicogenomics in risk assessment (Bercu et al., 2010; Currie, 2012; Waters et al., 2010).
3.2.1 Proposed mode of action
Our proposed genotoxic MOA for BaP is schematically represented in Figure 6, summarized in Table 6 (including evidence for human relevance), evaluated based on modified Bradford-Hill criteria (Table 7) and the BMDLs for the key events are presented in Table 8. Based on our data and review of the literature, this MOA is relevant to the three rodent tissues under examination in this risk assessment: liver, lung and (fore)stomach. BaP's MOA consists of six key events: (1) AHR binding; (2) induction of cytochrome P450 enzymes; (3) metabolism of BaP to DNA-reactive genotoxic compounds; and (4) formation of DNA adducts and DNA damage, which, if unrepaired, cause (5) mutation via erroneous replication. If these mutations occur in proto-oncogenes or tumor suppressor genes (e.g. p53), they can cause the last key event (6) altered cell signaling, proliferation and apoptosis, which leads to the adverse outcome tumorigenesis. Below, we use published information and our own toxicogenomics data to provide evidence supporting this MOA and, where possible, provide insight into the human relevance of each key event.
Figure 6.
RA2: Postulated genotoxic MOA of BaP in animals (shaded rectangles). BaP binds to the AHR and activates transcription of AHR-controlled genes. These include xenobiotic metabolism enzymes that convert BaP to a variety of products that are subsequently conjugated to water-soluble moieties and excreted. BaP metabolites escaping detoxication (Section 2.3.3, e.g. BPDE) and reactive oxygen species (ROS) are genotoxic, leading to DNA adducts and oxidative damage. If unrepaired, genotoxic damage may cause mutations, leading to uncontrolled cell proliferation and tumor formation. In addition, BaP-mediated immunosuppression may provide a favorable environment for tumor growth, and activation of other signaling pathways that may favor tumorigenesis (See Section 3.2.1.7 for details). The double lined box illustrates the dual role of BaP metabolism by CYPs: their beneficial role in activating BaP for further conjugation and removal that becomes apparent from CYP knock-out studies of mice, exposed to BaP (e.g., Nebert et al., 2004). Metabolism by CYPs leads to efficient detoxication of BaP metabolites. However, a small portion of genotoxic metabolites escapes the detoxication to lead to DNA damage and mutations (Nebert et al., 2004).
Table 6.
RA2. Key events in the proposed mode of action of benzo(a)pyrene for rodents and their human relevance.
| Key event | Evidence in rodents |
References | Evidence in humans |
References | ||
|---|---|---|---|---|---|---|
| Toxicogenomic endpoints | Apical endpoints | Toxicogenomic endpoints | Apical endpoints | |||
| 1.BaP binding to AHR (molecular initiating event) | Indirect evidence: upregulation of AHR-responsive genes. | YES: BaP is a well-established AHR ligand. | Malik et al. (2013); Ovesen et al. (2011). Table 5. | Cyp1b1 gene expression is induced by BaP in normal mammary epithelial cells. | YES: BaP binds to human AHR and activates AHR-dependent gene expression. | DiNatale et al. (2010; Gwinn et al. (2005) |
| 2.Induction of cytochrome P450 enzymes (CYPs) | YES: Multiple P450 genes (e.g. Cyp1a1, Cyp1a7, Cyp1b1, Cyp2b10, Cyp2b13, Cyp3a44, Cyp2b9, Cyp2c38 and Cyp2c40) are induced by BaP in mouse liver, lung and forestomach. | YES: induction of EROD in diverse tissues following BaP exposure. | Halappanavar et al. (2011); Malik et al. (2012); Yauk et al. (2011); IARC (2010) | YES: BaP induces genes coding for P450 enzymes (Cyp1a1, Cyp1a2, Cyp2b6, Cyp2e1) in BaP-treated primary human hepatocytes. | YES: BaP increases the activity of P450 enzymes (EROD, ECOD assays) in BaP-treated primary human hepatocytes. | Wilkening et al. (2003) |
| 3. BaP metabolism to reactive metabolites | YES: BaP induced phase II enzymes (e.g. Nrf2, Nqo1, Ugdh, Srxn1, Akr1b15, Ugt2b15, Gstm3, Gstm4 and Gstm7) in mouse liver, lung and forestomach to remove reactive metabolites. | YES: BaP metabolites have been isolated from animals exposed to BaP and studied in great detail. | Halappanavar et al. (2011); IARC (2010); Malik et al. (2012); Yauk et al. (2011) | YES: BaP induces Ephx1 and Nqo1, genes, coding for the enzymes involved in BaP metabolism, in BaP-treated primary human hepatocytes. | YES: BaP metabolites, including BaP-7,8-diol (a precursor to BPDE), were detected in human liver and lung microsomes. | Shimada et al. (1989); Wilkening et al. (2003) |
| 4. DNA adducts and DNA damage | YES: Increased expression of genes regulated by p53 in lung, lung and forestomach from BaP-treated mice. Upregulation of Polκ and Ddit4 in mouse liver and lung indicates DNA damage repair. | YES: Adducts of BaP metabolites with DNA have been measured directly in several tissues in rats and mice. | Denissenko et al. (1996); Halappanavar et al. (2011); Labib et al. (2012); Malik et al. (2012); Wester et al. (2012) | YES: p53 pathway is activated by BaP in cultured human TK6 cells; stress response and DNA repair response are activated by BaP in human lymphoblastoidcell line. | YES: BaP metabolites (BPDE) form adducts with DNA, and BPDE–DNA adducts are detected in human white blood cells. | J. Buick et al. (Submitted); DeMarini et al. (2001); Liu et al. (2005); Perera et al. (1988) |
| 5. Mutations | Indirect evidence: the induction of gene expression changes in DNA damage response pathways is highly correlated with genotoxicity and mutagenicity in rodents. | YES: Mutations were reported in multiple studies in rodents following BaP exposure. | Sakai et al. (2014); Malik et al. (2012); Labib et al. (2012); IARC (2010) | Indirect evidence: the induction of gene expression changes in DNA damage response pathways is highly correlated with genotoxicity and mutagenicity in human TK6 cells. | YES: BPDE–DNA adducts detected at mutational “hotspots” of p53 (G in codons 157, 248 and 273). Proto-oncogene K-Ras and p53 mutations found in lung tumors of nonsmokers exposed to BaP-containing PAH mixtures. Mutations in tumor suppressor gene p53 in BaP-exposed nonsmokers were detected. | Derk et al. (2014); Waters et al. (2010); J. Buick et al. (unpublished data); Denissenko et al. (1996); Rojas et al. (2000); DeMarini et al. (2001) |
| 6. Altered cell signaling, proliferation and apoptosis | YES: A number of signaling pathways (including cancer pathways) were perturbed by BaP treatment in mice. Mdm2, an inhibitor of p53, was induced in the lung and liver of BaP-treated mice at high doses. | YES: Multiple in vitro studies using cultured and animal cells identified increased cell proliferation due to BaP or its metabolites. Mutations in BaP-treated mice were increased compared with controls. | Brandon et al. (2009); Burdick et al. (2003); IARC (2010); Labib et al. (2012); Wester et al. (2012) | YES: Anti-apoptotic signaling gene expression was increased in MCF-7 human breast carcinoma and HepG2 human hepatocarcinoma cells. | Data gap. | Hockley et al. (2006) |
| Tumorigenesis (adverse outcome) | YES: Gene expression of putative hepatocarcinogenes is markers were altered in liver, lung and forestomach of BaP-treated mice (e.g. Cdkn1a, Mdm2, Cbr1, Notch1, Nqo1, Nfe2l2 and Ephx1). | YES: Liver and lung tumors were observed in several other species. | Horikawa et al. (1991); IARC (2010); Lavoie et al. (1987); Park et al. (2011); Wester et al. (2012); Wislocki et al. (1986) | Alteration of Cdk, Mdm2, Nqo1transcript levels | NO: No direct evidence, but mutational pattern of p53 in human lung cancer is very similar to that of BPDE-treated human HeLa cells and bronchial epithelial cells. | Denissenko et al. (1996); Buick et al. (submitted); Hockley et al. (2006) |
ECOD, ethoxycoumarin O-deethylase; EROD, ethylresorufin O-dealkylase
Table 7.
RA2. Evaluation of proposed mode of action by Bradford Hill criteria. As originally proposed by Hill (1965) and simplified by Meek et al. (2003).
| Bradford Hill criteria | Questions asked | Evidence in animals | References |
|---|---|---|---|
| 1. Dose–response relationships | Is increasing exposure to the compound associated with increased magnitude of adverse outcome? | YES: increased BaP doses were associated with increased tumor incidence in mice and rats. | Lavoie et al. (1987); Wester et al. (2012); Wislocki et al. (1986) |
| Are key events observed at doses below or similar to those associated with the adverse outcome? | YES: total cumulative doses in the toxicogenomics experiments were below the cumulative doses in the cancer bioassay. | ||
| 2. Temporal association between key events and adverse outcome and reversibility | Is sequence of events logical? | YES | Halappanavar et al. (2011); Labib et al. (2012); Malik et al. (2012); Yauk et al. (2011) |
| Do key events and adverse outcome occur in expected order? | YES | ||
| Does cessation of exposure (or any interference with the proposed key events) ameliorate adverse outcome? | YES: cessation of exposure (sacrificing animals 24 hours after the exposure) markedly affected BaP-perturbed gene expression, restoring the expression of many genes affected 4 hours after the treatment in mouse lung and liver. | ||
| 3. Strength, consistency and specificity of association between key events and adverse outcome | Is the evidence linking exposure to a chemical, key events and adverse outcome strong? | YES | Reviewed in IARC (2010) |
| Is it consistent in repeated experiments in the same laboratory and other laboratories? | YES | ||
| Are the associations between exposure, key events and adverse outcome specific? | YES: the specificity of associations has been confirmed by several apical endpoints, including BaP metabolite detection in animal samples, DNA adducts, cell proliferation, immunoblotting and immunocytochemistry. | ||
| 4. Biological plausibility and coherence of the database | Does hypothesized MOA make sense based on broader knowledge about the chemical and the biology associated with adverse outcome (e.g. chemical properties, disease etiology)? | AHR-mediated metabolism of BaP is consistent with what is known about other PAH ligands of AHR. Genotoxic carcinogens are well known, and DNA–BaP adducts are well documented. BaP is positive in mutagenicity tests. | Reviewed in IARC (2010) |
Table 8.
BMD and BMDL values (in mg/kg-bw-day) at the 10th percentile of all affected genes in an IPA pathway for each proposed key event. A BMD/BMDL ratio greater than 10 indicates a poor model fit and is indicated as not applicable (N.A.). The number of genes modelled were 8 for lung, 5 for liver and 40 for forestomach. The BMDLs proposed as PODs are shaded and bolded.
| Liver | Lung | Forestomach | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3day+4hr | 3day+24hr | 28day+3day | 28day+3day | 28day+3day | |||||||
| Key Event | Pathway Title | BMD | BMDL | BMD | BMDL | BMD | BMDL | BMD | BMDL | BMD | BMDL |
| 1. BaP binding to AhR (Molecular Initiating Event). | |||||||||||
| 2. Induction of P450. 3. BaP Metabolism to Reactive Metabolites. | Aryl Hydrocarbon Signaling | 13.9 | 10.7 | 32.9 | 20.8 | 24.1 | 16.8 | 17.2 | 7.7 | 14.6 | 8.2 |
| Xenobiotic Metabolism Signaling | 13.6 | 10.1 | 20.0 | 4.3 | 27.5 | 14.1 | 29.8 | 13.9 | 13.5 | 7.9 | |
| NRF2-mediated Oxidative Stress Response | 40.1 | 18.6 | 29.7 | 6.9 | 31.4 | 21.3 | 28.0 | 18.6 | 15.4 | 9.0 | |
| Glutathione-mediated Detoxification | 70.3 | 49.6 | 82.1 | 54.4 | < 5 genes | < 5 genes | 15.3 | 9.0 | |||
| 4. DNA Adducts and DNA Damage. | p53 Signaling | 63.4 | 45.5 | 22.0 | 12.5 | 29.6 | 13.6 | 13.5 | 7.7 | 11.4 | 7.4 |
| Cell Cycle: G1/S Checkpoint Regulation | 75.7 | 43.8 | 8.1 | 1.0 | 24.6 | 12.4 | 11.7 | 8.9 | 18.5 | 7.7 | |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 77.1 | 29.9 | 79.7 | 43.3 | < 5 genes | 14.8 | 3.7 | 22.1 | 13.1 | ||
| Cell Cycle Control of Chromosomal Replication | 104.1 | 65.0 | 70.7 | 39.7 | 42.2 | 17.2 | Unperturbed | 12.8 | 8.0 | ||
| 5. Mutations. | |||||||||||
| 6. Altered Cell Signalling, Proliferation, and Apoptosis. | Apoptosis Signaling | 68.6 | 38.9 | 54.1 | 34.5 | 51.5 | 32.7 | 27.6 | 14.6 | 12.7 | 7.9 |
| Induction of Apoptosis by HIV1 | 64.6 | 36.8 | 22.0 | 7.2 | < 5 genes | 26.5 | 12.1 | 15.3 | 9.0 | ||
| LPS-stimulated MAPK Signaling | 26.9 | 15.6 | 23.0 | 14.9 | 35.0 | 20.2 | 24.3 | 12.1 | 15.0 | 8.8 | |
| Myc Mediated Apoptosis Signaling | 26.2 | 5.2 | 15.4 | 1.9 | < 5 genes | 30.6 | 7.0 | 15.0 | 8.9 | ||
| p38 MAPK Signaling | 53.4 | 35.3 | 26.5 | 19.3 | 25.3 | 11.9 | 23.3 | N.A. | 19.1 | 10.1 | |
| PPAR Signaling | 69.2 | 37.4 | 6.5 | N.A. | 42.9 | 28.6 | 27.0 | N.A. | 18.5 | 10.1 | |
| Retinoic acid Mediated Apoptosis Signaling | 73.8 | 50.3 | 58.2 | 36.3 | < 5 genes | < 5 genes | 19.6 | 9.1 | |||
| TGF-beta Signaling | 73.5 | 45.9 | 13.3 | 1.5 | 25.3 | 11.9 | 9.2 | N.A. | 12.8 | 8.0 | |
| UVA-Induced MAPK Signaling | 75.1 | 58.0 | 56.5 | 36.9 | 29.0 | 13.4 | 16.8 | N.A. | 15.9 | 8.0 | |
| VEGF Signaling | 63.4 | 31.7 | 9.9 | N.A. | 26.5 | 11.6 | 21.2 | 7.4 | 15.1 | 8.6 | |
| Wnt/beta-catenin Signaling | 15.8 | N.A. | 36.0 | 25.1 | 23.7 | 6.7 | 23.0 | 7.7 | 25.1 | 10.8 | |
| 7. Tumorigenesis (Adverse Outcome). | HER-2 Signaling in Breast Cancer | 61.5 | 39.3 | 31.9 | 16.6 | 33.7 | 19.5 | 22.9 | 10.1 | 16.1 | 8.2 |
| Prostate Cancer Signaling | 88.7 | 47.4 | 16.4 | 2.2 | 37.0 | 22.7 | 22.9 | N.A. | 15.8 | 9.0 | |
| Bladder Cancer Signaling | 78.4 | 47.8 | 6.7 | 2.3 | 44.9 | 29.6 | 17.2 | 9.5 | 21.6 | 8.6 | |
| Role of Tissue Factor in Cancer | 5.6 | N.A. | 20.3 | 2.3 | 24.7 | N.A. | 20.1 | 11.1 | 15.1 | 7.9 | |
| Thyroid Cancer Signaling | 60.8 | 46.5 | 45.7 | 28.4 | < 5 genes | 33.4 | 19.2 | 15.6 | 7.6 | ||
| Endometrial Cancer Signaling | 26.2 | 19.1 | 43.7 | 27.4 | 35.0 | 20.2 | 30.0 | 10.0 | 16.2 | 9.3 | |
| Estrogen-Dependent Breast Cancer Signaling | 38.7 | 24.5 | 59.7 | 38.4 | 37.0 | 22.0 | 18.7 | 7.6 | 13.0 | 7.5 | |
| Hereditary Breast Cancer Signaling | 75.4 | 53.2 | 11.1 | 2.2 | 28.7 | 13.3 | 26.6 | 17.3 | 14.7 | 8.8 | |
| Molecular Mechanisms of Cancer | 30.4 | 15.6 | 16.3 | 1.8 | 26.5 | 11.9 | 13.4 | 6.8 | 16.8 | 8.9 | |
| Colorectal Cancer Metastasis Signaling | 35.0 | 9.0 | 16.8 | 2.2 | 26.8 | 13.1 | 19.0 | 7.8 | 16.2 | 8.1 | |
| Non-Small Cell Lung Cancer Signaling | 62.8 | 37.8 | 20.4 | 5.9 | < 5 genes | 40.5 | 25.1 | 15.9 | 7.8 | ||
| Basal Cell Carcinoma Signaling | 21.5 | 13.0 | 29.0 | 20.7 | 43.9 | 27.1 | 17.3 | 10.0 | 18.2 | 9.7 | |
| Small Cell Lung Cancer Signaling | 38.9 | 24.2 | 59.2 | 36.6 | < 5 genes | 23.9 | 11.0 | 15.4 | 7.7 | ||
| Lowest BMDL 10th percentile regardless of MOA | 6.3 | 0.8 | 0.3 | 0.2 | 25.4 | 5.3 | 15.7 | 2.1 | 16.1 | 4.5 | |
| CXCR4 Signaling | Notch Signaling | Rac Signaling | Cellular Effects of Sildenafil (Viagra) | Phenylalanine Degradation IV (Mammalian, | |||||||
3.2.1.1 Key event 1: BaP binding to the AHR (molecular initiating event)
Multiple xenobiotic ligands (including BaP) bind to the AHR (Ovesen et al., 2011). Specific binding of BaP to the AHR has been measured in vitro directly using radiolabeled substrate (Monteiro et al., 2008), indirectly utilizing luciferase reporters containing the AHR response element (Malik et al., 2013) and by monitoring the expression of a known AHR target gene, Cyp1a1 (Ovesen et al., 2011; Table 5). Activation of the AHR by BaP leads to its own metabolism (Section 2.3.3) creating reactive metabolites that produce its subsequent genotoxic MOA. In addition, BaP–AHR activation causes transcriptional changes in genes related to carcinogenesis (Labib et al., 2012; Van Delft et al., 2012) and largely accounts for the nongenotoxic MOAs of BaP. Mice devoid of the AHR do not develop BaP-induced tumors, highlighting the receptor's role in BaP-induced carcinogenicity (Shimizu et al., 2000).
The relevance of this key event to humans has been demonstrated by photoaffinity ligand competition assays of BaP binding to the AHR in human primary hepatocytes (DiNatale et al., 2010). AHR polymorphisms can reduce ligand binding to the AHR (thereby reducing sensitivity to AHR ligands as measured by toxicity and the induction of AHR-dependent gene expression), but the effect of human AHR polymorphisms on the receptor's ligand binding function is poorly understood (Harper et al., 2002; Okey et al., 2005). The binding affinity of the AHR to its ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and AHR-mediated CYP1A1 inducibility varies greatly in humans (Harper et al., 2002). In addition, human cell lines with impaired CYP1A1 induction have been described, but the underlying mechanisms of these phenomena (e.g. genetic polymorphisms in AHR or other AHR signaling pathway genes) are unknown (Harper et al., 2002). Thus, it is reasonable to expect that this key event is relevant in humans, but that genetic variations in the human population may affect the susceptibility of humans to BaP.
