Abstract
Induced pluripotent stem cells (iPSCs) derived from reprogrammed somatic cells are emerging as one of the most versatile tools in biomedical research and pharmacological studies. Oncogenic transformation and somatic cell reprogramming are multistep processes that share some common features, and iPSCs generated from cancerous cells can help us better understand the molecular mechanisms underlying the initiation and progression of human cancers and overcome them. Aside from the mechanistic modeling of human tumorigenesis, immediate applications of this technology in cancer research include high-throughput drug screening, toxicological testing, early biomarker identification, and bioengineering of replacement tissues. Here, we review the current advances in generating iPSCs from cancer cell lines and patient-derived primary cancer tissues, and discuss their potential applications.
Keywords: induced pluripotent stem cells, cancer, tumorigenesis, disease modeling
Introduction
Human embryonic stem cells (ESCs) may still represent the most promising cell type for basic and translational applications. However, ESC derivation and use pose major ethical dilemmas. Reprogramming of somatic tissues is a more pragmatic alternative. The core technology of induced pluripotent stem cell (iPSC) generation centers on the ectopic expression of master reprogramming factors and epigenetic reactivation of endogenous pluripotency genes. Takahashi and Yamanaka first used Oct4, Sox2, Klf4, and c-Myc (OSKM) to reprogram murine fibroblasts in 2006,1 paving the way for reprogramming human somatic cells.2,3 Since then, research on iPSCs has grown exponentially, along with technological advances in reprogramming methodologies using a variety of reprogramming factors and enhancers. Many groups have been able to avoid the use of the proto-oncogene c-Myc by replacing it with less dangerous genes such as L-Myc4,5 or Glis1.6,7 To date, numerous reprogramming factor delivery strategies have been developed to increase the safety and efficiency of iPSC derivation, through the use of nonintegrating systems such as Sendai virus, episomal vectors, and mRNAs.8 The ongoing advancement of iPSC research could lead to a paradigm shift in how we study human cancers. Yet some challenges remain that must be surmounted in order to realize the far-reaching potential of this revolutionary technology.
The Reprogramming Paradox
New iPSC lines are now routinely generated from primary tissues obtained from healthy donors and various patients, as well as from different species, cell types, and developmental stages. Moreover, knockdown of tumor suppressor genes is known to enhance reprogramming efficiency.9–11 However, reprogramming human primary cancer cells has proven to be, paradoxically, inefficient. Despite significant interest in generating iPSCs from cancer cells to help elucidate the molecular mechanisms of cancer progression, there have been relatively few reports demonstrating successful reprogramming of malignant human cells. The difficulties in reprogramming cancer cells are thought to stem in part from biological barriers, including cancer-specific genetic mutations, epigenetic modifications, accumulation of DNA damage, and reprogramming-triggered cellular senescence.12–14
In spite of these difficulties, several published studies have reported the generation of novel iPSC lines from existing cancer cell lines. Reports of iPSC lines derived from human cancer cell lines are summarized in Table 1 and cover a range of cancers such as melanoma,15,16 prostate cancer,15 gastrointestinal cancers,17 chronic myeloid leukemia,18 lung cancer,19 breast cancer,20 glioblastoma,21 and sarcomas.22 Lin et al reported successful reprogramming by expression of the microRNA miR-302 in Colo and PC-3 cells, which are cell lines established from melanoma and prostate cancer, respectively.15 Miyoshi et al reprogrammed several gastrointestinal cancer cell lines, and, interestingly, they had to optimize the transduction method using a combination of retroviral or lentiviral OSKM, NANOG, LIN28, BCL2, KRAS, and shRNA for tumor suppressors for each cell line. They obtained iPSC-like cells that re-expressed NANOG and were less tumorigenic compared to their parental cell line.17 Carette et al generated iPSCs from the chronic myeloid leukemia (CML) cell line KBM7 carrying the BCR–ABL fusion oncogene by expressing OSKM factors.18 Recently, an acute myeloid leukemia (AML) mouse model was established by retrovirally overexpressing the human mixed-lineage leukemia–AF9 (MLL–AF9) fusion gene in hematopoietic cells harvested from transgenic mice that carry four OSKM factors under the control of doxycycline (Dox).23 Upon addition of Dox to the culture, the MLL–AF9-expressing leukemia cells were efficiently converted into iPSCs that were capable of forming teratomas and producing chimeras. Most of the chimeric mice spontaneously developed AML, which showed that development had recreated the conditions for developing leukemia. The authors noted that their retroviral approach to induce pluripotency did not work for Notch1-initiated T-acute lymphoblastic leukemia.23 As these papers show, reprogramming of cells with cancer genomes is not an impossible feat. Yet, certain types of cancers may be refractory to commonly used reprogramming factors and the combination of factors will have to be carefully chosen depending on the type of cancer cell one is attempting to reprogram.
