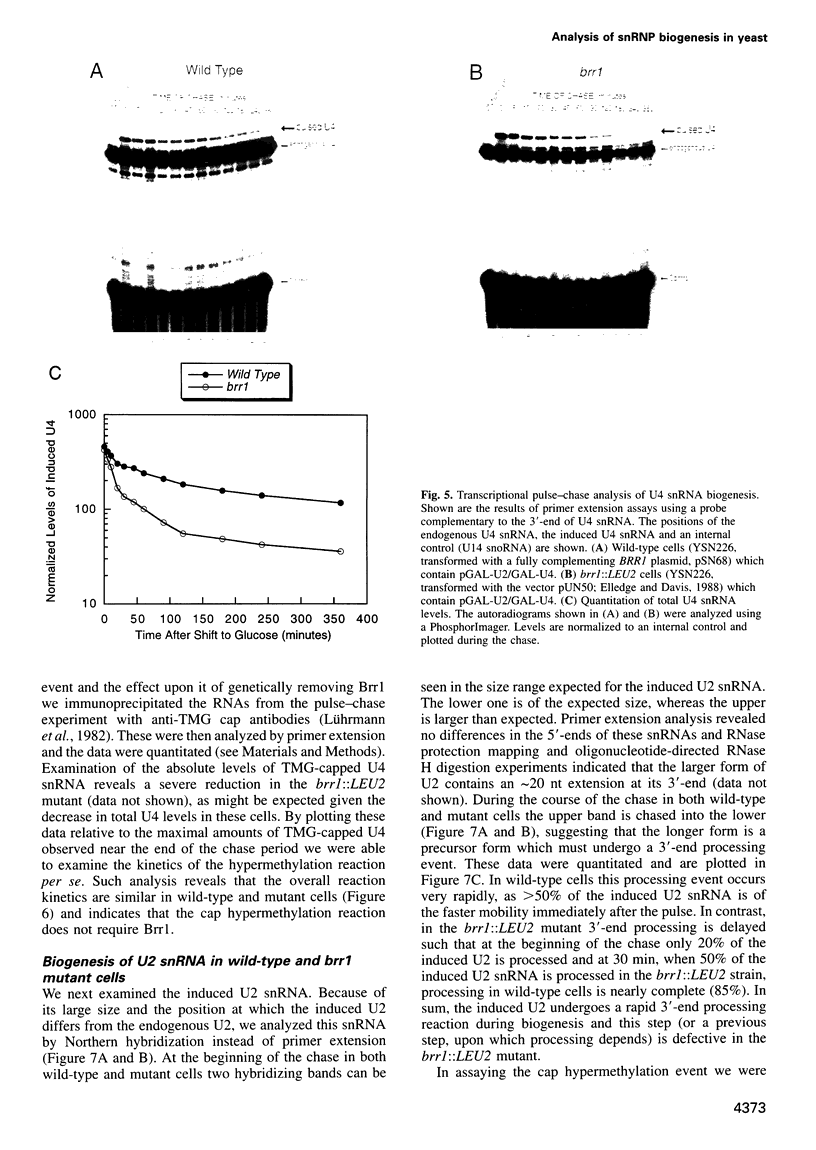
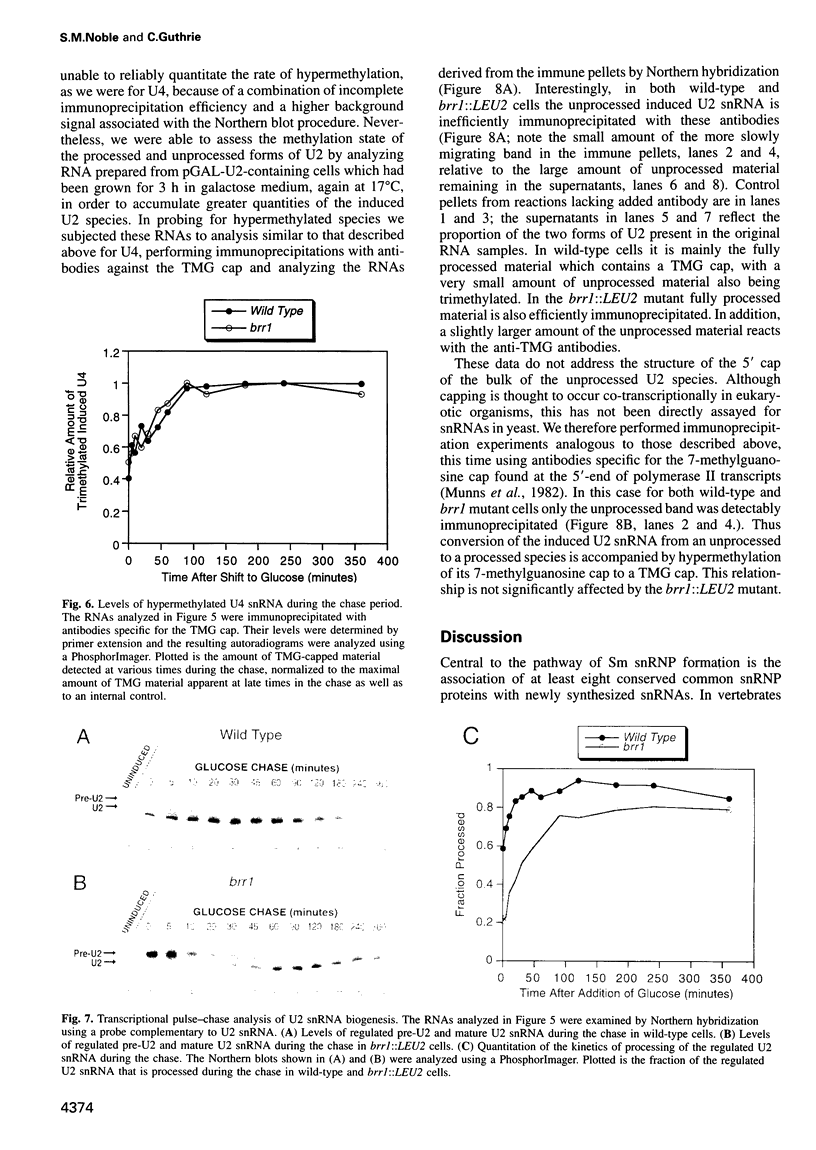
Abstract
Vertebrate spliceosomal snRNAs associate with a conserved set of proteins, the Sm proteins, via a conserved RNA sequence, the Sm site. Assembly of this complex is required for the accumulation of stable snRNPs, hypermethylation of the 5' cap structure and nuclear import of the resultant particles. The function of individual core snRNP proteins is poorly understood, in part because of the difficulty of selectively inactivating individual polypeptides in vivo. Using a transcriptional pulse-chase method we have defined for the first time the steps of snRNP biogenesis in Saccharomyces cerevisiae. We describe a novel component of spliceosomal snRNPs, Brr1, which is distinct in sequence from Sm core proteins and yet which shares many of their properties, as well as a genetic interaction with the yeast homolog of Sm D1 core protein. Through a kinetic analysis of snRNP formation in wild-type and brr1 mutant cells we demonstrate specific defects in a subset of steps in the brr1 mutant: newly synthesized snRNAs are destabilized and 3'-end processing is slowed, whereas the cap hypermethylation reaction is unaffected. Notably, the stability of mature particles, as measured by promoter shut-off experiments, is normal in the absence of the Brr1 snRNP protein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanton S., Srinivasan A., Rymond B. C. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol Cell Biol. 1992 Sep;12(9):3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cooper M., Johnston L. H., Beggs J. D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995 May 1;14(9):2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993 Aug;7(8):1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988 Oct 30;70(2):303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hackl W., Fischer U., Lührmann R. A 69-kD protein that associates reversibly with the Sm core domain of several spliceosomal snRNP species. J Cell Biol. 1994 Feb;124(3):261–272. doi: 10.1083/jcb.124.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann H., Fabrizio P., Raker V. A., Foulaki K., Hornig H., Brahms H., Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995 May 1;14(9):2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lehmeier T., Foulaki K., Lührmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990 Nov 25;18(22):6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Brow D. A. A rapid assay for quantitative detection of specific RNAs. Nucleic Acids Res. 1993 Sep 25;21(19):4645–4646. doi: 10.1093/nar/21.19.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Luhrmann R., Appel B., Bringmann P., Rinke J., Reuter R., Rothe S., Bald R. Isolation and characterization of rabbit anti-m3 2,2,7G antibodies. Nucleic Acids Res. 1982 Nov 25;10(22):7103–7113. doi: 10.1093/nar/10.22.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992 Nov 27;71(5):803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Munns T. W., Liszewski M. K., Tellam J. T., Sims H. F., Rhoads R. E. Antibody-nucleic acid complexes. Immunospecific retention of globin messenger ribonucleic acid with antibodies specific for 7-methylguanosine. Biochemistry. 1982 Jun 8;21(12):2922–2928. doi: 10.1021/bi00541a018. [DOI] [PubMed] [Google Scholar]
- Nelissen R. L., Will C. L., van Venrooij W. J., Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994 Sep 1;13(17):4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Dahlberg J. E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990 Jul;10(7):3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
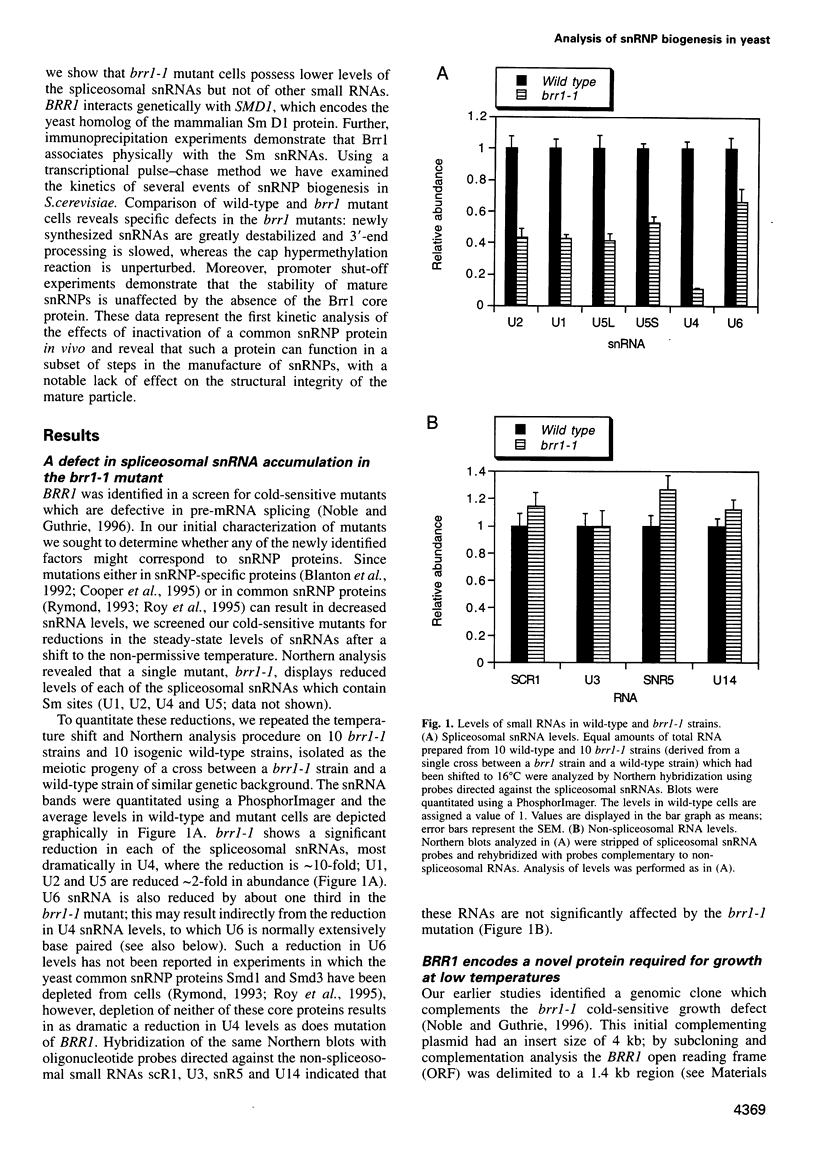