3.2.1.2 Key event 2: Induction of cytochrome P450 enzymes (CYPs)
CYPs are prototypical responders to treatment with AHR ligands (Ovesen et al., 2011). Some of the AHR's target genes code for the cytochrome P450 enzymes (e.g. CYP1A1 andCYP1B1) that effectively metabolize BaP. Indeed, a large majority of BaP is detoxified at the site of contact in the gastro-intestinal tract (Nebert et al., 2013a). However, metabolism can also cause the formation of genotoxic metabolites of BaP that can escape detoxication. Using our mouse data, we noted that the transcription of many P450 genes (e.g. Cyp1a1, Cyp7a1 Cyp1b1, Cyp2b10, Cyp2b13, Cyp3a44, Cyp2b9, Cyp2c38 and Cyp2c40) increased following treatment with BaP treatment in mouse liver and lung (Halappanavar et al., 2011; Malik et al., 2012; Yauk et al., 2011). Of these, Cyp1a1 responded at the earliest time points and with the highest fold change compared with controls. Table 5 summarizes the Cyp1a1 and Cyp1b1 gene expression changes in the lung and liver 4 and 24 hours post-exposure. We did not have access to the forestomach from the 3-day acute studies; however, Uno et al. (2008) have shown that following five days of oral BaP exposure, the mRNA and protein levels of CYP1A1 and CYP1B1 are elevated in the forestomach. Increased CYP1A1 activity, as measured by the Ethoxyresorufin-O-deethylase (EROD) assay, has been shown in the livers of mice exposed to BaP (Bjelogrlic et al., 1989) and in BaP exposed human HepG2 cells (Vakharia et al., 2001). The occurrence of this key event in humans is supported by toxicogenomics work on human cells in vitro. Toxicogenomics analysis of primary human hepatocytes and the hepatoma cell line HepG2 challenged with BaP revealed the induction of P450 enzymes, including CYP1A1, in both systems (Hockley et al., 2006; Wilkening et al., 2003).
3.2.1.3 Key event 3: BaP metabolism to reactive metabolites
On its own, BaP is chemically inert; however, P450 enzymes metabolize BaP to genotoxic carbon cations or epoxides, such as the ultimate carcinogenic BaP metabolite BPDE (Cavalieri and Rogan, 1995). This metabolism depends on CYP1A1 and CYP1B1 enzymes, as demonstrated by knockout studies in animals. For example, BaP blood levels in Cyp1a1−/− and Cyp1a1−/− Cyp1b1−/− knockout mice are 25- and 75-fold greater than in control animals, respectively, and these mice are less sensitive to many BaP-related toxicities (Uno et al., 2006).
BaP metabolites undergo further transformation by phase II enzymes in order to minimize cellular damage from reactive metabolites (e.g. BPDE) and ROS (Section 2.3.3). These reactions include BaP conjugation to glutathione, glucuronic acid or sulfates by GSTs, UGTs and SULTs, respectively. Many phase II enzymes are activated at the mRNA level by the transcription factor Nrf2 (Nfe2l2) in response to the presence of reactive metabolites and oxidative stress (Kaspar et al., 2009). We observed the transcriptional induction of Nrf2 and its target genes (Nqo1, Ugdh, Srxn1, Akr1b15, Ugt2b15, Gsta1, Gsta2, Gstm3, Gstm4, Gstm7 and Gsto1) in mouse lung, liver and forestomach in response to BaP, in at least one time point (3- or 28-day regimens), suggesting that detoxication mechanisms were activated in order to cope with BaP toxicity (Labib et al., 2013; Malik et al., 2012; Yauk et al., 2011).
There is clear evidence that BaP metabolism to reactive metabolites occurs in humans. For example, enzyme assays confirm that BaP treatment causes an increase in the activity of P450 enzymes in primary human hepatocytes (Wilkening et al., 2003). Induction of phase II enzymes in response to BaP also occurs in cultured human cells (Hockley et al., 2006; Wilkening et al., 2003). In addition, the BaP metabolite BaP-7,8-diol, a precursor to the genotoxic metabolite BPDE, is found in the microsomes of human liver and lung donors (Shimada et al., 1989). Similarly, 3-OH BaP, an excreted BaP metabolite discussed in Section 2.3.3, is readily detectable in the urine of occupationally exposed workers (Forster et al., 2008).
3.2.1.4 Key event 4: DNA adducts and DNA damage
BaP metabolites, such as BPDE, can form covalent adducts with DNA and proteins. In addition, ROS produced as by-products of BaP metabolism (from BaP catechol–quinone redox cycling) may oxidize DNA (Lan et al., 2004). BPDE–DNA adducts can be measured directly, and their levels are markedly elevated by BaP treatment in lung, liver and glandular stomach tissues in our studies (Figure 2; Halappanavar et al., 2011; Labib et al., 2012; Lemieux et al., 2011; Malik et al., 2012). Previous studies have established that levels of DNA adducts in the forestomach from mice orally exposed to BaP for 24 hours and five days are comparable to those formed in the glandular stomach (Arlt et al 2008). Thus, the forestomach has the metabolic capacity to form DNA reactive BaP metabolites that could influence the extent of DNA damage and the outcome following 28 days of repeated exposure. DNA polymerase kappa (Polκ), which encodes a DNA polymerase involved in the error-free bypass of BPDE adducts, was upregulated in mouse lung and liver, illustrating the ability of genomic data to identify specific repair mechanisms induced in response to BaP exposure. Gene polymorphisms in Polκ are associated incidence of head and neck cancer in humans (Michiels et al., 2007).
We also noted increased expression of p53 target genes (a proto-oncogene signaling pathway) in lung, liver and forestomach samples from BaP-treated mice, indicative of DNA damage, repair and cell cycle arrest. This key event is also supported by toxicogenomics data from the kidney of BaP-treated mice (M. Luijten et al., unpublished data). For example, BaP exposure led to decreased transcription of the neuronal PAS domain protein 2 (Npas2), a transcription factor involved in DNA repair (Hoffman et al., 2008) and circadian rhythm–related processes in the kidney (Zuber et al., 2009). Reduced Npas2 expression in response to BaP suggests a decrease in the ability of the kidney to repair DNA damaged by BaP metabolites, potentially resulting in renal toxicity (e.g. Knuckles et al., 2001).
Our toxicogenomics analysis of the expression changes in TK6 cells treated with BaP (in the presence of rat liver S9) supports that similar genotoxicity and DNA damage response pathways are invoked in humans as in our rodent models. We observed a gene expression pattern indicative of TP53 activation in TK6 cells treated with BaP and expression profiles of BaP-treated cells were consistent with genotoxic DNA-reactive chemicals (Figure 5) (J. Buick et al., submitted). Over 50 genes were expressed (upregulated or downregulated) that were consistent with TP53 activation. These findings suggest that the genotoxic hazard is likely relevant to human cells following exposure to metabolized BaP. However, there is insufficient information to allow extrapolation of the TK6 cell results to cancer risks in drinking water due to BaP exposure in humans. Please note that the human cell culture work was used to support the MOA derived from the in vivo studies and not to extrapolate to human ingestion.
3.2.1.5 Key event 5: Mutations
Following induction of DNA adducts there are various possible fates of DNA lesions and the cells that contain damage. This can include error-free repair of the DNA lesions and resumption of normal cellular function, or in severely damaged cells, cell death via apoptotic and necrotic processes. Adducts that escape repair can cause mutations upon replication or DNA strand breaks leading to chromosomal aberrations.
There is extensive evidence that BaP causes mutations in animal tissues and that BaP metabolites cause mutations in human cells in culture. BaP metabolites and DNA adducts have been detected in cells isolated from BaP-exposed humans (Rojas et al., 2000). Strikingly, BPDE–DNA adducts were detected at mutational “hotspots” of TP53 (G in codons 157, 248 and 273), which are the sites of mutations in human lung tumors (Denissenko et al., 1996). The association of TP53 gene polymorphisms and cancer in humans has been summarized by Whibley et al. (2009). This, along with mutations in the human proto-oncogene K-RAS and tumor suppressor gene TP53 in BaP-exposed nonsmokers (DeMarini et al., 2001), provides support that the animal MOA is likely operative in humans.
Levels of BPDE adducts reflect the relative contribution of the rates of: (1) BaP metabolism to BPDE; (2) BaP metabolite detoxication by phase II enzymes; (3) adduct repair; and (4) cell turnover (Lemieux et al., 2011). The presence of BPDE–DNA adducts increases errors occurring during DNA replication, providing a mechanism for passing mutations on to the next generations of cells and thus contributing to the onset and progression of cancer (Perlow et al., 2002). Thus, earlier key events (1–3) in this MOA represent adaptive responses, reflecting normal homeostatic responses to xenobiotic exposure that do not necessarily lead to tumorigenesis on their own. Similarly, while the subsequent key event, DNA adducts and DNA damage is undesirable, it is not in itself an adverse effect as certain types of DNA damage may or may not predispose to mutations. However, mutations represent an irreversible “committed” step in the MOA, and mutations are known to increase the likelihood of tumorigenesis. Therefore, we based our POD selection on the key event preceding mutations (i.e., key event 4 - DNA adducts and DNA damage) in order to adequately protect against carcinogenic outcome as it precedes the committed step.
3.2.1.6 Key event 6: Altered cell signaling, proliferation and apoptosis
In addition to mutations, we found prominent changes in cell signaling pathways, including altered transcription of genes in several “cancer pathways” (Gohlke et al., 2009) in the tissues of rodents treated with BaP. These pathways included TGF-beta signaling, p38 mitogen-activated protein kinase (MAPK) signaling, VEGF signaling, and apoptosis signaling in all three tissues and data points (Table 8). Altered intracellular signaling may result from BaP binding to the human β2-adrenergic receptor and increasing intracellular calcium levels to activate multiple signaling pathways, including protein kinase C (Mayati et al., 2012).
It is difficult to predict which signaling pathways of those noted above drive the underlying mechanisms associated with BaP-induced liver, lung and forestomach tumors in rodents, although the results suggest that multiple pathways may contribute. Toxicogenomics data can be used to guide future hypothesis-directed/targeted research by revealing the putative signaling pathways that may be associated with tumorigenesis. For example, lipid metabolism was identified as the top function in a network analysis conducted on the gene expression changes occurring in mouse liver following BaP exposure (3-day treatment, animals sacrificed 4 and 24 hours later; (Yauk et al., 2011; Halappanavar et al., 2011)). The BaP-altered lipid metabolism network consisted of several genes, including Hnf4α (hepatocyte nuclear factor 4 alpha) and Cyp7a1 (also known as cholesterol 7α-monooxygenase). Cyp7a1 affects serum total cholesterol levels in mice (Kojima et al., 2009). Dysregulated cholesterol metabolism may increase cell proliferation through increased mevalonate levels (Nesnow et al., 2011), farnesylation and activation of Ras leading to tumorigenesis (Murphy et al., 2012).
Several studies have reported increased cellular proliferation in cultured cells (reviewed in Burdick et al., 2003; IARC, 2010) and animal models (Brandon et al., 2009; Wester et al., 2012) in response to BaP exposure. Studies in murine and human cells have clearly shown that BaP induces apoptosis in these cell models (reviewed in IARC, 2010). Indeed, BPDE–DNA adducts correlate well with TP53 induction in cultured human lymphocytes as measured by immunocytochemical staining (Godschalk et al., 2001). This finding is in agreement with the robust activation of p53 target genes that we observed in the lung, liver and forestomach of BaP-treated mice (Labib et al., 2012, 2013; Malik et al., 2012). Similarly, BaP exposure of human TK6 cells increased DNA damage and apoptosis (Figure 4) in parallel with changes in the expression of TP53-regulated genes (e.g. CDKN1A, CCNG1 and TP53INP1) that are involved in DNA damage response, cell cycle arrest and apoptosis (Supplementary Table 6), in accordance with gene expression data from mouse tissues in our experiments.
Evidence suggests that BaP has similar effects on cellular signaling in humans as it does in rodents. In addition to the robust activation of TP53 target genes by BaP in our TK6 cells, the transcription of genes involved in anti-apoptotic signaling is increased in MCF-7 human breast carcinoma and HepG2 human hepatocarcinoma cells exposed to BaP (Hockley et al., 2006) in a pattern largely consistent with rodents. Thus, we believe that the overall data strongly suggest that alteration of cellular signaling, especially related to p53 signaling and apoptosis is an important part of BaP genotoxicity. More studies are required to verify whether perturbation of single specific signaling pathways, or a group of pathways, leads to deregulation in cellular signaling and tumorigenicity following BaP exposures.
3.2.1.7 Tumorigenesis (adverse outcome)
BaP is a known rodent carcinogen, and various studies have demonstrated liver, lung and forestomach tumors in rodents following BaP exposure using standard apical tests (IARC, 2012; Lavoie et al., 1987; Wislocki et al., 1986). We observed alterations in the expression of various putative markers of chemically induced liver carcinogenesis (Park et al., 2011), including Cdkn1a, Mdm2, Ces5, Trp53inp1, Ccng1, Cbr1, Notch1, Nqo1, Nfe2l2 and Ephx, in the livers, lungs and forestomachs of mice treated with BaP in our 3- and 28-day experiments. Some of these genes (Cdkn1a, Mdm2 and Nqo1) have also been shown to be upregulated by BaP in human cells (J. Buick et al., submitted; Hockley et al., 2006). Additional global gene expression analyses using Cyp1a1 and Cyp1b1 knock-out mice, fed BaP, stress the importance of detoxication enzymes and identify potential cancer biomarker genes (Shi et al. 2010; Gálvez-Peralta et al. 2013). However, more work is required to determine which gene expression changes are cancer biomarkers that can be predictive of tumorigenesis using subchronic bioassays and in vitro testing. We believe that the toxicogenomics data point to strong candidates for hypothesis-based testing. Moreover, the findings suggest that toxicogenomics profiling has the potential to become a valuable predictive tool for hazard identification and risk assessment of chemical carcinogens.
3.2.1.8 Other potential modes of action and modulating factors
It is widely accepted that toxicants may affect multiple cellular processes and operate via more than one MOA, potentially leading to several adverse outcomes (Woodruff et al., 2008). In keeping with this model, our toxicogenomics data provide insight into other putative MOAs associated with BaP exposure that may be relevant to both cancer and non-cancer adverse outcomes as well as modulating factors that may exacerbate the effects of our proposed MOA. These include perturbation of circadian rhythm, pro-immune-mediated cellular transformation, other receptor-mediated signaling pathways, and immunosuppression. These significantly enriched biological processes may be working independently of our proposed MOA to induce cancer or non-cancer endpoints, or they may be involved in amplifying the effects of our proposed MOA. The toxicogenomics data revealed multiple signaling pathways affected by BaP, which would be tedious and impractical to identify using standard experimental approaches exploring individual apical endpoints. The relevance of these signaling pathways to BaP-mediated tumorigenesis must be examined in more detail in order to determine their relevance in toxicological response.
3.2.1.9 Uncertainties, inconsistencies and data gaps
As BaP is a well-known genotoxic carcinogen (IARC group 1), there are relatively few uncertainties relating to the cancer MOA presented in this case study. BaP is genotoxic to a large number of tissues/species and to human cells in vitro. There are no significant inconsistencies noted in the literature relating to the required metabolic activation and genotoxicity/mutagenicity of BaP (reviewed in IARC 2010). Thus, the relationship between mutagenicity and cancer is well-established. A major limitation leading to uncertainty in RA1 and RA2 is the lack of available human epidemiological data to support genotoxicity and carcinogenicity in humans. This stems from the fact that BaP is generally present in mixtures and thus measured effects cannot be distinguished from the other mixture components and lifestyle factors. In addition, the human AHR is known to have a lower binding affinity for BaP (Harper et al., 2002; Okey et al., 2005); therefore, it is unclear how susceptible humans are to BaP-mediated carcinogenicity. Evaluation of toxicogenomics profiles in combination with genotoxicity/mutagenicity studies in human cell culture models derived from different donors would capture a broad array of potential inter-individual susceptibility and could be useful for addressing some of these data gaps and human relevance.
3.2.2 Confidence in the proposed mode of action and selection of the point of departure for risk assessment (RA2)
Our confidence in the proposed MOA is high because:
-
1)
Multiple studies support our proposed key events in the animal MOA, including our three genomics studies in vivo on different mouse strains and tissues and a large database of published apical studies.
-
2)
In general, all key events in the animal MOA are plausible in humans and are supported by our human cell culture study and published information.
-
3)
Apical and toxicogenomics data satisfy the Bradford Hill criteria (Table 7). Increasing exposure to BaP results in an increased magnitude of genotoxic, toxicogenomic and carcinogenic effects. The highest dose used in the toxicogenomics studies (300 mg/kg bw per day for 3 days) represents a total BaP dose of 900 mg/kg bw, which is less than the total BaP dose of ~1800 mg/kg bw that causes tumors in rat livers (2-year cancer bioassay, 5 days per week treatment with 3 mg/kg bw per day; Wester et al., 2012). The toxicogenomics data support the temporal association between key events and adverse outcome in the proposed MOA. The proposed MOA is consistent with knowledge in the literature about AHR agonists and the well-established effects induced by BaP.
To summarize, the genomics-informed MOA consists of AHR activation, induction of AHR target genes, BaP metabolism, DNA adducts and DNA damage. Replication of unrepaired DNA damage can yield mutations; if mutations occur in proto-oncogenes or tumor suppressor genes, they can cause tumorigenesis. Thus, earlier key events (1–3) in this MOA represent adaptive responses that reflect normal homeostatic responses to xenobiotic exposure do not necessarily lead to tumorigenesis on their own. Similarly, DNA adducts and damage are not necessarily adverse effects because of cellular stress response and DNA repair mechanisms that cope with such damge. However, mutations are irreversible and increase the likelihood of tumorigenesis. Therefore, our POD selection was based on DNA adducts and damage because it precedes the committed step in order to adequately protect against carcinogenic outcome. This approach was applied in order to be conservative and protect against subsequent adversity.
Table 8 provides BMD/BMDLs at the 10th percentile for all affected genes in the IPA pathways deemed relevant for each key event. While no formal guidelines exist for considering transcriptional BMDs in risk assessment, the BMD/BMDL ratio for traditional toxicity endpoints is an indication of uncertainty of a BMD estimate (Muri et al., 2009). It has been suggested that BMDL values reflect uncertainty of the data (e.g. due to measurement errors) and that BMD/BMDL ratios of < 2 indicate little uncertainty (Muri et al., 2009). We selected a BMD/BMDL ratio of < 10 as being acceptable to be conservative and also as an attempt to capture the relatively dynamic (and hence, perhaps, more prone to measurement errors) nature of gene expression data, compared to apical data. Further investigations are required to establish appropriate BMD/BMDL ratios for transcriptomics data.
The lowest transcriptional IPA pathway BMDL for key event 4 for the liver was “Cell Cycle: G1/S Checkpoint Regulation” at 1.0 mg/kg bw per day (Table 8). The lowest BMDLs for key event 4 in the lung and forestomach were the IPA pathways “Cell Cycle: G2/M DNA Damage Checkpoint Regulation” and “p53 Signaling” at 3.7 and 7.4 mg/kg bw per day, respectively (Table 8). For the liver, lung and forestomach, 18, 5 and 40 genes were modeled (i.e., had adequate BMD fits), respectively (Supplementary Table 7).