Table 1.
Human cancer cell-derived iPSC lines.
| CANCER TYPES | CELL LINE OR PRIMARY CELLS | REPROGRAMMING METHOD | CELLULAR PHENOTYPES | REFERENCE |
|---|---|---|---|---|
| Melanoma | Colo | Retroviral mir-302s | Reduced migration ability in iPSCs; reduced cell cycle-related gene expression in iPSCs | 15 |
| Prostate cancer | PC-3 | |||
| Melanoma | R545 | Lentiviral OCT4, KLF4, and c-MYC | SOX2 was dispensable for reprogramming melanocytes and melanoma cells | 16 |
| Chronic myeloid leukemia (blast crisis stage) | KBM7 | Retroviral OSKM | Acquired insensitivity to imatinib; loss of BCR-ABL dependency in iPSCs but restored after differentiation | 18 |
| Colorectal cancer | DLD-1, HT-29 | Combination of retroviral or lentiviral OSKM, NANOG, LIN28, BCL2, KRAS, and shRNA for tumor suppressors optimized for each cell line | Acquired sensitivity to chemotherapy in embryoid body cells; reduced invasion and tumorigenicity of embryoid body cells; higher expression of p16 and p53 of embryoid body cells compared to the parental cells | 17 |
| Esophageal cancer | TE-10 | |||
| Gastric cancer | MKN45 | |||
| Hepatocellular cancer | PLC | |||
| Pancreatic cancer | MIAPaCa-2, PANC-1 | |||
| Cholangiocellular cancer | HuCC-T1 | |||
| Chronic myeloid leukemia (chronic phase) | Patient-derived bone marrow cells | Episomal OSKM, NANOG, LIN28, SV40 LT | BCR–ABL fusion iPSCs maintained patient-specific complex karyotype | 24 |
| Lung cancer | A549 | Lentiviral OSNL and nondegradable HIFα | Increase tumorigenic properties of iPSCs in mice; more aggressive and invasive iPSCs | 19 |
| Chronic myeloid leukemia (chronic phase) | Patient-derived bone marrow cells | Retroviral OSKM | CML-iPSCs were insensitive to imatinib but still expressed BCR–ABL; recovered imatinib sensitivity after hematopoietic differentiation | 25 |
| Breast cancer | MCF-7 | Retroviral OSKM | MCF-7/Rep iPSCs did not fully reprogram, and overexpressed Sox2; displayed cancer stem cell features such as high ALDH activity and CD44 expression | 20 |
| Juvenile myelomonocytic leukemia (JMML) | Patient-derived mononuclear cells with E76K missense in PTPN11 gene | Lentiviral OSKM | Increased proliferative capacity, constitutive activation of GM-CSF, enhanced STAT5/ERK phosphorylation in myeloid cells differentiated from JMML iPSCs | 26 |
| Pancreatic ductal adenocarcinoma (PDAC) | Patient-derived pancreatic ductal adenocarcinoma | Lentiviral OSKM | Teratomas from PDAC iPSC-like cells undergo early to invasive stages of human cancer | 27 |
| Glioblastoma multiforme (GBM) | GBM neural stem (GNS) cell lines | PiggyBac driving OCT4 and KLF4 | GNS-iPSCs differentiated to neural progenitors displayed widespread resetting of GBM-associated epigenetic changes, but still remained malignant upon xenotransplantation | 21 |
| Osteosarcoma | SAOS2, HOS, MG63 | Lentiviral OSKM, NANOG, LIN28 | Sarcoma-iPSCs were less tumorigenic compared to parental sarcoma cell lines and could be terminally differentiated into mature connective tissue and red blood cells; terminal differentiation irreversibly abolished their tumorigenic potential | 22 |
| Liposarcoma | SW872 | |||
| Ewing’s sarcoma | SKNEP | |||
| Myelodysplastic syndromes (MDS) | Two patients with del(7q)-MDS | Lentiviral OSKM | MDS-iPSCs recapitulated disease-associated phenotypes, including impaired hematopoietic differentiation | 28 |
| Li Fraumeni syndrome (LFS) | Three patients with G245D missense in p53 gene | Sendai viral OSKM | LFS-iPSCs recapitulated features of osteosarcoma associated with LFS, including defective osteoblastic differentiation as well as tumorigenic ability | 29 |
| Ewing sarcoma (EWS) | CHLA-10 | Episomal OKSM | EWS-iPS cells sustained expression of the EWS-FLI1 fusion transcript; gave rise to tumors with characteristic Ewing histopathology and demonstrated recovery of drug sensitivity following differentiation | 30 |