- Noble S. M., Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996 May;143(1):67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Herrick D., Peltz S. W., Jacobson A. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- Plessel G., Fischer U., Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994 Jun;14(6):4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J., Zheng B., Rymond B. C., Woolford J. L., Jr Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995 Jan;15(1):445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. C. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond B. C., Rokeach L. A., Hoch S. O. Human snRNP polypeptide D1 promotes pre-mRNA splicing in yeast and defines nonessential yeast Smd1p sequences. Nucleic Acids Res. 1993 Jul 25;21(15):3501–3505. doi: 10.1093/nar/21.15.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauterer R. A., Feeney R. J., Zieve G. W. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA's in L929 mouse fibroblasts. Exp Cell Res. 1988 Jun;176(2):344–359. doi: 10.1016/0014-4827(88)90336-9. [DOI] [PubMed] [Google Scholar]
- Schneider J. C., Guarente L. Vectors for expression of cloned genes in yeast: regulation, overproduction, and underproduction. Methods Enzymol. 1991;194:373–388. doi: 10.1016/0076-6879(91)94028-b. [DOI] [PubMed] [Google Scholar]
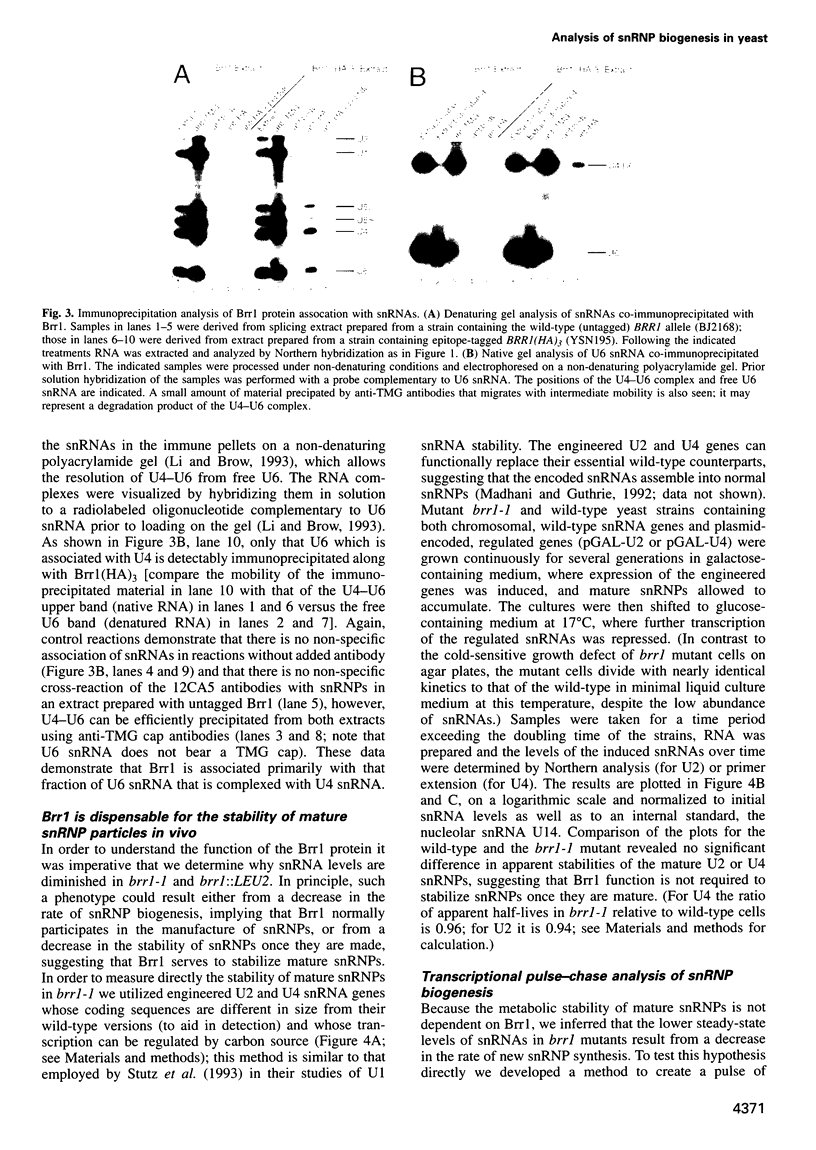