3.3 Genomics-only approach (RA3)
The goal of RA3 is to determine whether toxicogenomics information (in the absence of any other toxicological data) can be used to develop an MOA and inform a POD for risk assessment. This approach is particularly useful for data-poor chemicals. To this end, in RA3 we assume that we have no knowledge of BaP other than its structure and solubility. We propose an MOA and select a POD for this uncharacterized chemical using only the available genomics data. We analyzed our toxicogenomics data using a commercially available pathway analysis tool, IPA, but other publicly available data analysis tools (e.g. the freely available DAVID) can also be used. Bioinformatics tools were used to identify the most enriched pathways and biological processes associated with exposure to BaP. The development of our putative MOA was based on the degree of enrichment (i.e. the top affected pathways/processes), published literature on the biology of these pathways, the doses and time points at which they were affected and their BMDs. Support from other published BaP genomics studies were also used to support the findings.
3.3.1 Proposed mode of action
Our proposed MOA for BaP using exclusively genomics information (Figure 7) was analyzed for dose-response and temporal concordance (Table 9), and BMD/BMDLs at the 10th percentile of all affected genes in an IPA pathway for each proposed key event was determined (Table 9). The genomics information used to construct this MOA includes fold change in transcriptional response of BaP-exposed compared to control rodents or human cells for individual genes, as well as functional pathway analysis of all significantly differentially expressed genes at each dose and time point. Forestomach data were not included in the initial analysis because only samples from the 28 + 3 day regimen were available. However, forestomach data were used to support the MOA generated from the lung and liver data, and genes were modeled to derive BMDLs.
Figure 7.
RA3: Carcinogenic MOA of BaP in rodent liver, lung and forestomach developed using toxicogenomics data exclusively. In addition to the main genotoxic MOA (gray boxes), other factors are plausible based on the data. Redox reactions are unbalanced, showing conversion of molecular oxygen to reactive oxygen species (ROS; Section 2.3.3). Key events are numbered 1–4 and schematically represented.
Table 9.
RA3: Dose-response, temporal concordance and lowest BMDL1SDs (mg/kg bw per day) at the 10th perecentile for all affected genes in an IPA pathway for each proposed key event in benzo(a)pyrene's mode of action.
| Key event 1: Activation of AHR signaling |
Key event 2: Nrf2-mediated oxidative stress response |
Key event 3: DNA damage, repair and apoptosis |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Dose (mg/kg bw per day) |
Liver | Lung | Forestomach | Liver | Lung | Forestomach | Liver | Lung | Forestomach |
| 5 | Cyp1a1 25.5/ns | Cyp1a1 46.9/–7.5 | Nqo1 5.4/ns | ||||||
| Gpr97 1.8/ns | Cyp1b1 10.4/1.5 | Srxn1 1.8/ns | |||||||
| Slc46a3 1.9/ns | |||||||||
| 50 | Cyp1a1 110/ns/ns | Cyp1a1 103/–1.5/ns | Gsta1 2.5 | Nqo1 1.9/ns/ns | Nqo1 27.0/ns/ns | Bax ns/ns/2.5 | Bax ns/ns/2.5 | Bst2 2.7 | |
| Gpr97 2.4/ns/ns | Cyp1b1 23.5/1.7/ns | Ces5 1.6 | Srxn1 1.5/ns/ns | Srxn1 7.1/ns/ns | Ccnd1 ns/ns/1.8 | Ccnd1 ns/ns/1.8 | Mgmt 1.9 | ||
| Slc46a3 2.3/ns/ns | Gsta1 ns/2.4/ns | Ugt2b15 2.5/ns/ns | Ccng1 ns/ns/3.0 | Ccnf1 ns/ns/3.0 | Pmaip1 1.8 | ||||
| AHRR 1.5/ns/ns | Ugdh 2.0/ns/ns | Cdkn1a ns/ns/6.8 | Cdkn1a 2.2/ns/8.1 | Trp53inp1 1.7 | |||||
| Akr1b151.6/ns/ns | Gadd45g 1.8/ns/ns | Gadd45g 1.8/ns/ns | |||||||
| Polκ ns/ns/1.6 | Polκ ns/ns/1.6 | ||||||||
| Apaf1 ns/ns/1.8 | Serpine1 ns/ns/3.2 | ||||||||
| Zmat3 ns/ns/1.6 | Zmat3 ns/ns/1.7 | ||||||||
| Pmaip1 ns/ns/1.7 | Pmaip1 ns/ns/1.7 | ||||||||
| 150 | Cyp1a1 146.7/ns | Cyp1a1 179.3/14.7 | Nqo1 4.5/2.3 | Nqo1 48.2/1.9 | Gadd45g 4.6/ns | Gadd45g 1.5/ns | |||
| Gpr97 1.9/ns | Cyp1b1 70.9/2.4 | Srxn1 3.2/ns | Srxn1 12.6/ns | Cdkn1a 1.7/ns | Cdkn1a 7.0/2.4 | ||||
| Slc46a3 2.5/ns | Gsta2 1.8/ns | Ugt2b15 3.2/ns | Ugt1a9 ns/–2.3 | Ccng1 ns/1.5 | Ccng1 1.5/1.6 | ||||
| AHRR 2.3/ns | Gsta1 1.8/1.6 | Ugdh 3.4/ns | Mdm2 1.6/ns | ||||||
| Akr1b15 2.6/ns | Polκ 1.5/ns | ||||||||
| Bax 1.8/ns | |||||||||
| Sesn2 1.6/1.7 | |||||||||
| 300 | Cyp1a1 165.4/9.0 | Cyp1a1 210.1/42.9 | Nqo1 8.6/4.3 | Nqo1 52.6/6.6 | Gadd45a 1.5/ns | ||||
| Gpr97 ns/ns | Cyp1b1 161.2/21.8 | Srxn1 6.3/ns | Srxn1 16.2/1.6 | Gadd45g 12.6/ns | Gadd45g 2.9/–1.5 | ||||
| Slc46a3 2.4/1.7 | Gsta2 2.2/ns | Ugt2b15 3.7/ns | Cdkn1a 5.0/ns | Cdkn1a 15.8/4.5 | |||||
| AHRR 3.0/ns | Gsta1 2.5/ns | Ugdh 4.3/ns | Ccng1 2.0/1.7 | Ccng1 2.2/2.3 | |||||
| Akr1b15 3.0/1.5 | Ccnd1 –1.5/2.2 | Mdm2 2.0/1.6 | |||||||
| Mdm2 1.5/ns | Polκ 2.1/1.9 | ||||||||
| Ddit4 3.9/ns | Bax 2.6/2.6 | ||||||||
| Sesn2 2.8/2.3 | |||||||||
| Lowest BMDL pathway | 4.3 Xenobiotic Metabolism Signaling | 7.7 Aryl Hydrocarbon Signaling | 7.9 Xenobiotic Metabolism Signaling | 6.9 NRF2-mediated Oxidative Stress Response | 18.6 NRF2-mediated Oxidative Stress Response | 9 NRF2-mediated Oxidative Stress Response | 1 Cell Cycle: G1/S Checkpoint Regulation | 3.7 Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 7.4 p53 signaling |
The values shown with each gene represent fold changes (> 1.5 fold change from control and FDR-adjusted p-value < 0.05; ns indicates no statistical significance) at that dose for: (3 days + 4 hours / 3 days + 24 hours). An additional timepoint is indicated for the 50 mg/kg bw/day dose (3 days + 4 hours / 3 days + 24 hours / 28 days + 3 days). The darkening of the cells indicates the expected progession of gene expression responses from early time points to later time points and with increasing dose as support for progression of proposed key events.
The top canonical pathways in liver and lung tissues (identified using IPA) perturbed in mice exposed to the highest doses of BaP in our experiments (150–300 mg/kg bw per day) included Nrf2-mediated response to oxidative stress, glutathione metabolism, AHR signaling and xenobiotic metabolism (Figures 8 and 9). To determine the appropriate order of these pathways to construct a putative MOA, we examined the dose-response and temporal concordances for changes in the transcriptional responses of genes within these pathways. At the lowest dose and time point we found changes in AHR regulated genes, while at the next highest dose we detected changes in genes related to Nrf2-mediated responses to oxidative stress and DNA damage response (occurring at the later time point, 28 days, and at higher doses). Thus, we proposed the four key events described below.
Figure 8.
RA3: Top eight canonical pathways affected by BaP in the livers of mice. The data shown are for mice treated with 300 mg/kg bw per day for 3 days and sacrificed 4 hours after the last exposure (Yauk et al., 2011). The left Y-axis represents the percentage of genes in each pathway, the numbers at the top of the graph indicate the number of genes in each pathway and the right Y-axis shows the negative log (p-value) of the Fisher's exact test performed by Ingenuity Systems Analysis software.
Figure 9.
Heatmap analysis showing Ingenuity® Pathway Analysis (IPA) canonical pathways significantly affected in BaP-exposed TK6 cells and lungs, liver and forestomach of mice exposed to BaP. The left panel (A) compares in vitro TK6 pathways with lung and liver tissue from acute exposures (3 days + 4 hours), whereas the right panel (B) compares the in vitro TK6 pathways with lung, liver and forestomach tissue from the subchronic exposures (28 + 3 days). Pathways highlighted in yellow are consistent with the genotoxic MOA presented in this document. P-value scores (indicated by gradation of purple colors) are a measure of the significance of the pathway's association with the dataset.
3.3.1.1 Key event 1: Activation of AHR signaling
At our lower BaP doses (5 and 50 mg/kg bw per day), significant changes in transcription were found in genes involved in AHR signaling in both the liver (Cyp1a1, Gpr97, Slc46a3 and AHRR) and lung (Cyp1a1 and Cyp1b1). Activation of AHR signaling plays two roles in response to a toxin that depends on the individual ligands: (1) a transcriptomic response that alters cell signaling; and (2) an adaptive response resulting in metabolism of endogenous and exogenous chemicals. Microarray studies have significantly contributed to the identification of gene products that are regulated by the AHR (reviewed in Denison et al., 2011). The transcriptomic response plays a role in halogenated aromatic hydrocarbon-associated carcinogenesis, whereas metabolism of toxicants to reactive metabolites is implicated in PAH-induced carcinogenesis. CYP1A1and CYP1B1 show different substrate preferences and can catalyze different stereospecific oxidations (Murray et al., 2001). These enzymes are known to metabolize certain xenobiotics into reactive metabolites. Upregulation of the AHR-regulated genes Cyp1a1, Cyp1b1, Gpr97 and Slc46a3 at the lowest dose strongly suggests that key event 1 is the activation of AHR signaling. The function of Slc46a3 (solute carrier family 46, member 3) is unknown, but it appears to be regulated by Cyp1a1 (Dragin et al., 2008). Increased Gpr97 (G protein-coupled receptor 97) has also been observed in mice treated with the prototypical AHR inducer TCDD (Tijet et al., 2006). Unlike Cyp1a1, the relevance of Gpr97 and Slc46a3 to the MOA of BaP is not obvious, and the expression of these genes lacked a dose–response relationship (Table 9B). The AHR repressor (AHRR) is a negative regulator of the transcriptional activity of AHR. AHRR is also under the regulation of the AHR, and its expression was also increased in a dose-dependent manner in the liver. Therefore, our data suggest that AHR signaling, and the subsequent induction of xenobiotic metabolism, is an early key event in the MOA of BaP-induced toxicity.
There is support for the plausibility of this key event in humans using genomics data. For example, transcriptomic profiles from cultured human HepG2 and MCF-7 cells demonstrate the upregulation of CYP1A1 and CYP1B1 in response to BaP treatment (Hockley et al., 2006; Van Delft et al., 2010).
Thus, activation of the AHR, and subsequent xenobiotic metabolism signaling, is a logical early key event in BaP-mediated toxicity that was identifiable using genomics data in the absence of apical data.
3.3.1.2 Key event 2: Nrf2-mediated response to oxidative stress
“Glutathione metabolism” and “Nrf2-mediated oxidative stress response” (~50% of the 192 genes in this pathway responded) were the top two pathways affected by BaP treatment in mouse liver and lung (Figure 8). Expression of high levels of P450 enzymes, such as CYP1A1 and CYP1B1, results in increased metabolism of chemicals and the subsequent generation of ROS and oxidative stress. Nrf2 is a transcription factor that mediates cellular response to oxidative stress and xenobiotics in what is known as the AHR–Nrf2 gene battery (Yeager et al., 2009), including Nqo1 and Ugt (Kaspar et al., 2009; Thimmulappa et al., 2002). Many of the Nrf2-controlled genes aid in the repair of macromolecules damaged by ROS and produced during xenobiotic metabolism, as well as detoxication processes, through the conjugation of water-soluble moieties to xenobiotic metabolites produced by phase I enzymes. Maintenance of the appropriate cellular redox status is crucial, and this is achieved primarily by a high (millimolar) cellular content of glutathione. Downstream targets of Nrf2-dependent gene expression were altered at 50 mg/kg bw per day in liver and 5 mg/kg bw per day in lung, consistent with the idea that AHR-mediated CYP1A1 induction is required to initiate BaP metabolism. At the second lowest dose (50 mg/kg bw per day), 5 of the 34 genes affected 4 hours post-treatment were upregulated Nrf2-dependent genes (Nqo1, Ugdh, Srxn1, Akrb1b15 and Ugt2b15), in addition to upregulation of Nrf2 itself. The transcriptional levels of all five of these genes in the liver, as well as Nqo1 and Srxn1 in the lung, increased with increasing BaP dose. Nrf2-mediated response to oxidative stress is thus proposed to be key event 2 in both the lung and liver, based on the mouse genomics data alone.
Toxicogenomics profiling of human cells supports this key event. Indeed, strong upregulation of phase II metabolism genes is observed in cultured human HepG2 and MCF-7 cells (Hockley et al., 2006; Van Delft et al., 2010). Furthermore, the Nrf2 signaling pathway was among the main upregulated gene clusters in BaP-treated HepG2 cells (Van Delft et al., 2010). Thus, genomics data in human cells in culture support the relevance of this key event to humans.
Epidemiological research has shown that certain Nrf2 gene polymorphisms are associated with an increase in the incidence of breast cancer and decreased survival outcome in breast cancer patients (Hartikainen et al., 2012). This finding suggests that certain perturbations in the Nrf2 signaling pathway are relevant to carcinogenesis in humans.
Overall, in the absence of apical endpoints, the genomics data suggest that the induction of the AHR initiates xenobiotic metabolism of the chemical to a toxic metabolite or produces ROS, which leads to the induction of the Nrf2-mediated signaling. In the absence of data on tissue levels of the chemical and its metabolites and cellular ROS levels, it is not possible to discern between these two mediating factors. Thus, we have assigned the generic name “Nrf2-mediated response to oxidative stress” to this key event. Quantitative structure–activity relationship (QSAR) modeling in parallel with analysis of the genomics data would be beneficial to support the genomics data analysis for data-poor chemicals.
3.3.1.3 Key event 3: DNA damage, repair and apoptosis
Following the induction of an oxidative stress response, it is logical to suspect that DNA damage response and apoptosis may ensue. We previously reported a robust response in the p53 signaling pathway in gene expression profiles from the livers (Malik et al., 2012), lungs (Labib et al., 2012) and forestomachs (Labib et al., 2013) of mice treated for 28 days with BaP. Multiple p53-dependent genes (including Cdkn1a, Ccng1 and Ccnd1) were similarly dose-dependently upregulated in mouse liver and lung at the 3-day time point. In addition, transcript levels of Mdm2 (a known p53 inhibitor), Ddit4 and Gadd45g (growth arrest and DNA damage inducible 45 gamma) were significantly and dose-dependently upregulated in mouse liver and lung after 3 days of BaP exposure. p53 is a transcription factor that is referred to as “the guardian of genome” and mediates cellular processes, including cell cycle arrest, DNA repair and apoptosis (reviewed in Meek, 2009). Several genes implicated in these three events were differentially expressed in the 28-day lung, liver and forestomach gene expression profiles (Table 9), such as Cdkn1a and Ccng1 (cell cycle arrest), Polκ (DNA repair), Bax and Pmaip1 (apoptosis). The upregulation of these genes strongly suggests activation of p53 in response to DNA damage. Taken together, these results suggest that BaP exposure causes DNA damage through the generation of DNA-reactive metabolites by AHR-regulated metabolic enzymes. Based on these findings, we would speculate that the chemical operates through a genotoxic MOA and thus has a high probability of being carcinogenic.
Our work in cultured human TK6 cells supports that BaP is genotoxic in these human cells. Our DNA microarray analysis revealed that over 60% of the genes in the TP53 pathway responded to BaP treatment in TK6 cells (J. Buick et al., submitted). In addition, the toxicogenomics profiles of treated TK6 cells classified BaP as a genotoxic compound that clusters with other known DNA-reactive mutagenic carcinogens (J. Buick et al., submitted). Toxicogenomics analyses of HepG2 cells have also shown activation of the TP53 signaling pathway and perturbations in apoptosis-related gene sets as a result of exposure to BaP (Van Delft et al., 2010). More recently, activation of oxidative stress and DNA damage response pathways by BaP was confirmed by an independent toxicogenomics RNA-Seq technology (Van Delft et al., 2012). We note that our toxicogenomics data are consistent with the observed increases in cytotoxicity, apoptosis and induction of micronuclei that we measured in the TK6 cells treated with BaP (Figure 4).
3.3.1.4 Key event 4: Mutations
As described in RA2, there are various possible fates of cells with DNA damage including error-free DNA repair or cell death via apoptotic processes. DNA lesions that escape repair may cause mutations or chromosomal aberrations. In contrast to DNA damage, a mutation is a permanent change in the DNA sequence that cannot be repaired. Thus, mutation induction can be considered a committed step in this MOA as described in RA2. Although induction of DNA damage response pathways is not a direct measure of mutation or chromosome aberrations, induction of these pathways is a well-established response to pre-mutagenic lesions and thus provides evidence that mutagenicity is a potential outcome of the exposure. Indeed, many studies have used omics approaches to show that the induction of gene expression changes in DNA damage response pathways is highly informative of genotoxicity (e.g., Sakai et al., 2014; Derks et al., 2014; Waters et al., 2010, our own work in TK6 cells). As noted for the previous proposed key event, our case study data clearly show induction of expression changes in DNA damage response pathways in the tissues of exposed mice and human TK6 cells, inferring that the agent may be mutagenic. Thus, even though published reports of BaP's in vivo mutagenicity are not considered here, we argue that our genomics analysis provides evidence that the toxicant is mutagenic in vivo in rodents and that this is likely a key event in the MOA.
3.3.1.5 Other potential modes of action and modulating factors
Other pathways affected by BaP treatment may represent biological responses that result from complex interplays between signaling pathways affected by BaP and its metabolites. Circadian rhythm and lipid metabolism are examples of BaP-inducible pathways (also affected by BaP in other toxicogenomics studies; Hockley et al., 2006; Van Delft et al., 2010) that are difficult to fit into a carcinogenic MOA. In addition, gene signatures related to immunotoxicity were prevalent in the lung and forestomach datasets, indicative of immunosuppression and proinflammatory responses, respectively (Halappanavar et al., 2011; Labib et al., 2012; Labib et al., 2013). Therefore, our genomics data identify immunosuppression as a possible hazard that is a plausible modulating factor in the MOA for BaP-induced carcinogenicity in the lung and the recognition of excessively accumulated metabolites as potential pathogens in the forestomach. These perturbations may impact a diverse array of toxicities outside of cancer. However, their expression was affected at higher doses compared with those activated in key events 1 and 2.
Although simplistic, our analysis suggests that the genotoxic MOA is the most relevant for health risk assessment of BaP-induced carcinogenicity, especially for extrapolations to low doses.
3.3.1.6 Uncertainties, inconsistencies and data gaps
Unlike RA2, there are many uncertainties in the genotoxic MOA that we proposed for BaP based solely on toxicogenomics data. This is due to many reasons including: (a) there were only a handful of toxicogenomics studies that were conducted on BaP; (b) the studies were conducted in only a few different cell culture and animal models; (c) the studies were focused on different tissues; and (d) the experimental designs were not optimal for risk assessment (e.g., because of suboptimal sampling time and high doses). Moreover, in the complete absence of any ADME, PBPK or apical data, RA3 is limited. However, we expect that toxicogenomics data alone will be invaluable in prioritization, hazard identification, and determination of other important apical tests, as we discuss in Section 4.4 below.