Furthermore, reports of iPSCs generated from human primary malignant cells (Table 1) have been few and far between, and are limited to cancers such as leukemia24–26,28 and pancreatic cancer.27 Hu et al reported reprogramming primary human lymphoblasts from a BCR−ABL+CML patient using transgene-free iPSC technology to ectopically express OSKM, NANOG, LIN28, and the SV40 large T gene.24 Kim et al were successful in deriving a single iPSC-like line that contained the KRAS G12D mutation in the parental pancreatic ductal adenocarcinoma (PDAC) cancer cultures and an isogenic control line without the mutations from the noncancerous marginal tissue.27 The paucity of success stories demonstrates the difficulty of reprogramming primary cancer cells to iPSCs. Although technical limitations, such as difficulties in keeping primary cancer tissues in culture, cannot be excluded, fundamental biological hurdles may also directly undermine the reprogramming processes in cancer cells. The Jaenisch lab performed a very interesting study where they transplanted nuclei from melanoma, leukemia, lymphoma, and breast cancer cells into enucleated oocytes.31 The nuclei derived from primary leukemia and lymphoma cells failed to reprogram completely. A modest percentage of the transplanted nuclei from all cancer cell lines and transplanted tumors were reprogrammed, and the surviving oocytes went on to develop into blastocysts. However, only blastocysts derived from the melanoma cell line yielded ESC lines, demonstrating that not all cancer genomes can be epigenetically reprogrammed to full developmental pluripotency by nuclear transplantation.31 Furthermore, when chimeras were generated using the melanoma nuclear transfer ESCs, the chimeras developed cancer earlier on with a higher penetrance and expanded tumor variety than the original nuclear donor mouse model. These studies indicate that reprogramming is indeed more difficult in primary tumor cells and that further technological development is needed to be able to generate reliable iPSC models of cancers.
Re-Creating Cancer Progression with iPSCs
Many model organisms have been generated in an attempt to recapitulate human disease phenotypes, and yet these models are at best approximations of human disease. Primary tumors resected from patients and their derivative cell lines can help us understand late-stage markers and cellular phenotypes, but they are an inadequate system to study the early stages of cancer progression. On the other hand, patient-derived iPSCs quite possibly represent the earliest stage of disease. Using patient-derived iPSCs to recapitulate the conditions of differentiation could help identify significant molecular events responsible for disease initiation and progression directly in a susceptible cell type. Also, understanding the niche in which cancers develop will be crucial for re-creating the cancer-initiating context and building a more physiologically relevant disease model. Therefore, iPSC cancer models have numerous potential practical applications, including the identification of early biomarkers to stage disease progression and better stratification of patients.
The PDAC-derived iPSC study illustrates the power of the iPSC approach. When the PDAC iPSCs were injected into immunodeficient mice, they generated pancreatic intraepithelial neoplasia (PanIN), precursors to PDAC that progressed to the invasive stage.27 Furthermore, proteomic analysis revealed that PanIN-like cells cultured as organoids secreted proteins from many genes that were known to be expressed in human pancreatic cancer progression. Further pathway analysis of many newly identified proteins showed that these proteins were part of a network centered on the transcription factor HNF4a, a gene not previously implicated in PDAC. Indeed, when a human PDAC tissue array was probed, HNF4a was specifically detected in early to intermediate-stage lesions, but not in more advanced PDAC tissues. Thus, even though only a single PDAC iPSC line was derived, this approach was able to provide not only a model of early human pancreatic cancer progression but also new insights into disease progression.