- Shannon K. W., Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991 May;5(5):773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- Shuster E. O., Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988 Oct 7;55(1):41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Liao X. C., Rosbash M. U1 small nuclear ribonucleoprotein particle-protein interactions are revealed in Saccharomyces cerevisiae by in vivo competition assays. Mol Cell Biol. 1993 Apr;13(4):2126–2133. doi: 10.1128/mcb.13.4.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségault V., Will C. L., Sproat B. S., Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995 Aug 15;14(16):4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
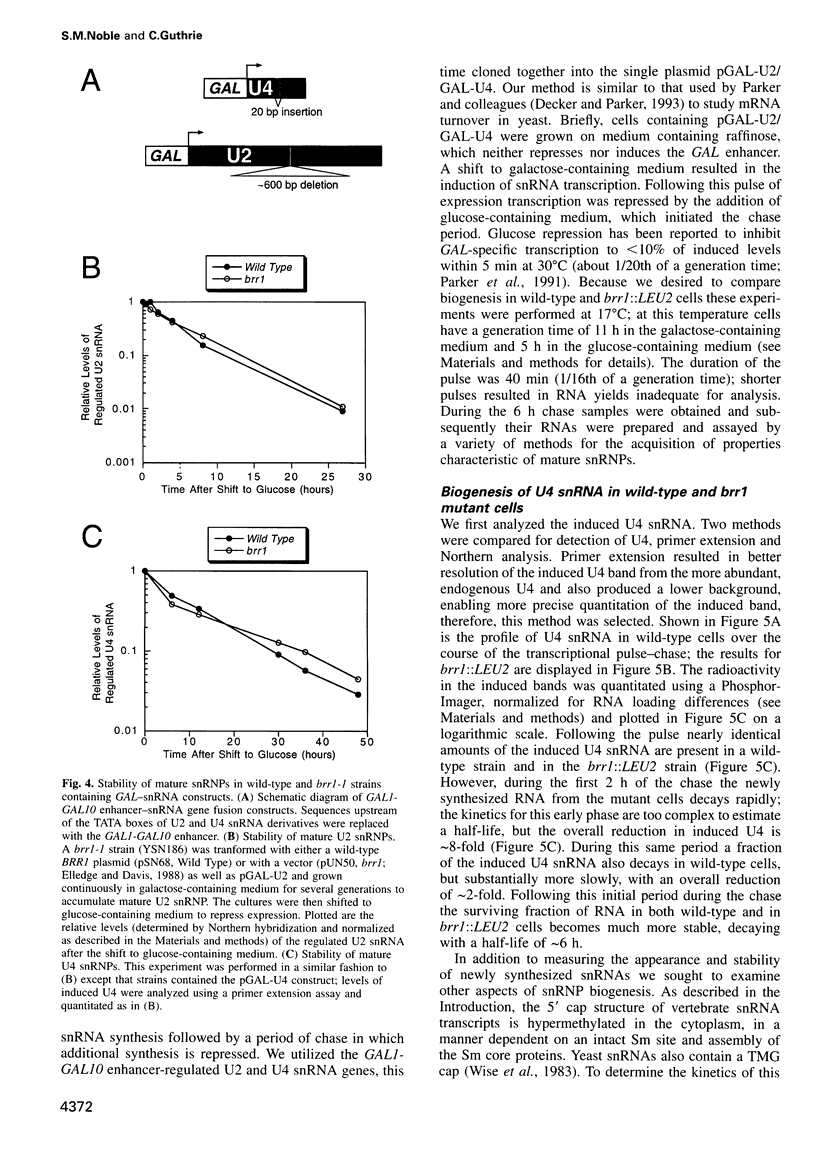
- Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995 May 1;14(9):2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J. A., Tollervey D., Maloney D., Swerdlow H., Dunn E. J., Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983 Dec;35(3 Pt 2):743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- Zieve G. W., Sauterer R. A. Cell biology of the snRNP particles. Crit Rev Biochem Mol Biol. 1990;25(1):1–46. doi: 10.3109/10409239009090604. [DOI] [PubMed] [Google Scholar]