For example, although it is clear that the chemical agent induces AHR signaling followed by oxidative stress and a DNA damage response, it is unclear whether the metabolism/detoxication occurs, whether the parent compounds, metabolites or ROS alone are the genotoxic agents and whether the DNA damage indeed leads to DNA mutations (as DNA damage can be repaired). Given our understanding of the relationship between mutagenicity and carcinogenicity, complementary knowledge on the ability of the chemical to cause mutations will always be critical.
One very general uncertainty lies in the application of mRNA profiling on its own since biological processes are carried out at higher levels of organization. Validation at the phenotype level (anchoring to some apical endpoint(s)) is still required at this time and thus we would recommend targeted testing following a toxicogenomics study. We expect that the predictive capabilities of toxicogenomics will improve as more work is completed in the field to link pathway perturbations measured at the RNA level to apical endpoints and adverse effects. The use of toxicogenomics profiles is promising, as there is a great deal of consistency across toxicogenomics studies in various species. These support the MOA proposed here. Nevertheless, at this time the uncertainties are great.
A major gap in the toxicogenomics approach was the limited number of tissues examined in rodents. Chemicals can and do demonstrate tissue-specificities (and interaction occurs among tissues in MOAs), thus it must be established that the tissues selected are representative of target tissues (see Section 4.3 for further discussion). Given that we observed clear evidence of genotoxicity in every tissue studied in mice, and in cultured human cells, we can fairly confidently say that this chemical induces genotoxicity, and thus most likely tumors, in many tissues. Moreover, our TK6 signatures for the chemical are aligned with highly genotoxic agents, predicting that the chemical is genotoxic. This signature is currently undergoing extensive validation through the Health and Environmental Sciences Institutes.
Overall, there were few toxicogenomics studies and thus it is clear that the gaps and uncertainties will be much larger for RA3. A required next step in a toxicogenomics risk assessment would be targeted testing of the associated adverse effects predicted from the signature, and this was not conducted here.
3.3.2 Quantitative structure–activity relationship (QSAR)
We explored whether in silico approaches could provide support to our toxicogenomics analysis for RA3. As noted, exposure of mice to BaP resulted in significantly increased expression of CYP1A1and a robust DNA damage response in lung and liver, indicative of the genotoxicity of BaP. To provide supporting data to demonstrate that BaP is a substrate of CYP1A1 that is metabolized to genotoxic reactive products, an analysis of the correlation between the structural parameters of BaP and other similar compounds with different abilities to be activated by CYP1A1 and inflict DNA damage would be helpful. Indeed, such a correlation analysis was conducted previously for several PAHs, excluding BaP (Lewis, 1987; Wan et al., 2006). The models successfully predicted the ability of PAHs to induce CYP1A1 via the AHR, their hydroxylation to reactive intermediates, their carcinogenic potential (Lewis, 1987) and their ability to induce apoptosis (Wan et al., 2006). The latter model was validated by a flow cytometry–based apoptosis assay, which yielded positive results for BaP and negative results for 2-methylanthracene, which lacks a bay region. The bay region was demonstrated to be important for the aforementioned biological activities of PAHs. In our case, had we had no other genotoxic endpoints (e.g. DNA adduct data), QSAR could confirm our gene expression data, pointing to BaP-mediated DNA damage. Therefore, QSAR methods could provide strong support for toxicogenomics data and may aid in extending toxicogenomics findings to other related compounds.
3.3.3 Rationale for the proposed mode of action and selection of the point of departure for risk assessment (RA3)
Using toxicogenomics-derived data in combination with supporting evidence from QSAR models, we propose a genotoxic MOA that includes: (1) the induction of xenobiotic metabolism genes via activation of the AHR pathway; (2) metabolic activation leading to reactive metabolites and ROS (Nrf2-mediated response to oxidative stress); and (3) DNA damage, repair and apoptosis (p53 signaling) and (4) mutations. While this genomics approach did not identify the full mechanism underlying BaP toxicity, we believe that genomics identified the major key events in BaP-induced tumor formation. The proposed MOA is supported by in-house as well as published animal and human genomics data. The key events in the MOA passed the Bradford Hill criteria (Table 7): (1) dose–response relationship for all key events were consistent with expectations; (2) key events occurred in a logical sequence; (3) the results were consistent across various experiments; (4) the events are biologically plausible; and (5) the key events are supported by human data. Based on the same logic used in RA2 (Section 3.2.2), we chose key event 4 as the “committed step,” increasing the likelihood of achieving an adverse outcome in the genotoxic MOA. Therefore, the preceding key event was selected as the POD. The lowest BMDL values for the key event preceding the committed step in the liver, lung and forestomach were 1.0, 3.7 and 7.4 mg/kg bw per day, respectively (representing the IPA pathways “Cell Cycle: G1/S Checkpoint Regulation”, “Cell Cycle:G2/M DNA Damage Checkpoint Regulation” and “p53 Signaling” respectively; Table 9).
3.4 Alternative genomics approaches
Thomas and colleagues (Thomas et al., 2007a, 2011, 2012) propose that the pathways or biological functions with the lowest median gene expression BMD values correlate well with BMDs for apical cancer and noncancer endpoints. This approach is especially useful for chemicals that act via nonselective mechanisms, perturbing multiple different biological pathways (Thomas et al., 2013). If we use an approach similar to Thomas et al. (2011), whereby the BMDL for the most sensitive pathway or process (i.e. the one with the lowest BMDL value at the 10th percentile) is selected for establishing the POD (after removing pathways in which fewer than five genes were significant), we achieve the following BMDLs: (1) liver IPA pathway “Notch Signaling” BMDL of 0.2 mg/kg bw/day (seven genes were modeled); (2) lung IPA pathway “Cellular Effects of Sildenafil (Viagra)” BMDL of 2.1 mg/kg bw/day (nine genes were modeled); and (3) forestomach IPA pathway “Phenylalanine Degradation IV (Mammalian, via Side Chain)” BMDL of 4.5 mg/kg bw/day (six genes were modelled) (Table 10). Thus, using an approach similar to Thomas et al. (2011) our POD (assuming no knowledge of the MOA) would be 0.2 mg/kg bw per day.
Table 10.
Summary of calculated BMD/BMDLs for local (forestomach) and systemic (liver and lung) effects using traditional and toxicogenomics approaches. Benchmark response is BMD10/BMDL10 for quantal tumor data and BMD1SD/BMDL1SD for continuous mutations and transcriptional data (mg/kg bw per day).
| Liver | Lung | Forestomach | ||||
|---|---|---|---|---|---|---|
| Approach | BMD/BMDL | Response | BMD/BMDL | Response | BMD/BMDL | Response |
| RA1: Traditional | 1.8 /1.2 | tumors | 0.8* | tumors | 0.8/0.5 | tumors |
| RA1: Mutations | 7.2/4.8 | mutations | 2.2/1.4 | mutations | 0.5/0.3 | mutations |
| RA2/RA3: Toxicogenomics-key event preceeding the committed step | 8.1/1 | DNA damage | 14.8/3.7 | DNA damage | 11.4/7.4 | p53 signaling |
| Toxicogenomics-lowest MOA-associated pathway | 8.1/1 | DNA damage | 14.8/3.7 | DNA damage | 11.4/7.4 | p53 signaling |
| Toxicogenomics-Lowest pathway# | 0.3/0.2 | notch signaling | 15.7/2.1 | cellular effects of sildenafil | 16.1/4.5 | phenylalanine degradationIV |
Caution, based on a LOAEL from coal tar data (Culp et al., 1998)
Similar to Thomas et al., (2011) but examined BMD/BMDLs at the 10th percentile rather than median.
An additional toxicogenomics approach would select the lowest POD associated with the MOA without defining a committed key event. Using this approach the PODs for liver, lung and forestomach would be 1.0, 3.7 and 7.4 mg/kg bw per day (Table 10). These are the same PODs and pathways identified using the RA2 and RA3 approach (selecting the key event preceeding the committed key event as the POD).
4. Discussion and conclusions
Our objective was to compare toxicogenomics and traditional approaches to inform MOA and POD for the risk assessment of BaP in drinking water. Overall, we demonstrate that the PODs derived from the three approaches were highly similar (Table 10). However, we note that RA1 was based on the collection of decades of research on BaP and integrated two full 2-year cancer bioassays. In comparison, RA3 used fewer than 100 mice in subchronic and acute exposure scenarios, in parallel with analysis of human cells in culture. The approximate cost for these experiments using the applied technologies was $100,000. However, we note that with improving technologies (e.g. multiplex RNA-sequencing approaches, similar to wha has been described by Li and co-workers (2012)), and with integration of toxicogenomics in acute or sub-chronic repeat-dose studies, tissue profiling could be done much more quickly and cost effectively. Thus, for data-poor chemicals, our RA3 data support that implementing a genomics-based risk assessment approach may provide an effective means to reduce cost, time and animal use in preliminary human health risk assessment. RA2 demonstrates the utility of integrating toxicogenomics data with data from traditional toxicity testing to support POD selection and MOA development. Below, we compare the RA approaches and describe the existing value-added and limitations of toxicogenomics in human health risk assessment.
As a practical case study, we derived tissue-specific MOA-related transcriptional PODs for RA2 and RA3. RA2 and RA3 PODs added value to the MOA by: (1) comparison of the expression profiles of genotoxic and non-genotoxic compounds, which predicted that BaP is genotoxic in human cell lines; and (2) mining the expression signatures to identify measurable key events that are necessary (but not necessarily sufficient) for induction of apical toxicity (phenotypic anchoring) based on the IPCS/ILSI MOA framework (Boobis et al., 2006; Meek et al., 2003; Sonich-Mullin et al., 2001) and Bradford Hill test for causation (Hill, 1965). Once the key events in the MOA were proposed and their BMDLs calculated, the “committed step” was proposed, and the key event immediately preceding it was selected as the most relevant POD for risk assessment. This analysis revealed PODs of 1.0, 3.7 and 7.4 mg/kg bw per day for transcriptional changes in liver, lung and forestomach, respectively, for both RAs. In comparison, a comprehensive literature review (RA1) derived apical PODs of 1.2, 0.8 and 0.5 mg/kg bw per day for tumors for each tissue, respectively (Table 10). Analysis of tissue-specific mutagenicity (lacZ transgene mutant frequency) also yielded similar PODs of 4.8, 1.4 and 0.3 mg/kg bw per day for liver, lung and glandular stomach, respectively (Table 10). Thus, the transcriptional PODs are similar to the tumor and mutation PODs, with the largest discrepancy observed for the stomach. However, PODs for stomach are still within an order of magnitude. We believe that these numbers would be more aligned had the samples been collected immediately following the last exposure rather than following a 3-day break for transcriptional profiling, which is supported by the lower BMDLs found in liver (derived 4 and 24 h post exposure). It is also important to note that the PODs for local effects (forestomach) are within an order of magnitude of systemic effects (liver and lung). This provides some support for the use of surrogate tissues (liver and lung) to predict response in other tissues (forestomach). However, we caution that our analysis is on a genotoxic carcinogen and this may not apply to chemicals that have a tissue-specific MOA.
In contrast to expectations, MOA-related PODs for transcriptional changes were higher than apical endpoints. These higher transcriptional BMDLs may be due to: (1) doses used in the studies producing apical data were more likely to be within the BMR range, whereas transcriptional study doses were high (only one dose was in the BMR range in one study only); (2) shorter exposure duration of the transcriptional studies may require a higher dose of BaP (resulting in a higher BMD) relative to studies in which exposure doses are accumulated over a longer duration (lower BMD) to achieve the same effect; (3) BMD modeling of apical endpoints is slightly more sensitive (settings more customizable) than the BMDExpress modeling of transcriptional endpoints; and (4) genes are part of dynamic and interconnected networks and a gene expression signature is a single snapshot in time representing the complex response of that network, which may not effectively represent each gene's time-dependent response. It is interesting to note that one hesitation to the use of genomics data in risk assessment has been the concern that genomics endpoints would be too sensitive, leading to extremely low proposed exposure thresholds; The PODs within this case study demonstrate that this is unlikely.
The toxicogenomics data used in this case study revealed that early key events that take place within days of initial BaP exposure and that may contribute to carcinogenesis can be detected and used for MOA development and POD estimation. Nesnow et al. (2009, 2011) used toxicogenomics to identify key events in the carcinogenicity of propiconazole in mice following short-term 4-day and 30-day exposure (Nesnow et al., 2009) and 4-day exposure periods (Nesnow et al., 2011). These experiments demonstrate that when analyzing toxicogenomics data for chemicals that perturb a broad spectrum of pathways, an MOA approach may be used as a filter to reveal the most meaningful and practical information occurring at low doses. In general, these MOA-centric BMD values were similar to those derived using an approach applying the most sensitive pathways associated with the MOA or regardless of the MOA (Section 3.3; Table 10) (Thomas et al., 2007a, 2011, 2012). Indeed, the latter approach may be useful in preliminary analyses when the MOA is unknown or unclear. However, we propose that estimating BMDL values based on a putative MOA is biologically relevant and provides greater confidence in the use of gene expression data and in selecting a gene expression–based POD.
4.1 Advantages of a genomics approach
While all three risk assessment approaches identified similar PODs, the genomics approach offers numerous advantages. These include the following:
-
1)
Genomics data can be obtained within a much shorter time frame and at a greatly reduced cost relative to conducting a full battery of standardized tests (a major goal of Toxicity Testing in the 21st Century; NRC, 2007b).
-
2)
The approach is highly effective in decreasing animal use (another major goal of Toxicity Testing in the 21st Century; NRC, 2007b).
-
3)
Potential hazards can be identified using acute and subchronic assays and/or by analyzing toxicogenomics data from cultured human cells. Importantly, the top pathways shared between human in vitro and mouse in vivo data are AHR signaling, Nrf2, and p53 signaling and cell cycle signaling, which is in excellent agreement with the genotoxic MOA proposed here (Figure 9). An important lesson from this is that one can significantly minimize costly animal experiments and rapidly identify genotoxic chemicals (and, perhaps, other health hazards) by obtaining toxicogenomics information from cells cultured in vitro. For example, our TK6 study correctly identified BaP's genotoxic MOA rapidly (experiments could be completed within 2 weeks) and did not require any animal use. Such an approach (i.e. screening chemicals for hazards using cultured human cells in vitro) would be in excellent agreement with the two-tiered approach to toxicity testing proposed by Thomas and colleagues (2013) and would help streamline hazard identification and MOA development for chemicals with little or no toxicity information. Our findings also support the utility of in vitro data as an additional line of evidence.
-
4)
Target tissue(s) for specific toxicities can be identified at relatively early time points by assessing global gene expression changes in a small battery of tissues that includes the sites of contact and the most frequent targets for chemically induced cancer, neurotoxicity, developmental toxicity and reproductive toxicity.
-
5)
Since the approach does not target a specific toxicity, inclusion of toxicogenomics tests across an informative set of tissues early in a test battery can relatively rapidly inform a large suite of potential adverse effects. Additional case studies on noncancer endpoints are needed to confirm this.
-
6)
Cross-species extrapolations are problematic in risk assessment in general, as well as in approaches applying toxicogenomics. However, we note that as increased understanding of the degree of conservation in pathways between species is gained, toxicogenomics can be used to provide greater insight into human relevance. This is one of the strengths of mechanistic-based approaches. Indeed, this is the rationale behind the use of the MOA/human relevance framework that we applied as described in Section 3.1.3. In addition, comparison of gene expression and pathway changes in animals and human cells provides plausibility of an animal MOA in humans.
-
7)
Toxicogenomics data can be used to fill in gaps in the MOA and minimize uncertainties in the dose–response relationships for intermediary steps, as well as to identify new, previously unknown MOAs for a toxicant.
-
8)
Toxicogenomics is unbiased, thus preselection of pathways or genes of interest are not necessary.
-
9)
The approach may be applicable to mixtures, other chemicals and other exposure scenarios.
-
10)
The expression signatures can be used to derive preliminary MOAs to identify key events in a chemical's risk assessment and derive PODs based on these, rather than excluding that chemical while waiting for lengthy and costly studies, ultimately increasing the number of published risk assessments.
4.2 Obstacles to incorporation of toxicogenomics into risk assessment
While the advantages of genomics approaches are numerous, many concepts in this field need to be refined, and many obstacles stand in the way of the incorporation of toxicogenomics into risk assessment:
-
1)
Clear guidelines are necessary for the identification of appropriate toxicogenomics datasets that are suitable for risk assessment purposes.
-
2)
Determination of which BMDL should be selected as a POD has not been clearly defined. Selecting the lowest transcriptional BMDL from all pathways and tissues is the most conservative approach, but may not logically relate mechanistically to the adverse effect and may be unnecessarily conservative. We propose that an additional option would be to select the lowest BMDL from a pathway that is known to be associated with a preliminary MOA derived from gene expression data or a known MOA/adverse effect.
-
3)
Some toxicants perturb a large number of pathways, and thus it can be difficult to know which to select for POD calculations in the absence of apical data.
-
4)
Genes without annotations or assignment to a pathway are exluded from toxicogenomic approaches. In addition, the relationship between specific pathways and disease/apical outcomes is not well defined. As further annotation of gene functions and linkages to apical outcomes are completed this will become less of an obstacle.
-
5)
A large number of data must be analyzed, and thus some specialized training and software packages are needed.
-
6)
Further biological validation is needed to confirm that expression changes truly reflect adverse phenotypes.
-
7)
The technologies and bioinformatics approaches are rapidly evolving.
-
8)
There is no OECD guideline in place for any gene expression assay.
-
9)
There is a need for additional validation. More exercises such as this one are needed to fully develop a framework for implementing toxicogenomics in risk assessment, especially for chemicals with hazards other than genotoxicity or carcinogenicity.
4.3 Limitations of the data used in this case study
Overall, it is clear that further work is required before transcriptional pathway–based risk assessments can be integrated into traditional approaches. In addition to these obstacles, there were several limitations in our experiments, as they were not designed for the purpose of integrating genomics into the risk assessment of BaP, but do reflect real-world data a risk assessor would have access to:
-
1)
Our case study reflects one of the major challenges faced by risk assessors – the availability of relevant data. We did not generate any new data and relied on previous experiments that were not specifically designed to address risk assessment issues. Thus, the transcriptomics datapoints were not necessarily collected using an optimal experimental design. Indeed, a gene expression profile is a single snapshot in time from which it may be hard to elucidate the dynamic nature of genes operating in networks. This is clearly seen by the fact that transcriptomics PODs for lung and forestomach (that were sampled three days after the last exposure) were several fold higher than their respective cancer PODs, while the transcriptomics POD from liver data (sampled 4 or 24 hours or three days after the last exposure) was more aligned with the cancer POD (1.0 vs. 1.2 mg/kg bw per day, respectively). We considered the most appropriate dataset for BMD modeling in our experiment to be the liver 3-day BaP exposure, sampled 4 and 24 hours after the last exposure since: (1) samples collected at an early time point would appropriately capture the initial effects of the toxicant; (2) the 3-day dataset included a broader range of doses, including the lower dose range; and (3) the transcriptional responses for the initiating and early key events may be diminished or not captured after the 3 day time-lag in sample collection after our 28-day exposure (instead capturing responses resulting from toxicity). However, toxicogenomics signatures taken after 90 days of exposure (sampling immediately after final dose) are proposed to be more predictive of carcinogenesis than earlier time points (two or 14 days) (Auerbach et al., 2010; Thomas et al., 2013). Samples collected at the 90-day time point are postulated to express initiating and early key events, as well as early changes involved in the transformation of a tissue to a disease phenotype.
-
2)
The number of dose groups was limited. Inclusion of some lower doses within the BMR would have been preferred.
-
3)
The use of a single non-metabolically competent human cell culture model was a limitation and can only be used for hazard identification to support the genotoxic MOA. An improved approach would be to apply tissue-relevant metabolically competent cell types for additional information.
-
4)
The true target tissue, the forestomach, was not assayed at the acute exposure time points. Forestomach tissue was available only from the 28 + 3 day regimen, at which time most of the BaP in the forestomach has been cleared. However, we note that our data support that the evaluation of gene expression changes in key target tissues may also be predictive/protective of potential changes in other tissues.
-
5)
Only two tissues were assayed in both the acute and subchronic regimens. However, the toxicogenomics-derived POD is within an order of magnitude of the traditionally derived POD; thus, in the short term, the integration of toxicogenomics into standard risk assessments (such as this one) will continue to drive this field forward and will begin to clarify the precise means by which toxicogenomics data should be used.
-
6)
We acknowledge that because there were only three tissues analyzed in RA3 that this could indicate bias relating to target tissue selection. However, we believe that liver is an obvious first-pass target tissue for preliminary toxicogenomic analyses of a chemical delivered orally as the site of metabolism. Although such an assumption may not hold for chemicals with rapid metabolism, hence the need for two or more time points. For example, CYP1A1 induction by BaP in the mouse liver would be missed by analyzing 24h data alone (See Table 5). A similar argument can be extended to (fore)stomach as a point of contact for oral exposures, and lung (exposure via circulation). Analysis of a compendium of cancer bioassay data revealed that 92% (354/384) of mouse and 82% (388/471) of rat carcinogens showed positive response in at least one of the eight tissues: liver, lung, mammary gland, stomach, vascular system kidney, hematopoietic system, and urinary bladder (Gold et al., 2001). Thus, we recommend that these eight tissues comprise the starting point to analyze global gene expression for chemical assessments as has been suggested by others (Thomas et al., 2013). Additional tissues and experimental setups could be considered to address other toxicity endpoints.
-
7)
It is worth emphasizing that this proof-of-principle case study illustrates the utility of toxicogenomics in the risk assessment of a genotoxic carcinogen. The selection of a target organ/tissue for genotoxic carcinogens is more straightforward than for the assessment of non-genotoxic or non-cancer effects. This is illustrated by the fact that adducts and mutations were detected in all three tissues that we examined (Figure 2). We note that if the eight tissues proposed above for cancer are profiled exclusively with toxicogenomics, some important toxicity information may be missed. However, collecting all of the target tissues for detailed toxicological evaluations is challenging even for established toxicity endpoints. For example, the OECD guideline 407 for repeat dose 28-day oral toxicity in rodents examines a wide variety of tissues for a diverse array of toxicological effects related to nervous, immune, endocrine, and reproductive systems. We anticipate that examination of various tissues for a variety of toxicological effects may benefit risk assessment by providing rich mechanistic information that could be otherwise missed by traditional testing strategies. Additional case studies like the present one will be required to verify this for nongenotoxic and non-cancer toxicants compounds. It is also worth considering that toxicogenomics approaches are becoming increasingly higher throughput and less expensive (e.g., Li et al., 2012). Thus, it is feasible that in the near future all of the tissues collected in repeat-dose studies can be profiled for gene expression changes. Integration of toxicogenomics into current standard in vivo toxicity tests would provide information that could ultimately lead to the appropriate selection of target tissues for toxicogenomics studies in the future to address non-cancer effects. We believe that selection of the appropriate tissues that encompass a large portion of potential toxicities is an important avenue for further research.
-
8)
Genomics approaches only capture transcriptional changes; other toxicity mechanisms may not be captured using this approach alone.
4.4 Next steps
We believe that there are many take-home lessons from this collaborative project involving both risk assessors and researchers (summarized in greater detail in a separate publication (N. Chepelev et al., accompanying paper)). Briefly, we demonstrated the potential applications of toxicogenomics to informing MOAs and PODs in chemical evaluations. We encourage government agencies to more actively develop and implement applied guidelines to support these applications. This will require a coordinated effort from policymakers, risk managers, risk assessors and research scientists. In addition, enhanced communication and understanding of risk assessment needs would enable researchers to design more effective studies for regulatory applications. For example, research experiments are often conducted with an insufficient number of doses, outside the environmentally relevant range or in inappropriate target tissues required for risk assessment. Time-course experiments would benefit risk assessment in establishing whether transcriptional effects are adaptive or trigger adverse endpoints. Closer interaction between regulators and research scientists will facilitate the development of methodologies for integration into risk assessment. Such collaborations will be critical for the development of approved guidelines for integration of toxicogenomics data in risk assessment.
Clearly, additional exercises such as the present case study will help advance this field. This was a retrospective assessment, based on real-world data a risk assessor would have access to. An important next step is to apply toxicogenomics to evaluate data-poor chemicals and use targeted testing follow-up approaches to validate the use of global gene expression profiling in hazard identification, MOA and POD determination. For validation and wider acceptance, toxicogenomics-based MOAs will continue to rely on phenotypic anchoring rather than being used as a stand-alone approach for the time being. Clear pathway annotations, established linkage between expression changes and apical changes, standardized protocols, validated expression signatures that predict a diverse number of toxicities and an approach to deal with interactions or differences among tissues are required before toxicogenomics can be routinely incorporated into risk assessment.
In conclusion, this case study has shown that toxicogenomics can correctly identify BaP's genotoxic MOA and derive PODs that are similar to those derived using current approaches. As the field of toxicogenomics continues to evolve, we believe that risk assessors should use a new approach (Figure 10) that incorporates toxicogenomics information to support the determination of MOA and derivation of PODs for both data-poor and data-rich chemicals. This new approach should integrate all existing data, which may include in vitro genetic toxicity (e.g., Ames assay) and kinetics/metabolism tests, high throughput screening (HTS), in silico predictions and QSAR. For data-poor chemicals requiring testing and assessment, we support that toxicogenomics be used in the first tier of toxicity testing to: 1) prioritize chemicals (e.g. if an environmentally-relevant dose doesn't perturb toxicity-associated pathways it could be considered of low priority for risk assessment, allowing greater resources for chemicals identified as higher concern), 2) determine if targeted toxicity testing is required and if so, which traditional toxicological assays are relevant for hazard identification and derivation of PODs, and 3) inform hazard and derive a genomic POD (when standard toxicity tests are unavailable) for screening assessments. While it may be desirable to use toxicogenomics changes as an alternative to traditionally obtained toxicity information when standard testing strategies are unavailable, we caution that until the application of toxicogenomics in supporting health risk assessment is fully developed, genomic PODs should be used in the presence of additional lines of evidence supporting the overall hazard potential of the chemical being assessed.
Figure 10.
Proposed role and integration of toxicogenomics in human health risk assessment.Toxicogenomics can benefit data-poor as well as data-rich chemicals by rapidly and inexpensively providing data that are useful for risk assessment. Toxicogenomics can be the first tier of toxicity testing to inform hazard identification and prioritization and identify relevant tests for further targeted testing. In addition, toxicogenomics can be integrated with existing data from empirical and alternative approaches (high-throughput screening [HTS], quantitative structure–activity relationship [QSAR]) to derive genomics PODs for data-poor chemicals. Knowledge of chemical's metabolism and kinetics are essential and should be developed utilizing in vitro systems in parallel with the genomics approach. For data-poor chemicals, toxicogenomics could rapidly generate genomics PODs as well as identify important endpoints for more thorough POD derivation by standard approaches, if applicable. Similarly, if no obvious toxicity is identified in toxicogenomics (e.g. no pathways relevant to toxicity are perturbed at certain doses), data-poor chemicals may be considered as “lower priority” for risk assessment. Based on the available toxicity information and other considerations, genomics PODs can be used either to inform targeted testing or to reach screening-level risk assessment and management decisions. For data-rich chemicals, toxicogenomics can provide mechanistic data to support the development of detailed MOAs and hence the risk assessment approach. The current approach is indicated in gray boxes, and the new approach is in red.
For data-rich chemicals, toxicogenomics data derived from in vivo studies should be considered to: 1) help identify the most relevant endpoint (i.e. the most sensitive pathways and processes perturbed by a chemical) for hazard identification; 2) provide mechanistic data to support the development of a detailed MOA; and subsequently to 3) support the risk assessment approach (e.g. linear or threshold). As in the standard approach, the relevance of an animal MOA to humans can then be evaluated based on epidemiological data; alternatively, this can be done with toxicogenomics experiments in cultured human cells, deriving mechanistic information that is rarely achieved in epidemiological studies.
In our case study, we acknowledge that the genotoxicity of BaP would be detected by standard in vitro tests. Our work was not done to suggest that standard assays should be discarded in favor of toxicogenomics. Indeed, we believe that these in vitro assays are powerful early predictors of potential toxicity and should continue to be used. We note that toxicogenomics does not indicate that a mutation has occurred, but instead indicates that a DNA damage response has been initiated, which is consistent with the presence of genotoxicity.However, toxicogenomics also provides insight into the MOA through which BaP causes genotoxicity (i.e., activation of AHR, metabolism, induction of p53 and DNA damage response, in addition to the effects on cell cycle and other signaling pathways). In addition, the approach can be used to provide insight into the relevance of the MOA in humans and inform the RA approach. This information can be used to support positive results from an Ames test or a standard genetic toxicity assay that address the ability of the compound to reach the tissue, be activated and interact with DNA in vivo. Through the analysis of multiple tissues it offers insight into tissue-specificity, distribution and metabolism. Moreover, the analysis reveals non-genotoxic carcinogenic (and non-carcinogenic) MOAs that would be missed through application of genetic toxicology assays alone. We have previously published a study on furan, a hepatocarcinogen that operates primarily through the induction of cytotoxicity followed by regenerative proliferation, which demonstrates this point (Jackson et al., 2014). Although detailed genomics information (e.g. the activation of certain DNA-damage response pathways) in the absence of other information is currently not very informative in risk assessment, we believe that development of specific genomic biomarkers will become invaluable in the near future as we move towards MOA-based, mechanistic approaches in toxicity testing.
A tiered testing paradigm whereby first order margins of exposure (MOEs) are calculated using data from in vitro high-throughput screening assays has been recently proposed (Thomas et al., 2013). Given the similarity between the PODs derived by the traditional and the toxicogenomics-based approaches, we propose that PODs derived from toxicogenomics data can also be used to inform regulatory guidance on MOEs (historically used for non-genotoxic carcinogens and non-cancer effects) or maximum acceptable concentrations. As per any risk assessment, the specific exposure information and the total uncertainty would be agency-specific.
Overall, our research illustrates the utility of toxicogenomics to inform MOA and PODs in risk assessment and provides two examples (RA2 and RA3) of how toxicogenomics could be applied in human health risk assessment in the future.
Supplementary Material
Figure 3.
General overview of BaP metabolism. Cytochrome P450s (CYPs) and other enzymes and cellular oxidants can convert BaP to radical cations. These can be further metabolized by CYPs to epoxide and then to diols by epoxide hydrolase (EH). BaP diols are converted to catechols by aldo-keto reductases (AKRs) or to BaP-7,8-diol-9,10-epoxide (BPDE) by CYPs. BPDE, BaP quinones, reactive oxygen species (ROS) (generated during BaP catechol–quinone redox cycling or from CYPs’ uncoupling) and BaP cations react with DNA (forming adducts) and leading to DNA mutations (predominantly G to T transversions). EH can further metabolize BPDE to tetraols that, along with diols, phenols (not shown) and quinines, are conjugated to glucuronosyl by uridine diphosphate–glucuronosyl transferases (UGTs). Diols, phenols and quinones can be also converted to sulfate esters by sulfotransferases (SULTs). Glutathione S-transferases (GSTs) conjugate BaP epoxides, diol epoxides and quinones to glutathione (Ramesh et al., 2004). Only representative BaP metabolites (i.e. modified at positions 7, 8, 9 and 10) are shown. In addition, BaP metabolism can yield many other hydroxy-, oxide-, dihydroxy- and quinone-related compounds at each of the 12 carbon atoms and it has been estimated that 709 oxygenated metabolites of BaP exist (reviewed in Nebert et al., 2013a); similar BaP metabolites are formed at other positions (Ramesh et al., 2004). See Section 2.3.3 for additional details.
Acknowledgements
We thank Amanda Green, Kim Shepard and Phillip Garibaldi for technical support and Paul White and Francesco Marchetti for helpful discussions and insight. We thank Alexandra Long and Paul White for their insights into the metabolism of BaP and its subsequent induction of various genotoxic endpoints across multiple tissues and doses. We also thank the Water and Air Quality Bureau of Health Canada for its continued support in exploring new methodologies in risk assessment. We gratefully acknowledge the helpful comments and suggestions provided by Anne Vézina, Véronique Morisset, Lynn Berndt-Weis, Guosheng Chen and Tara Barton-MacLaren.
Footnotes
Declaration of interest
The authors’ affiliations are as shown on the cover page. The authors have sole responsibility for the writing and content of the paper. The authors declare no conflicts of interest. This research was funded by the Genomics Research and Development Initiative of Health Canada and the Chemicals Management Plan of the Government of Canada.
References
- Aboutabl ME, Zordoky BN, El-Kadi AO. 3-Methylcholanthrene and benzo(a)pyrene modulate cardiac cytochrome P450 gene expression and arachidonic acid metabolism in male Sprague Dawley rats. Br J Pharmacol. 2009;158(7):1808–1819. doi: 10.1111/j.1476-5381.2009.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboutabl ME, Zordoky BN, Hammock BD, El-Kadi AO. Inhibition of soluble epoxide hydrolase confers cardioprotection and prevents cardiac cytochrome P450 induction by benzo(a)pyrene. J Cardiovasc Pharmacol. 2011;57(3):273–281. doi: 10.1097/FJC.0b013e3182055baf. [DOI] [PubMed] [Google Scholar]
- Allan LL, Schlezinger JJ, Shansab M, Sherr DH. CYP1A1 in polycyclic aromatic hydrocarbon–induced B lymphocyte growth suppression. Biochem Biophys Res Commun. 2006;342(1):227–235. doi: 10.1016/j.bbrc.2006.01.131. [DOI] [PubMed] [Google Scholar]
- Anderson KE, Kadlubar FF, Kulldorff M, et al. Dietary intake of heterocyclic amines and benzo(a)pyrene: Associations with pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(9):2261–2265. doi: 10.1158/1055-9965.EPI-04-0514. [DOI] [PubMed] [Google Scholar]
- Arafa HM, Aly HA, Abd-Ellah MF, El-Refaey HM. Hesperidin attenuates benzo[α]pyrene-induced testicular toxicity in rats via regulation of oxidant/antioxidant balance. Toxicol Ind Health. 2009;25(6):417–427. doi: 10.1177/0748233709106624. [DOI] [PubMed] [Google Scholar]
- Armstrong B, Thériault G. Compensating lung cancer patients occupationally exposed to coal tar pitch volatiles. Occup Environ Med. 1996;53(3):160–167. doi: 10.1136/oem.53.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong BG, Tremblay CG, Cyr D, Theriault GP. Estimating the relationship between exposure to tar volatiles and the incidence of bladder cancer in aluminum smelter workers. Scand J Work Environ Health. 1986;12(5):486–493. doi: 10.5271/sjweh.2109. [DOI] [PubMed] [Google Scholar]
- ATSDR (Agency for Toxic Substances and Disease Registry) Toxicological Profile for Polycyclic Aromatic Hydrocarbons. US Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry; Atlanta, GA: 1995. [PubMed] [Google Scholar]
- Auerbach SS, Shah RR, Mav D, et al. Predicting the hepatocarcinogenic potential of alkenylbenzene flavoring agents using toxicogenomics and machine learning. Toxicol Appl Pharmacol. 2010;243(3):300–314. doi: 10.1016/j.taap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Avlasevich SL, Bryce SM, Cairns SE, Dertinger SD. In vitro micronucleus scoring by flow cytometry: Differential staining of micronuclei versus apoptotic and necrotic chromatin enhances assay reliability. Environ Mol Mutagen. 2006;47(1):56–66. doi: 10.1002/em.20170. [DOI] [PubMed] [Google Scholar]
- Bjelogrlic N, Iscan M, Raunio H, Pelkonen O, Vahakangas K. Benzo[a]pyrene-DNA-adducts and monooxygenase activities in mice treated with benzo[a]pyrene, cigarette smoke or cigarette smoke condensate. Chem Biol Interact. 1989;70(1-2):51–61. doi: 10.1016/0009-2797(89)90062-8. [DOI] [PubMed] [Google Scholar]
- Bercu JP, Jolly RA, Flagella KM, Baker TK, Romero P, Stevens JL. Toxicogenomics and cancer risk assessment: A framework for key event analysis and dose–response assessment for nongenotoxic carcinogens. Regul Toxicol Pharmacol. 2010;58(3):369–381. doi: 10.1016/j.yrtph.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Black MB, Budinsky RA, Dombkowski A, et al. Cross-species comparisons of transcriptomic alterations in human and rat primary hepatocytes exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2012;127(1):199–215. doi: 10.1093/toxsci/kfs069. [DOI] [PubMed] [Google Scholar]
- Bock KW, Bock-Hennig BS. UDP-glucuronosyltransferases (UGTs): From purification of Ah-receptor-inducible UGT1A6 to coordinate regulation of subsets of CYPs, UGTs, and ABC transporters by nuclear receptors. Drug Metab Rev. 2010;42(1):5–12. doi: 10.3109/03602530903205492. [DOI] [PubMed] [Google Scholar]
- Bolognesi C, Rossi L, Barbieri O, Santi L. Benzo[a]pyrene-induced DNA damage in mouse fetal tissues. Carcinogenesis. 1985;6(8):1091–1095. doi: 10.1093/carcin/6.8.1091. [DOI] [PubMed] [Google Scholar]
- Boobis AR, Cohen SM, Dellarco V, et al. IPCS framework for analyzing the relevance of a cancer mode of action for humans. Crit Rev Toxicol. 2006;36(10):781–792. doi: 10.1080/10408440600977677. [DOI] [PubMed] [Google Scholar]
- Bouayed J, Bohn T, Tybl E, Kiemer AK, Soulimani R. Benzo[α]pyrene-induced anti-depressive-like behaviour in adult female mice: Role of monoaminergic systems. Basic Clin Pharmacol Toxicol. 2012;110(6):544–550. doi: 10.1111/j.1742-7843.2011.00853.x. [DOI] [PubMed] [Google Scholar]
- Brandon JL, Conti CJ, Goldstein LS, DiGiovanni J, Gimenez-Conti IB. Carcinogenic effects of MGP-7 and B[a]P on the hamster cheek pouch. Toxicol Pathol. 2009;37(6):733–740. doi: 10.1177/0192623309344203. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Brinkmann J, Stolpmann K, Trappe S, et al. Metabolically competent human skin models: Activation and genotoxicity of benzo[a]pyrene. Toxicol Sci. 2013;131(2):351–359. doi: 10.1093/toxsci/kfs316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RP, Delp MD, Lindstedt SL, et al. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health. 1997;13:407–484. doi: 10.1177/074823379701300401. [DOI] [PubMed] [Google Scholar]
- Bui QQ, Tran MB, West WL. A comparative study of the reproductive effects of methadone and benzo[a]pyrene in the pregnant and pseudopregnant rat. Toxicology. 1986;42(2–3):195–204. doi: 10.1016/0300-483x(86)90009-0. [DOI] [PubMed] [Google Scholar]
- Burdick AD, Davis JW, II, Liu KJ, et al. Benzo(a)pyrene quinones increase cell proliferation, generate reactive oxygen species, and transactivate the epidermal growth factor receptor in breast epithelial cells. Cancer Res. 2003;63(22):7825–7833. [PubMed] [Google Scholar]
- Burstyn I, Kromhout H, Partanen T, et al. Polycyclic aromatic hydrocarbons and fatal ischemic heart disease. Epidemiology. 2005;16(6):744–750. doi: 10.1097/01.ede.0000181310.65043.2f. [DOI] [PubMed] [Google Scholar]
- Busbee D, Joe C, Rankin P. Benzo(a)pyrene uptake by lymph: A possible transport mode for immunosuppressive chemicals. J Toxicol Environ Health. 1984;13(1):43–51. doi: 10.1080/15287398409530480. [DOI] [PubMed] [Google Scholar]
- Butler LM, Sinha R, Millikan RC, et al. Heterocyclic amines, meat intake, and association with colon cancer in a population-based study. Am J Epidemiol. 2003;157(5):434–445. doi: 10.1093/aje/kwf221. [DOI] [PubMed] [Google Scholar]
- Cal/EPA (California Environmental Protection Agency) Public Health Goals for Chemicals in Drinking Water: Benzo(a)pyrene. California Environmental Protection Agency, Office of Environmental Health Hazard Assessment; Sacramento, CA: 2010. [Google Scholar]
- Cao L, Zhou X, Sens M, et al. Keratin 6 expression correlates to areas of squamous differentiation in multiple independent isolates of As(+3)-induced bladder cancer. J Appl Toxicol. 2011;30(5):416–430. doi: 10.1002/jat.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson GP, Fossa AA, Morse MA, Weaver PM. Binding and distribution studies in the SENCAR mouse of compounds demonstrating a route-dependent tumorigenic effect. Environ Health Perspect. 1986;68:53–60. doi: 10.1289/ehp.866853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri E, Rogan E. The approach to understanding aromatic hydrocarbon carcinogenesis. The central role of radical cations in metabolic activation. Pharmacol Ther. 1992;55(2):183–199. doi: 10.1016/0163-7258(92)90015-r. [DOI] [PubMed] [Google Scholar]
- Cavalieri E, Rogan E. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25(7):677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- CCME (Canadian Council of Ministers of the Environment) Canadian Soil Quality Guidelines: Carcinogenic and Other Polycyclic Aromatic Hydrocarbons (PAHs) (Environmental and Human Effects). Scientific Supporting Document. 2010:218. [Google Scholar]
- Chakravarti D, Pelling JC, Cavalieri EL, Rogan EG. Relating aromatic hydrocarbon–induced DNA adducts and c-H-ras mutations in mouse skin papillomas: The role of apurinic sites. Proc Natl Acad Sci USA. 1995;92(22):10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEMFATE 2013 http://esc.syrres.com/scripts/CASCFcgi.exe?CASNUM=50328.
- Chen C, Tang Y, Jiang X, et al. Early postnatal benzo(a)pyrene exposure in Sprague-Dawley rats causes persistent neurobehavioral impairments that emerge postnatally and continue into adolescence and adulthood. Toxicol Sci. 2012;125(1):248–261. doi: 10.1093/toxsci/kfr265. [DOI] [PubMed] [Google Scholar]
- Chen X, An H, Ao L, et al. The combined toxicity of dibutyl phthalate and benzo(a)pyrene on the reproductive system of male Sprague Dawley rats in vivo. J Hazard Mater. 2011;186(1):835–841. doi: 10.1016/j.jhazmat.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Chepelev N, Moffat I, Bowers W, Yauk C. Neurotoxicity may be an overlooked consequence of benzo(a)pyrene exposure that is relevant to human health risk assessment. (Submitted to Chem Res Toxicol) [DOI] [PubMed]
- Crowell SR, Amin SG, Anderson KA, et al. Preliminary physiologically based pharmacokinetic models for benzo[a]pyrene and dibenzo[def,p]chrysene in rodents. Toxicol Appl Pharmacol. 2011;257(3):365–376. doi: 10.1016/j.taap.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp S, Gaylor D, Sheldon W, Goldstein L, Beland F. A comparison of the tumors induced by coal tar and benzo[a]pyrene in a 2-year bioassay. Carcinogenesis. 1998;19(1):117–124. doi: 10.1093/carcin/19.1.117. [DOI] [PubMed] [Google Scholar]
- Currie RA. Toxicogenomics: The challenges and opportunities to identify biomarkers, signatures and thresholds to support mode-of-action. Mutat Res. 2012;746(2):97–103. doi: 10.1016/j.mrgentox.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Davila D, Romero D, Burchiel S. Human T cells are highly sensitive to suppression of mitogenesis by polycyclic aromatic hydrocarbons and this effect is differentially reversed by alpha-naphthoflavone. Toxicol Appl Pharmacol. 1996;139(2):333–341. doi: 10.1006/taap.1996.0173. [DOI] [PubMed] [Google Scholar]
- Davis JA, Gift JS, Zhao QJ. Introduction to benchmark dose methods and U.S. EPA's benchmark dose software (BMDS) version 2.1.1. Toxicol Appl Pharmacol. 2011;254(2):181–191. doi: 10.1016/j.taap.2010.10.016. [DOI] [PubMed] [Google Scholar]
- De Jong W, Kroese E, Vos J, Van Loveren H. Detection of immunotoxicity of benzo[a]pyrene in a subacute toxicity study after oral exposure in rats. Toxicol Sci. 1999;50(2):214–220. doi: 10.1093/toxsci/50.2.214. [DOI] [PubMed] [Google Scholar]
- Derks KW, Hoeijmakers JH, Pothof J. The DNA damage response: The omics era and its impact. DNA Repair (Amst). Advance online publication. 2014 doi: 10.1016/j.dnarep.2014.03.008. doi: 10.1016/j.dnarep.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani E, Boffetta P, Deneo-Pellegrini H, et al. Meat intake, meat mutagens and risk of lung cancer in Uruguayan men. Cancer Causes Control. 2009;20(9):1635–1643. doi: 10.1007/s10552-009-9411-2. [DOI] [PubMed] [Google Scholar]
- Dean JH, Luster MI, Boorman GA, Lauer LD, Leubke RW, Lawson L. Selective immunosuppression resulting from exposure to the carcinogenic congener of benzopyrene in B6C3F1 mice. Clin Exp Immunol. 1983;54(1):199–206. [PMC free article] [PubMed] [Google Scholar]
- DeMarini DM, Landi S, Tian D, et al. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61(18):6679–6681. [PubMed] [Google Scholar]
- Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol Sci. 2011;124(1):1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in p53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- DiGiovanni J, Beltran L, Rupp A, Harvey RG, Gill RD. Further analysis of c-Ha-ras mutations in papillomas initiated by several polycyclic aromatic hydrocarbons and papillomas from uninitiated, promoter-treated skin in SENCAR mice. Mol Carcinog. 1993;8(4):272–279. doi: 10.1002/mc.2940080410. [DOI] [PubMed] [Google Scholar]
- DiNatale BC, Murray IA, Schroeder JC, et al. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci. 2010;115(1):89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragin N, Shi Z, Madan R, et al. Phenotype of the Cyp1a1/1a2/1b1−/− triple-knockout mouse. Mol Pharmacol. 2008;73(6):1844–1856. doi: 10.1124/mol.108.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bayoumy K, Chae Y-H, Upadhyaya P, et al. Comparative tumorigenicity of benzo[a]pyrene, 1-nitropyrene and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine administered by gavage to female CD rats. Carcinogenesis. 1995;16(2):431–434. doi: 10.1093/carcin/16.2.431. [DOI] [PubMed] [Google Scholar]
- Environment Canada . Toxic Chemicals in the Great Lakes and Associated Effects: Volume I. Contaminant Levels and Trends. Government of Canada; Ottawa, ON: 1991. [Google Scholar]
- Fang AH, Smith WA, Vouros P, Gupta RC. Identification and characterization of a novel benzo[a]pyrene-derived DNA adduct. Biochem Biophys Res Commun. 2001;281(2):383–389. doi: 10.1006/bbrc.2000.4161. [DOI] [PubMed] [Google Scholar]
- Fischer A, Koeper L, Vohr H. Specific antibody responses of primary cells from different cell sources are able to predict immunotoxicity in vitro. Toxicology In Vitro. 2011;25(8):1966–1973. doi: 10.1016/j.tiv.2011.06.020. [DOI] [PubMed] [Google Scholar]
- Forster K, Preuss R, Rossbach B, Bruning T, Angerer J, Simon P. 3-Hydroxybenzo[a]pyrene in the urine of workers with occupational exposure to polycyclic aromatic hydrocarbons in different industries. Occup Environ Med. 2008;65(4):224–229. doi: 10.1136/oem.2006.030809. [DOI] [PubMed] [Google Scholar]
- Friesen M, Demers P, Spinelli J, Eisen E, Lorenzi M, Le N. Chronic and acute effects of coal tar pitch exposure and cardiopulmonary mortality among aluminum smelter workers. Am J Epidemiol. 2010;172(7):790–799. doi: 10.1093/aje/kwq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez-Peralta M, Shi Z, Chen J, Miller ML, Nebert DW. Oral benzo(a)pyrene in Cyp1a1/1b1(−/−) double-knockout mice: microarray analysis during squamous cell carcinoma formation in preputial gland duct. Int J Cancer. 2012;132(9):2065–2075. doi: 10.1002/ijc.27897. [DOI] [PubMed] [Google Scholar]
- Garg R, Gupta S, Maru GB. Dietary curcumin modulates transcriptional regulators of phase I and phase II enzymes in benzo[a]pyrene-treated mice: Mechanism of its anti-initiating action. Carcinogenesis. 2008;29(5):1022–1032. doi: 10.1093/carcin/bgn064. [DOI] [PubMed] [Google Scholar]
- Garner R, Stanton C, Martin C, Harris C, Grafstrom R. Rat and human explant metabolism, binding studies, and DNA adduct analysis of benzo(a)pyrene and its 6-nitro derivative. Cancer Res. 1985;45(12):6225–6231. [PubMed] [Google Scholar]
- Geacintov N, Broyde S, Buterin T, et al. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65(3):202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- Gibbs GW, Sevigny M. Mortality and cancer experience of Quebec aluminum reduction plant workers, part 4: Cancer incidence. J Occup Environ Med. 2007;49(12):1351–1366. doi: 10.1097/JOM.0b013e318156ecbc. [DOI] [PubMed] [Google Scholar]
- Godschalk RWL, Ostertag JU, Zandsteeg AMG, et al. Impact of GSTM1 on aromatic–DNA adducts and p53 accumulation in human skin and lymphocytes. Pharmacogenetics. 2001;11(6):537–543. doi: 10.1097/00008571-200108000-00008. [DOI] [PubMed] [Google Scholar]
- Gohlke JM, Thomas R, Zhang Y, et al. Genetic and environmental pathways to complex diseases. BMC Syst Biol. 2009;3:46. doi: 10.1186/1752-0509-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold LS, Manley NB, Slone TH, Ward JM. Compendium of chemical carcinogens by target organ: results of chronic bioassays in rats, mice, hamsters, dogs, and monkeys. Toxicol Pathol. 2001;29(6):639–652. doi: 10.1080/019262301753385979. [DOI] [PubMed] [Google Scholar]
- Goodsaid FM, Amur S, Aubrecht J, et al. Voluntary exploratory data submissions to the US FDA and the EMA: Experience and impact. Nat Rev Drug Discov. 2010;9(6):435–445. doi: 10.1038/nrd3116. [DOI] [PubMed] [Google Scholar]
- Grova N, Schroeder H, Farinelle S, Prodhomme E, Valley A, Muller CP. Sub-acute administration of benzo[a]pyrene (B[a]P) reduces anxiety-related behaviour in adult mice and modulates regional expression of N-methyl-d-aspartate (NMDA) receptors genes in relevant brain regions. Chemosphere. 2008;73(1 Suppl):S295–S302. doi: 10.1016/j.chemosphere.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Grova N, Valley A, Turner JD, Morel A, Muller CP, Schroeder H. Modulation of behavior and NMDA-R1 gene mRNA expression in adult female mice after sub-acute administration of benzo(a)pyrene. Neurotoxicology. 2007;28(3):630–636. doi: 10.1016/j.neuro.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Gwinn M, Keshava C, Olivero O, Humsi J, Poirier M, Weston A. Transcriptional signatures of normal human mammary epithelial cells in response to benzo[a]pyrene exposure: A comparison of three microarray platforms. OMICS. 2005;9(4):334–350. doi: 10.1089/omi.2005.9.334. [DOI] [PubMed] [Google Scholar]
- Habs M, Jahn S, Schmähl D. Carcinogenic activity of condensate from coloquint seeds (Citrullus colocynthis) after chronic epicutaneous administration to mice. J Cancer Res Clin Oncol. 1984;108(1):154–156. doi: 10.1007/BF00390988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad S, Withey J, Laparé S, Law F, Krishnan K. Physiologically-based pharmacokinetic modeling of pyrene in the rat. Environ Toxicol Pharmacol. 1998;5(4):245–255. doi: 10.1016/s1382-6689(98)00008-8. [DOI] [PubMed] [Google Scholar]
- Hakami R, Mohtadinia J, Etemadi A, et al. Dietary intake of benzo(a)pyrene and risk of esophageal cancer in north of Iran. Nutr Cancer. 2008;60(2):216–221. doi: 10.1080/01635580701684831. [DOI] [PubMed] [Google Scholar]
- Halappanavar S, Wu D, Williams A, et al. Pulmonary gene and microRNA expression changes in mice exposed to benzo(a)pyrene by oral gavage. Toxicology. 2011;285:133–141. doi: 10.1016/j.tox.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg R. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harper PA, Wong JMY, Lam MSM, Okey AB. Polymorphisms in the human AH receptor. Chem Biol Interact. 2002;141(1–2):161–187. doi: 10.1016/s0009-2797(02)00071-6. [DOI] [PubMed] [Google Scholar]
- Hartikainen JM, Tengström M, Kosma VM, Kinnula VL, Mannermaa A, Soini Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012;72(21):5537–5546. doi: 10.1158/0008-5472.CAN-12-1474. [DOI] [PubMed] [Google Scholar]
- Hattemer-Frey HA, Travis CC. Benzo-a-pyrene: Environmental partitioning and human exposure. Toxicol Ind Health. 1991;7(3):141–157. doi: 10.1177/074823379100700303. [DOI] [PubMed] [Google Scholar]
- Hazardous Substances Data Bank (HSDB) Available at: http://toxnet.nlm.nih.gov/cgi-bin/sis/htmlgen?HSDB.
- He X, Chen W, Liu Z, Chapman RS. An epidemiological study of lung cancer in Xuan Wei County, China: Current progress. Case–control study on lung cancer and cooking fuel. Environ Health Perspect. 1991;94:9–13. doi: 10.1289/ehp.94-1567943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia-Ortiz R, Bouchard M, Marie-Desvergne C, Viau C, Maître A. Modeling of the internal kinetics of benzo(a)pyrene and 3-hydroxybenzo(a)pyrene biomarker from rat data. Toxicol Sci. 2011;122(2):275–287. doi: 10.1093/toxsci/kfr135. [DOI] [PubMed] [Google Scholar]
- Hernandez LG, Slob W, Van Steeg H, Van Benthem J. Can carcinogenic potency be predicted from in vivo genotoxicity data?: A meta-analysis of historical data. Environ Mol Mutagen. 2011;52(7):518–528. doi: 10.1002/em.20651. [DOI] [PubMed] [Google Scholar]
- Hill AB. The environment and disease: Association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach K, Van Leeuwen D, Gottschalk R, et al. Transcriptomic fingerprints in human peripheral blood mononuclear cells indicative of genotoxic and non-genotoxic carcinogenic exposure. Mutat Res. 2012;746(2):124–134. doi: 10.1016/j.mrgentox.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Hockley S, Arlt VM, Brewer D, Giddings I, Phillips DH. Time- and concentration-dependent changes in gene expression induced by benzo(a)pyrene in two human cell lines, MCF-7 and HepG2. BMC Genomics. 2006;7:260. doi: 10.1186/1471-2164-7-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6(9):1461–1468. doi: 10.1158/1541-7786.MCR-07-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holladay SD. Prenatal immunotoxicant exposure and postnatal autoimmune disease. Environ Health Perspect. 1999;107:687–691. doi: 10.1289/ehp.99107s5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K, Sera N, Otofuji T, et al. Pulmonary carcinogenicity of 3,9- and 3,7-dinitrofluoranthene, 3-nitrofluoranthene and benzo[a]pyrene in F344 rats. Carcinogenesis. 1991;12(6):1003–1007. doi: 10.1093/carcin/12.6.1003. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer) Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monogr Eval Carcinog Risks Hum. 2010:92. [PMC free article] [PubMed] [Google Scholar]
- IARC (International Agency for Research on Cancer) A review of human carcinogens: Chemical agents and related occupations. IARC Monogr Eval Carcinog Risks Hum. 2012;100F:111–144. [Google Scholar]
- Ide M, Kato T, Ogata K, Mochiki E, Kuwano H, Oyama T. Keratin 17 expression correlates with tumor progression and poor prognosis in gastric adenocarcinoma. Ann Surg Oncol. 2012;19(11):3506–3514. doi: 10.1245/s10434-012-2437-9. [DOI] [PubMed] [Google Scholar]
- Intera Technologies Ltd. W.M.S. Associates Ltd. National Water Research Institute (Canada) Investigation of Suspected PAH Contamination of Groundwater, Newcastle, New Brunswick. W.M.S. Associates Ltd; 1989. [Google Scholar]
- Jackson AF, Williams A, Recio L, Waters MD, Lambert IB, Yauk CL. Case study on the utility of hepatic global gene expression profiling in the risk assessment of the carcinogen furan. Toxicol Appl Pharmacol. 2014;274(1):63–77. doi: 10.1016/j.taap.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Jeng HA, Pan CH, Diawara N, et al. Polycyclic aromatic hydrocarbon–induced oxidative stress and lipid peroxidation in relation to immunological alteration. Occup Environ Med. 2011;68(9):653–658. doi: 10.1136/oem.2010.055020. [DOI] [PubMed] [Google Scholar]
- Jules GE, Pratap S, Ramesh A, Hood DB. In utero exposure to benzo(a)pyrene predisposes offspring to cardiovascular dysfunction in later-life. Toxicology. 2012;295(1–3):56–67. doi: 10.1016/j.tox.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerley-Hamilton JS, Trask HW, Ridley CJ, et al. Inherent and benzo[a]pyrene-induced differential aryl hydrocarbon receptor signaling greatly affects life span, atherosclerosis, cardiac gene expression, and body and heart growth in mice. Toxicol Sci. 2012;126(2):391–404. doi: 10.1093/toxsci/kfs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Park SS, Nam BH, Kim IH, Kim SY. Reversal of drug resistance in breast cancer cells by transglutaminase 2 inhibition and nuclear factor-kappaB inactivation. Cancer Res. 2006;66(22):10936–10943. doi: 10.1158/0008-5472.CAN-06-1521. [DOI] [PubMed] [Google Scholar]
- Knaapen AM, Curfs DM, Pachen DM, et al. The environmental carcinogen benzo[a]pyrene induces expression of monocyte-chemoattractant protein-1 in vascular tissue: A possible role in atherogenesis. Mutat Res. 2007;621(1–2):31–41. doi: 10.1016/j.mrfmmm.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Knuckles ME, Inyang F, Ramesh A. Acute and subchronic oral toxicities of benzo[a]pyrene in F-344 rats. Toxicol Sci. 2001;61(2):382–388. doi: 10.1093/toxsci/61.2.382. [DOI] [PubMed] [Google Scholar]
- Kojima M, Ashino T, Yoshida T, Iwakura Y, Sekimoto M, Degawa M. IL-1 regulates the Cyp7a1 gene and serum total cholesterol level at steady state in mice. Biochem Biophys Res Commun. 2009;379(2):239–242. doi: 10.1016/j.bbrc.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Kossoy G, Ben-hur H, Elhayany A, Schneider D, Zusman I. The morphological pathway for mouse forestomach cancer. Oncol Rep. 2006;15:479–483. [PubMed] [Google Scholar]
- Krewski D, Acosta D, Andersen M, et al. Toxicity Testing in the 21st Century: A Vision and a Strategy. J Toxicol Environ Health B Crit Rev. 2010;13(2-4):51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen P, Eilertsen E, Einarsdottir E, Haugen A, Skaug V, Ovrebo S. Fertility in mice after prenatal exposure to benzo[a]pyrene and inorganic lead. Environ Health Perspect. 1995;103(6):588–590. doi: 10.1289/ehp.95103588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib S, Guo C, Williams A, Yauk C, White P, Halappanavar S. Toxicogenomic outcomes predictive of forestomach carcinogenesis following exposure to benzo(a)pyrene: Relevance to human cancer risk. [30 August 2013];Toxicol Appl Pharmacol. 2013 00:1–12. doi: 10.1016/j.taap.2013.05.027. [DOI] [PubMed] [Google Scholar]
- Labib S, Yauk C, Williams A, et al. Sub-chronic oral exposure to benzo(a)pyrene leads to distinct transcriptomic changes in the lungs that are related to carcinogenesis. Toxicol Sci. 2012;129(1):213–224. doi: 10.1093/toxsci/kfs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q, Mumford JL, Shen M, et al. Oxidative damage–related genes AKR1C3 and 0GG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25(11):2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- Lavoie EJ, Braley J, Rice JE, Rivenson A. Tumorigenic activity of non-alternant polynuclear aromatic hydrocarbons in newborn mice. Cancer Lett. 1987;34(1):15–20. doi: 10.1016/0304-3835(87)90068-1. [DOI] [PubMed] [Google Scholar]
- Le Hégarat L, Huet S, Fessard V. A co-culture system of human intestinal Caco-2 cells and lymphoblastoid TK6 cells for investigating the genotoxicity of oral compounds. Mutagenesis. 2012;27(6):631–636. doi: 10.1093/mutage/ges028. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Douglas G, Gingerich J, et al. Simultaneous measurement of benzo[a]pyrene-induced Pig-a and lacZ mutations, micronuclei and DNA adducts in Muta™Mouse. Environ Mol Mutagen. 2011;52(9):756–765. doi: 10.1002/em.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DF. Molecular orbital calculations and quantitative structure–activity relationships for some polyaromatic hydrocarbons. Xenobiotica. 1987;17(11):1351–1361. doi: 10.3109/00498258709047165. [DOI] [PubMed] [Google Scholar]
- Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012;98:4.13.1–4.13.9. doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Muehlbauer KR, Schmeiser HH, Hergenhahn M, Belharazem D, Hollstein MC. P53 mutations in benzo(a)pyrene-exposed human P53 knock-in murine fibroblasts correlate with P53 mutations in human lung tumors. Cancer Res. 2005;65(7):2583–2587. doi: 10.1158/0008-5472.CAN-04-3675. [DOI] [PubMed] [Google Scholar]
- Lodovici M, Akpan V, Giovannini L, Migliani F, Dolara P. Benzo[a]pyrene diol-epoxide DNA adducts and levels of polycyclic aromatic hydrocarbons in autoptic samples from human lungs. Chem Biol Interact. 1998;116(3):199–212. doi: 10.1016/s0009-2797(98)00091-x. [DOI] [PubMed] [Google Scholar]
- Lu LJW, Disher RM, Reddy MV, Randerath K. 32P-postlabeling assay in mice of transplacental DNA damage induced by the environmental carcinogens safrole, 4-aminobiphenyl, and benzo(a)pyrene. Cancer Res. 1986;46(6):3046–3054. [PubMed] [Google Scholar]
- Lu X, Shao J, Li H, Yu Y. Early whole-genome transcriptional response induced by benzo[a]pyrene diol epoxide in a normal human cell line. Genomics. 2009;93(4):332–342. doi: 10.1016/j.ygeno.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Luo W, Fan W, Xie H, et al. Phenotypic anchoring of global gene expression profiles induced by N-hydroxy-4-acetylaminobiphenyl and benzo[a]pyrene diol epoxide reveals correlations between expression profiles and mechanism of toxicity. Chem Res Toxicol. 2005;18(4):619–629. doi: 10.1021/tx049828f. [DOI] [PubMed] [Google Scholar]
- Mackay D, Paterson S. Evaluating the multimedia fate of organic chemicals: A level III fugacity model. Environ Sci Technol. 1991;25(3):427–436. [Google Scholar]
- Mackenzie KM, Angevine DM. Infertility in mice exposed in utero to benzo(a)pyrene. Biol Reprod. 1981;24(1):183–191. doi: 10.1095/biolreprod24.1.183. [DOI] [PubMed] [Google Scholar]
- Maier M, Maier D, Lloyd BJ. Factors influencing the mobilisation of polycyclic aromatic hydrocarbons (PAHs) from the coal-tar lining of water mains. Water Res. 2000;34(3):773–786. [Google Scholar]
- Malik A, Williams A, Lemieux C, White P, Yauk C. Hepatic mRNA, microRNA, and miR-34a-Target responses in mice after 28 days exposure to doses of benzo(a)pyrene that elicit DNA damage and mutation. Environ Mol Mutagen. 2012;53(1):10–21. doi: 10.1002/em.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AI, Rowan-Carroll A, Williams A, Lemieux CL, Long AS, et al. Hepatic genotoxicity and toxicogenomic responses in Muta™Mouse males treated with dibenz[a,h]anthracene. Mutagenesis. 2013;28(5):543–54. doi: 10.1093/mutage/get031. [DOI] [PubMed] [Google Scholar]
- Marie C, Bouchard M, Heredia-Ortiz R, Viau C, Maître A. A toxicokinetic study to elucidate 3-hydroxybenzo(a)pyrene atypical urinary excretion profile following intravenous injection of benzo(a)pyrene in rats. J Appl Toxicol. 2010;30(5):402–410. doi: 10.1002/jat.1511. [DOI] [PubMed] [Google Scholar]
- Mass MJ, Jeffers AJ, Ross JA, et al. Ki-ras oncogene mutations in tumors and DNA adducts formed by benz[j]aceanthrylene and benzo[a]pyrene in the lungs of strain A/J mice. Mol Carcinog. 1993;8(3):186–192. doi: 10.1002/mc.2940080309. [DOI] [PubMed] [Google Scholar]
- Mayati A, Levoin N, Paris H, et al. Induction of intracellular calcium concentration by environmental benzo(a)pyrene involves a β2-adrenergic receptor/adenylyl cyclase/Epac-1/inositol 1,4,5-trisphosphate pathway in endothelial cells. J Biol Chem. 2012;287(6):4041–4052. doi: 10.1074/jbc.M111.319970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9(10):714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- Meek ME, Bucher JR, Cohen SM, et al. A framework for human relevance analysis of information on carcinogenic modes of action. Crit Rev Toxicol. 2003;33(6):591–653. doi: 10.1080/713608373. [DOI] [PubMed] [Google Scholar]
- Meek ME, Palermo CM, Bachman AN, North CM, Jeffrey Lewis R. Mode of action human relevance (species concordance) framework: Evolution of the Bradford Hill considerations and comparative analysis of weight of evidence. J Appl Toxicol. 2014;34(6):595–606. doi: 10.1002/jat.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinl W, Ebert B, Glatt H, Lampen A. Sulfotransferase forms expressed in human intestinal Caco-2 and TC7 cells at varying stages of differentiation and role in benzo[a]pyrene metabolism. Drug Metab Dispos. 2008;36(2):276–283. doi: 10.1124/dmd.107.018036. [DOI] [PubMed] [Google Scholar]
- Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J, Trump S, Rudzok S, et al. Transcriptional signatures of regulatory and toxic responses to benzo-[a]-pyrene exposure. BMC Genomics. 2011;12:502. doi: 10.1186/1471-2164-12-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels S, Danoy P, Dessen P, et al. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28(8):1731–1739. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- Mitsuishi Y, Motohashi H, Yamamoto M. The Keap1-Nrf2 system in cancers: Stress response and anabolic metabolism. Front Oncol. 2012;2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D, Viau A, Chu I, Withey J, McMullen E. Pharmacokinetics of benzo[a]pyrene in the rat. J Toxicol Environ Health. 1998;53(7):507–530. doi: 10.1080/009841098159114. [DOI] [PubMed] [Google Scholar]
- Monteiro P, Gilot D, Langouet S, Fardel O. Activation of the aryl hydrocarbon receptor by the calcium/calmodulin-dependent protein kinase kinase inhibitor 7-oxo-7H-benzimidazo[2,1-a]benz[de] isoquinoline-3-carboxylic acid (STO-609). Drug Metab Dispos. 2008;36(12):2556–2563. doi: 10.1124/dmd.108.023333. [DOI] [PubMed] [Google Scholar]
- Moore CJ, Pruess-Schwartz D, Mauthe RJ, Gould MN, Baird WM. Interspecies differences in the major DNA adducts formed from benzo(a)pyrene but not 7,12-dimethylbenz(a)anthracene in rat and human mammary cell cultures. Cancer Res. 1987;47(16):4402–4406. [PubMed] [Google Scholar]
- Mosca E, Alfieri R, Merelli I, Viti F, Calabria A, Milanesi L. A multilevel data integration resource for breast cancer study. BMC Syst Biol. 2010;4:76. doi: 10.1186/1752-0509-4-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaart E, Buytenhek M, Brouwer A, Lohman P, Vijg J. Genotoxic effects of intragastrically administered benzo[a]pyrene in rat liver and intestinal cells. Carcinogenesis. 1989;10(2):393–395. doi: 10.1093/carcin/10.2.393. [DOI] [PubMed] [Google Scholar]
- Muri SD, Schlatter JR, Brüschweiler BJ. The benchmark dose approach in food risk assessment: is it applicable and worthwhile? Food Chem Toxicol. 2009;47(12):2906–25. doi: 10.1016/j.fct.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Murphy LA, Moore T, Nesnow S. Propiconazole-enhanced hepatic cell proliferation is associated with dysregulation of the cholesterol biosynthesis pathway leading to activation of Erk1/2 through Ras farnesylation. Toxicol Appl Pharmacol. 2012;260(2):146–154. doi: 10.1016/j.taap.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Murray GI, Melvin WT, Greenlee WF, Burke MD. Regulation, function, and tissue-specific expression of cytochrome P450 CYP1B1. Annu Rev Pharmacol Toxicol. 2001;41:297–316. doi: 10.1146/annurev.pharmtox.41.1.297. [DOI] [PubMed] [Google Scholar]
- N'Diaye M, Le Ferrec E, Kronenberg F, Dieplinger H, Le Vee M, Fardel O. TNFα- and NF-κB-dependent induction of the chemokine CCL1 in human macrophages exposed to the atherogenic lipoprotein(a). Life Sci. 2009;84(13–14):451–457. doi: 10.1016/j.lfs.2009.01.012. [DOI] [PubMed] [Google Scholar]
- N'Diaye M, Le Ferrec E, Lagadic-Gossmann D, et al. Aryl hydrocarbon receptor– and calcium-dependent induction of the chemokine CCL1 by the environmental contaminant benzo[a]pyrene. J Biol Chem. 2006;281(29):19906–19915. doi: 10.1074/jbc.M601192200. [DOI] [PubMed] [Google Scholar]
- Nakamura BN, Mohar I, Lawson GW, et al. Increased sensitivity to testicular toxicity of transplacental benzo[a]pyrene exposure in male glutamate cysteine ligase modifier subunit knockout (Gclm−/−) mice. Toxicol Sci. 2012;126(1):227–241. doi: 10.1093/toxsci/kfs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal J, Rigdon R. Gastric tumors in mice fed benzo(a)pyrene: A quantitative study. Tex Rep Biol Med. 1967;25(4):553–557. [PubMed] [Google Scholar]
- Neal MS, Mulligan Tuttle AM, Casper RF, Lagunov A, Foster WG. Aryl hydrocarbon receptor antagonists attenuate the deleterious effects of benzo[a]pyrene on isolated rat follicle development. Reprod Biomed Online. 2010;21(1):100–108. doi: 10.1016/j.rbmo.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Nebert D, Dalton T. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Shi Z, Galvez-Peralta M, Uno S, Dragin N. Oral benzo(a)pyrene: understanding pharmacokinetics, detoxication, and consequences—Cyp1 knockout mouse lines as a paradigm. Mol Pharmacol. 2013a;84(3):304–313. doi: 10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Zhang G, Vesell ES. Genetic risk prediction: individualized variability in susceptibility to toxicants. Annu Rev Pharmacol Toxicol. 2013b;53:355–375. doi: 10.1146/annurev-pharmtox-011112-140241. [DOI] [PubMed] [Google Scholar]
- Nesnow S, Padgett WT, Moore T. Propiconazole induces alterations in the hepatic metabolome of mice: Relevance to propiconazole-induced hepatocarcinogenesis. Toxicol Sci. 2011;120(2):297–309. doi: 10.1093/toxsci/kfr012. [DOI] [PubMed] [Google Scholar]
- Nesnow S, Ward W, Moore T, Ren H, Hester SD. Discrimination of tumorigenic triazole conazoles from phenobarbital by transcriptional analyses of mouse liver gene expression. Toxicol Sci. 2009;110(1):68–83. doi: 10.1093/toxsci/kfp076. [DOI] [PubMed] [Google Scholar]
- Niu Q, Zhang H, Li X, Li M. Benzo[a]pyrene-induced neurobehavioral function and neurotransmitter alterations in coke oven workers. Occup Environ Med. 2010;67(7):444–448. doi: 10.1136/oem.2009.047969. [DOI] [PubMed] [Google Scholar]
- NRC (National Research Council) Applications of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment. The National Academies Press; Washington, DC: 2007a. [PubMed] [Google Scholar]
- NRC (National Research Council) Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press; Washington, DC: 2007b. [Google Scholar]
- Oesterling E, Toborek M, Hennig B. Benzo[a]pyrene induces intercellular adhesion molecule-1 through a caveolae and aryl hydrocarbon receptor mediated pathway. Toxicol Appl Pharmacol. 2008;232(2):309–316. doi: 10.1016/j.taap.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okey A, Franc M, Moffat I, et al. Toxicological implications of polymorphisms in receptors for xenobiotic chemicals: The case of the aryl hydrocarbon receptor. Toxicol Appl Pharmacol. 2005;207(2):43–51. doi: 10.1016/j.taap.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Ovesen JL, Schnekenburger M, Puga A. Aryl hydrocarbon receptor ligands of widely different toxic equivalency factors induce similar histone marks in target gene chromatin. Toxicol Sci. 2011;121(1):123–131. doi: 10.1093/toxsci/kfr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens EO, Toborek M, Hennig B. Flavonoids protect against intercellular adhesion molecule-1 induction by benzo[a]pyrene. Bull Environ Contam Toxicol. 2009;83(1):4–7. doi: 10.1007/s00128-009-9664-1. [DOI] [PubMed] [Google Scholar]
- Pan G, Hanaoka T, Yamano Y, et al. A study of multiple biomarkers in coke oven workers—A cross-sectional study in China. Carcinogenesis. 1998;19(11):1963–1968. doi: 10.1093/carcin/19.11.1963. [DOI] [PubMed] [Google Scholar]
- Park HJ, Oh JH, Park SM, et al. Identification of biomarkers of chemically induced hepatocarcinogenesis in rasH2 mice by toxicogenomic analysis. Arch Toxicol. 2011;85(12):1627–1640. doi: 10.1007/s00204-011-0715-0. [DOI] [PubMed] [Google Scholar]
- Payan JP, Lafontaine M, Simon P, et al. 3-Hydroxybenzo(a)pyrene as a biomarker of dermal exposure to benzo(a)pyrene. Arch Toxicol. 2009;83(9):873–883. doi: 10.1007/s00204-009-0440-0. [DOI] [PubMed] [Google Scholar]
- Perera FP, Hemminki K, Young TL, Brenner D, Kelly G, Santella RM. Detection of polycyclic aromatic hydrocarbon–DNA adducts in white blood cells of foundry workers. Cancer Res. 1988;48(8):2288–2291. [PubMed] [Google Scholar]
- Perera FP, Rauh V, Whyatt RM, et al. Molecular evidence of an interaction between prenatal environmental exposure and birth outcomes in a multiethnic population. Environ Health Perspect. 2004;112(5):626–630. doi: 10.1289/ehp.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera FP, Rauh V, Whyatt RM, et al. Effect of prenatal exposure to airborne polycyclic aromatic hydrocarbons on neurodevelopment in the first 3 years of life among inner-city children. Environ Health Perspect. 2006;114(8):1287–1292. doi: 10.1289/ehp.9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera FP, Li TY, Zhou ZJ, et al. Benefits of reducing prenatal exposure to coal-burning pollutants to children's neurodevelopment in China. Environ Health Perspect. 2008;116(10):1396–1400. doi: 10.1289/ehp.11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlow RA, Kolbanovskii A, Hingerty BE, Geacintov NE, Broyde S, Scicchitano DA. DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J Mol Biol. 2002;321(1):29–47. doi: 10.1016/s0022-2836(02)00593-4. [DOI] [PubMed] [Google Scholar]
- Plazar J, Hreljac I, Pirih P, Filipič M, Groothuis GMM. Detection of xenobiotic-induced DNA damage by the comet assay applied to human and rat precision-cut liver slices. Toxicol In Vitro. 2007;21(6):1134–1142. doi: 10.1016/j.tiv.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Podechard N, Le Ferrec E, Rebillard A, Fardel O, Lecureur V. NPC1 repression contributes to lipid accumulation in human macrophages exposed to environmental aryl hydrocarbons. Cardiovasc Res. 2009;82(2):361–370. doi: 10.1093/cvr/cvp007. [DOI] [PubMed] [Google Scholar]
- Proctor D, Gatto N, Hong S, Allamneni K. Mode-of-action framework for evaluating the relevance of rodent forestomach tumors in cancer risk assessment. Toxicol Sci. 2007;98(2):313–326. doi: 10.1093/toxsci/kfm075. [DOI] [PubMed] [Google Scholar]
- Qiu C, Cheng S, Xia Y, Peng B, Tang Q, Tu B. Effects of subchronic benzo(a)pyrene exposure on neurotransmitter receptor gene expression in the rat hippocampus related with spatial learning and memory change. Toxicology. 2011;289(3):83–90. doi: 10.1016/j.tox.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Ramesh A, Hood DB, Inyang F, et al. Comparative metabolism, bioavailability, and toxicokinetics of benzo[a]pyrene in rats after acute oral, inhalation, and intravenous administration. Polycyclic Aromat Compd. 2002;22(3–4):969–980. [Google Scholar]
- Ramesh A, Inyang F, Knuckles ME. Modulation of adult rat benzo(a)pyrene (BaP) metabolism and DNA adduct formation by neonatal diethylstilbestrol (DES) exposure. Exp Toxicol Pathol. 2004;56(3):129–38. doi: 10.1016/j.etp.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Recio L, Hsie AW. Glucuronide conjugation reduces the cytotoxicity but not the mutagenicity of benzo(a)pyrene in the CHO/HGPRT assay. Teratog Carcinog Mutagen. 1984;4(5):391–402. doi: 10.1002/tcm.1770040503. [DOI] [PubMed] [Google Scholar]
- Ridolfi R, Guidoboni M, Ridolfi L. Cancer immunoediting and dioxin-activating aryl hydrocarbon receptor: A missing link in the shift toward tumor immunoescape? J Nucleic Acids Invest. 2010;1(6):24–30. [Google Scholar]
- Rigdon R, Neal J. Gastric carcinomas and pulmonary adenomas in mice fed benzo(a)pyrene. Tex Rep Biol Med. 1966;24(2):195–207. [PubMed] [Google Scholar]
- Rigdon R, Neal J. Relationship of leukemia to lung and stomach tumors in mice fed benzo(a)pyrene. Proc Soc Exp Biol Med. 1969;130:146–148. doi: 10.3181/00379727-130-33508. [DOI] [PubMed] [Google Scholar]
- Ringuette S, Germain A, Gonthier C, Perron F. Presence of PAHs in the Canadian Environment: An Overview. Prepared for Environment Canada, Conservation and Protection, Quebec Region, Montreal, Quebec (Priority Substances List Supporting Document No. 2) 1993.
- Rojas M, Alexandrov K, Auburtin G, et al. Anti-benzo[a]pyrene diolepoxide–DNA adduct levels in peripheral mononuclear cells from coke oven workers and the enhancing effect of smoking. Carcinogenesis. 1995;16(6):1373–1376. doi: 10.1093/carcin/16.6.1373. [DOI] [PubMed] [Google Scholar]
- Rojas M, Cascorbi I, Alexandrov K, et al. Modulation of benzo[a]pyrene diolepoxide–DNA adduct levels in human white blood cells by CYP1A1, GSTM1 and GSTT1 polymorphism. Carcinogenesis. 2000;21(1):35–41. doi: 10.1093/carcin/21.1.35. [DOI] [PubMed] [Google Scholar]
- Rowlands CJ, Staskal DF, Gollapudi B, Budinsky R. The human AHR: Identification of single nucleotide polymorphisms from six ethnic populations. Pharmacogenet Genomics. 2010;20(5):283–290. doi: 10.1097/fpc.0b013e32833605f8. [DOI] [PubMed] [Google Scholar]
- Sadeu JC, Foster WG. Effect of in vitro exposure to benzo[a]pyrene, a component of cigarette smoke, on folliculogenesis, steroidogenesis and oocyte nuclear maturation. Reprod Toxicol. 2011;31(4):402–408. doi: 10.1016/j.reprotox.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Sakai R, Kondo C, Oka H, Miyajima A, Kubo K, Uehara T. Utilization of CDKN1A/p21 gene for class discrimination of DNA damage-induced clastogenicity. Toxicology 6. 2014;315:8–16. doi: 10.1016/j.tox.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Santodonato J, Howard P, Basu D. Health and ecological assessment of polynuclear aromatic hydrocarbons. J Environ Pathol Toxicol. 1981;5(1):1–364. [PubMed] [Google Scholar]
- Saunders CR, Das SK, Ramesh A, Shockley DC, Mukherjee S. Benzo(a)pyrene-induced acute neurotoxicity in the F-344 rat: Role of oxidative stress. J Appl Toxicol. 2006;26(5):427–438. doi: 10.1002/jat.1157. [DOI] [PubMed] [Google Scholar]
- Saunders CR, Shockley DC, Ramesh A. Kinetic behavior as a determinant of benzo(a)pyrene-caused neurobehavioral toxicity in F-344 rats. Res Commun Pharmacol Toxicol. 2003;8(1–2):1–15. [Google Scholar]
- Schaap MM, Zwart EP, Wackers PF, et al. Dissecting modes of action of non-genotoxic carcinogens in primary mouse hepatocytes. Arch Toxicol. 2012;86(11):1717–1727. doi: 10.1007/s00204-012-0883-6. [DOI] [PubMed] [Google Scholar]
- Schneider K, Roller M, Kalberlah F, Schuhmacher-Wolz U. Cancer risk assessment for oral exposure to PAH mixtures. J Appl Toxicol. 2002;22(1):73–83. doi: 10.1002/jat.828. [DOI] [PubMed] [Google Scholar]
- Shertzer H, Clay C, Genter M, et al. Uncoupling-mediated generation of reactive oxygen by halogenated aromatic hydrocarbons in mouse liver microsomes. Free Radic Biol Med. 2004;36(5):618–631. doi: 10.1016/j.freeradbiomed.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Shi Z, Dragin N, Gálvez-Peralta M, et al. Organ-specific roles of CYP1A1 during detoxication of dietary benzo[a]pyrene. Mol Pharmacol. 2010a;78(1):46–57. doi: 10.1124/mol.110.063438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Dragin N, Miller ML, et al. Oral benzo(a)pyrene-induced cancer : two distinct types in different target organs depend on the mouse Cyp1 genotype. Int J Cancer. 2010b;127(10):2334–2350. doi: 10.1002/ijc.25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Satake S, Itoh S, Hattori C, Hayashi M, Ishidate MJ. Multiple-dosing effects of benzo[a]pyrene in the mouse bone marrow micronucleus test. Mutat Res. 1990;234(3–4):179–181. doi: 10.1016/0165-1161(90)90012-d. [DOI] [PubMed] [Google Scholar]
- Shimada T, Martin MV, Pruess-Schwartz D, Marnett LJ, Guengerich FP. Roles of individual human cytochrome P-450 enzymes in the bioactivation of benzo(a)pyrene, 7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene, and other dihydrodiol derivatives of polycyclic aromatic hydrocarbons. Cancer Res. 1989;49(22):6304–6312. [PubMed] [Google Scholar]
- Shimizu Y, Nakatsuru Y, Ichinose M, et al. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2000;97(2):779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugart L, Matsunami R. Adduct formation in hemoglobin of the newborn mouse exposed in utero to benzo[a]pyrene. Toxicology. 1985;37(3–4):241–245. doi: 10.1016/0300-483x(85)90087-3. [DOI] [PubMed] [Google Scholar]
- Shum S, Jensen NM, Nebert DW. The murine Ah locus: In utero toxicity and teratogenesis associated with genetic differences in benzo[a]pyrene metabolism. Teratology. 1979;20(3):365–376. doi: 10.1002/tera.1420200307. [DOI] [PubMed] [Google Scholar]
- Sivak A, Niemeier R, Lynch D, Beltis K, Simon S, Salomon R, Latta R, Belinky B, Menzies K, Lunsford A, Cooper C, Ross A, Bruner R. Skin carcinogenicity of condensed asphalt roofing fumes and their fractions following dermal application to mice. Cancer Lett. 1997;117:113–123. doi: 10.1016/s0304-3835(97)00214-0. [DOI] [PubMed] [Google Scholar]
- Somji S, Bathula C, Zhou X, Sens M, Sens D, Garrett S. Transformation of human urothelial cells (UROtsa) by As and Cd induces the expression of keratin 6a. Environ Health Perspect. 2008;116(4):434–440. doi: 10.1289/ehp.10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonich-Mullin C, Fielder R, Wiltse J, et al. IPCS conceptual framework for evaluating a mode of action for chemical carcinogenesis. Regul Toxicol Pharmacol. 2001;34(2):146–152. doi: 10.1006/rtph.2001.1493. [DOI] [PubMed] [Google Scholar]
- Spinelli JJ, Demers PA, Le ND, et al. Cancer risk in aluminum reduction plant workers (Canada). Cancer Causes Control. 2006;17(7):939–948. doi: 10.1007/s10552-006-0031-9. [DOI] [PubMed] [Google Scholar]
- Tang D, Li TY, Liu JJ, Chen YH, Qu L, Perera F. PAH-DNA adducts in cord blood and fetal and child development in a Chinese cohort. Environ Health Perspect. 2006;114:1297–1300. doi: 10.1289/ehp.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- Thomas R, Allen B, Nong A, et al. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci. 2007a;98(1):240–248. doi: 10.1093/toxsci/kfm092. [DOI] [PubMed] [Google Scholar]
- Thomas R, Philbert M, Auerbach S, et al. Incorporating new technologies into toxicity testing and risk assessment: Moving from 21st century vision to a data-driven framework. Toxicol Sci. 2013;136(1):4–18. doi: 10.1093/toxsci/kft178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Pluta L, Yang L, Halsey T. Application of genomic biomarkers to predict increased lung tumor incidence in 2-year rodent cancer bioassays. Toxicol Sci. 2007b;97(1):55–64. doi: 10.1093/toxsci/kfm023. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Clewell HJ, III, Allen BC, Yang L, Healy E, Andersen ME. Integrating pathway-based transcriptomic data into quantitative chemical risk assessment: A five chemical case study. Mutat Res. 2012;746(2):135–143. doi: 10.1016/j.mrgentox.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Clewell HJ, III, Allen BC, et al. Application of transcriptional benchmark dose values in quantitative cancer and noncancer risk assessment. Toxicol Sci. 2011;120(1):194–205. doi: 10.1093/toxsci/kfq355. [DOI] [PubMed] [Google Scholar]
- Thyssen J, Althoff J, Kimmerle G, Mohr U. Inhalation studies with benzo[a]pyrene in Syrian golden hamsters. J Natl Cancer Inst. 1981;66(3):575–577. [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69(1):140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Trush MA, Mimnaugh EG, Gram TE. Activation of pharmacologic agents to radical intermediates. Implications for the role of free radicals in drug action and toxicity. Biochem Pharmacol. 1982;31(21):3335–3346. doi: 10.1016/0006-2952(82)90609-8. [DOI] [PubMed] [Google Scholar]
- Turusov VS, Nikonova TV, Parfenov YD. Increased multiplicity of lung adenomas in five generations of mice treated with benz(a)pyrene when pregnant. Cancer Lett. 1990;55(3):227–231. doi: 10.1016/0304-3835(90)90123-f. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton TP, Derkenne S, et al. Oral exposure to benzo(a)pyrene in the mouse: detoxication by inducible cytochrome P450 is more important than metabolic activation. Mol Pharmacol. 2004;65(5):1225–1237. doi: 10.1124/mol.65.5.1225. [DOI] [PubMed] [Google Scholar]
- Uno S, Dalton T, Dragin N, et al. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol Pharmacol. 2006;69(4):1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- Urso P, Gengozian N. Alterations in the humoral immune response and tumor frequencies in mice exposed to benzo[a]pyrene and X-rays before or after birth. J Toxicol Environ Health. 1982;10(4–5):817–835. doi: 10.1080/15287398209530297. [DOI] [PubMed] [Google Scholar]
- Urso P, Gengozian N. Subnormal expression of cell-mediated and humoral immune responses in progeny disposed toward a high incidence of tumors after in utero exposure to benzo[a]pyrene. J Toxicol Environ Health. 1984;14(4):569–584. doi: 10.1080/15287398409530606. [DOI] [PubMed] [Google Scholar]
- US EPA (United States Environmental Protection Agency) Methods for determinations of organic compounds in drinking water - Supplement I. EPA/600/4-90/020. US Environmental Protection Agency, Office of Research and Development; Washington, DC: 1990. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Methods for determinations of organic compounds in drinking water - Supplement III. EPA/600/R-95/131. US Environmental Protection Agency, Office of Research and Development; Washington, DC: 1995. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Analytical feasibility support document for the six year review of existing National Primary Drinking Water Regulations (reassessment of feasibility for chemical contaminants). EPA/815/R-03/003. US Environmental Protection Agency, Office of Groundwater and Drinking Water; Washington, DC: 2003. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Guidelines for Carcinogen Risk Assessment. EPA/630/P-03/001F. US Environmental Protection Agency, Risk Assessment Forum; Washington, DC: 2005. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Benzo(a)pyrene (BaP). TEACH Chemical Summary. US Environmental Protection Agency, Toxicity and Exposure Assessment for Children's Health; Washington, DC: 2007. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Analytical feasibility support document for the second six-year review of existing National Primary Drinking Water Regulations. EPA/815/B-09/003. US Environmental Protection Agency, Office of Water; Washington, DC: 2009. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Benchmark dose technical guidance. EPA/100/R-12/001. US Environmental Protection Agency, Risk Assessment Forum; Washington, DC: 2012. [Google Scholar]
- US EPA (United States Environmental Protection Agency) Estimation Programs Interface Suite™ for Microsoft® Windows, v4.11. US Environmental Protection Agency; Washington, DC: 2013. [Google Scholar]
- Vakharia DD, Liu N, Pause R, et al. Polycyclic aromatic hydrocarbon/metal mixtures: effect on PAH induction of CYP1A1 in human HEPG2 cells. Drug Metab Dispos. 2001;29(7):999–1006. [PubMed] [Google Scholar]
- Van de Wiel J, Fijneman P, Duijf C, Anzion R, Theuws J, Bos R. Excretion of benzo[a]pyrene and metabolites in urine and feces of rats: Influence of route of administration, sex and long-term ethanol treatment. Toxicology. 1993;80(2–3):103–115. doi: 10.1016/0300-483x(93)90174-q. [DOI] [PubMed] [Google Scholar]
- Van Delft J, Gaj S, Lienhard M, et al. RNA-seq provides new insights in the transcriptome responses induced by the carcinogen benzo[a]pyrene. Toxicol Sci. 2012;130(2):427–439. doi: 10.1093/toxsci/kfs250. [DOI] [PubMed] [Google Scholar]
- Van Delft J, Mathijs K, Staal Y, et al. Time series analysis of benzo[a]pyrene-induced transcriptome changes suggests that a network of transcription factors regulates the effects on functional gene sets. Toxicol Sci. 2010;117(2):381–392. doi: 10.1093/toxsci/kfq214. [DOI] [PubMed] [Google Scholar]
- Volk D, Thiviyanathan V, Rice J, et al. Solution structure of a cis-opened (10R)-N6-deoxyadenosine adduct of (9S,10R)-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA duplex. Biochemistry. 2003;42(6):1410–1420. doi: 10.1021/bi026745u. [DOI] [PubMed] [Google Scholar]
- Wan B, Sayler GS, Schultz TW. Structure–activity relationships for flow cytometric data of smaller polycyclic aromatic hydrocarbons. SAR QSAR Environ Res. 2006;17(6):597–605. doi: 10.1080/10629360601033374. [DOI] [PubMed] [Google Scholar]
- Waters M, Jackson M, Lea I. Characterizing and predicting carcinogenicity and mode of action using conventional and toxicogenomics methods. Mutat Res. 2010;705(3):184–200. doi: 10.1016/j.mrrev.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Wester P, Muller J, Slob W, Mohn G, Dortant P, Kroese E. Carcinogenic activity of benzo[a]pyrene in a 2 year oral study in Wistar rats. Food Chem Toxicol. 2012;50(3–4):927–935. doi: 10.1016/j.fct.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Weyand EH, Chen YC, Wu Y, Koganti A, Dunsford HA, Rodriguez LV. Differences in the tumorigenic activity of a pure hydrocarbon and a complex mixture following ingestion: Benzo[a]pyrene vs manufactured gas plant residue. Chem Res Toxicol. 1995;8(7):949–954. doi: 10.1021/tx00049a008. [DOI] [PubMed] [Google Scholar]
- White KL, Jr, Holsapple MP. Direct suppression of in vitro antibody production by mouse spleen cells by the carcinogen benzo(a)pyrene but not by the noncarcinogenic congener benzo(e)pyrene. Cancer Res. 1984;44(8):3388–3393. [PubMed] [Google Scholar]
- WHO (World Health Organization) Polynuclear Aromatic Hydrocarbons in Drinking-water: Background Document for Development of WHO Guidelines for Drinking-water Quality. World Health Organization; Geneva, Switzerland: 2003. [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- Wielgosz S, Brauze D, Pawlak A. Ah locus–associated differences in induction of sister-chromatid exchanges and in DNA adducts by benzo[a]pyrene in mice. Mutat Res. 1991;246(1):129–137. doi: 10.1016/0027-5107(91)90115-5. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31(8):1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- Wislocki PG, Bagan ES, Lu AY, et al. Tumorigenicity of nitrated derivatives of pyrene, benz[a]anthracene, chrysene and benzo[a]pyrene in the newborn mouse assay. Carcinogenesis. 1986;7(8):1317–1322. doi: 10.1093/carcin/7.8.1317. [DOI] [PubMed] [Google Scholar]
- Wojdani A, Alfred L. Alterations in cell-mediated immune functions induced in mouse splenic lymphocytes by polycyclic aromatic hydrocarbons. Cancer Res. 1984;44(3):942–945. [PubMed] [Google Scholar]
- Woodruff TJ, Zeise L, Axelrad DA, et al. Meeting report: Moving upstream—Evaluating adverse upstream end points for improved risk assessment and decision-making. Environ Health Perspect. 2008;116(11):1568–1575. doi: 10.1289/ehp.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ramesh A, Nayyar T, Hood DB. Assessment of metabolites and AhR and CYP1A1 mRNA expression subsequent to prenatal exposure to inhaled benzo(a)pyrene. Int J Dev Neurosci. 2003;21(6):333–346. doi: 10.1016/s0736-5748(03)00073-x. [DOI] [PubMed] [Google Scholar]
- Xia Y, Cheng S, He J, et al. Effects of subchronic exposure to benzo[a]pyrene (B[a]P) on learning and memory, and neurotransmitters in male Sprague-Dawley rat. Neurotoxicology. 2011;32(2):188–198. doi: 10.1016/j.neuro.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Xu C, Chen JA, Qiu Z, et al. Ovotoxicity and PPAR-mediated aromatase downregulation in female Sprague-Dawley rats following combined oral exposure to benzo[a]pyrene and di-(2-ethylhexyl) phthalate. Toxicol Lett. 2010;199(3):323–332. doi: 10.1016/j.toxlet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Xu Z, Brown LM, Pan GW, et al. Cancer risks among iron and steel workers in Anshan, China, Part II: Case–control studies of lung and stomach cancer. Am J Ind Med. 1996;30(1):7–15. doi: 10.1002/(SICI)1097-0274(199607)30:1<7::AID-AJIM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhou L, Wang Z, et al. Overexpression of antioxidant enzymes in ApoE-deficient mice suppresses benzo(a)pyrene-accelerated atherosclerosis. Atherosclerosis. 2009;207(1):51–58. doi: 10.1016/j.atherosclerosis.2009.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Allen BC, Thomas RS. BMDExpress: A software tool for the benchmark dose analyses of genomic data. BMC Genomics. 2007;8:387–394. doi: 10.1186/1471-2164-8-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Jackson K, Malowany M, Williams A. Lack of change in microRNA expression in adult mouse liver following treatment with benzo(a)pyrene despite robust mRNA transcriptional response. Mutat Res. 2011;722(2):131–139. doi: 10.1016/j.mrgentox.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Introducing the “TCDD-inducible AhR-Nrf2 gene battery.”. Toxicol Sci. 2009;111(2):238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SJ, Tian HJ, Cao J, Gao YQ. Exposure to di(n-butyl)phthalate and benzo(a)pyrene alters IL-1β secretion and subset expression of testicular macrophages, resulting in decreased testosterone production in rats. Toxicol Appl Pharmacol. 2010;248(1):28–37. doi: 10.1016/j.taap.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Zhou J, Shephard E. Mutation, polymorphism and perspectives for the future of human flavin-containing monooxygenase 3. Mutat Res. 2006;612(3):165–171. doi: 10.1016/j.mrrev.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Zuber AM, Centeno G, Pradervand S, et al. Molecular clock is involved in predictive circadian adjustment of renal function. Proc Natl Acad Sci USA. 2009;106(38):16523–16528. doi: 10.1073/pnas.0904890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.