Pluripotency, reprogramming, lineage specification, and oncogenic transformation are fundamentally related processes that rely heavily on epigenetic regulation. Pluripotency-associated genes are often ectopically expressed in aggressive human tumors, suggesting that tumor formation involves a de-differentiation process. Epigenetic modifications control developmental processes, and their aberrant regulation may drive the oncogenic process in many cancers.32 For example, in transformed lung fibroblasts that spontaneously lost p16 (CDKN2A), reprogramming by OSKM was able to erase aberrant epigenetic marks associated with the repression of p16 and reactivate it in the iPSC stage.33 The derivation of iPSCs from cancer cell lines and primary tumors suggests that the cancer-specific epigenetic state may be reversible and that reprogramming processes can reset it.
A study by Ohnishi et al showed that incomplete reprogramming could lead to cancer development.34 They generated a mouse ESC line with Dox-inducible OSKM knocked into the ROSA 26 locus. Cultured embryonic fibroblasts from this mouse could be induced to become iPSCs in vitro by Dox treatment. Next, they generated chimeras using these ESCs to test whether reprogramming could also occur in vivo. Treatment of chimeric mice with Dox for 4 weeks resulted in the development of multiple teratomas. When they cultured the teratoma ex vivo, iPSC-like cells could be derived from them and these cells were able to generate chimeras. They also found that late expression of reprogramming factors by Dox administration in vivo for 3–9 days resulted in tumor development in various somatic tissues consisting of undifferentiated dysplastic cells, accompanied by global changes in DNA methylation patterns. The tumors arising in the kidney shared a number of characteristics with Wilms tumor, a common pediatric kidney cancer. Interestingly, the kidney tumor cells could be further reprogrammed to iPSCs in vitro by a 2-week treatment with Dox. When the reprogrammed cells were injected into blastocysts, they gave rise to non-neoplastic normal kidney cells in the chimeric mice, proving that they did not undergo irreversible genetic transformation.34 Their findings suggest that the same epigenetic processes associated with iPSC reprogramming may also drive the development of particular types of cancer and that these processes are bidirectionally reversible.
Li Fraumeni syndrome (LFS), a familiar form of cancer caused by mutations in the tumor suppressor p53 gene, has been modeled using patient-derived iPSCs.29 Mouse models of LFS do not fully recapitulate the human disease. Instead of site-specific cancers, LFS patients suffer from a variety of tumors of diverse cellular origins, including osteosarcoma (OS), soft tissue sarcoma, breast cancer, brain tumor, leukemia, and adrenocortical carcinoma. LFS iPSC-derived osteoblasts recapitulated hallmarks of OS, including defective osteoblastic differentiation and tumorigenicity. Remarkably, compared to wild-type osteoblasts, LFS osteoblasts did not demonstrate increased rates of cytogenetic alterations in 18 regions commonly associated with late-stage OS. LFS osteoblasts exhibited impaired upregulation of the imprinted gene H19 during osteogenesis, and restoring its expression in LFS osteoblasts improved osteoblastic differentiation and suppressed tumorigenicity. This study demonstrates the power of iPSC technology in generating a familial cancer model, which also happens to cover a wide spectrum of cancers. Thus, even though iPSCs are often thought of as complementary approaches to traditional cell line and animal models, they can also be applied as stand-alone model systems for research.
Potential Biomedical Applications
The reprogramming of human primary cancer cells to induce pluripotency is a transformative approach that has numerous potential biomedical applications. The cancer-cell-derived iPSC model is poised to become an important tool for studying human cancers originating from tissues and cell types that have a limited lifespan in tissue culture or cannot be easily obtained from live patients. Moreover, iPSCs can model tumors where the human cancer-associated genes have no clear mouse counterpart or have mutations that are too complex to engineer into the mouse genome. Human cancer-derived iPSCs can be used to preserve these unique genotypes by banking cells that can be differentiated into many cell types for later study. Generation of iPSCs from banked cord blood35 from newborns that may develop cancer later on will also offer a unique opportunity to understand the developmental and molecular mechanisms underlying the sequential progression from a precancerous to a cancerous cell.
The use of iPSCs presents both advantages and disadvantages compared to traditional approaches such as the use of cancer cell lines and animal models. First, iPSCs are species-specific and individual-specific, and thus cancer-causing mutations can be studied in the genomic context of the cancer patient. However, due to the stochastic nature of reprogramming and the resultant epigenetic variability, discerning whether the phenotype stems from individual clonal variability or from the general pathological mechanism may be difficult. Secondly, iPSCs are renewable and scalable systems that allow high-throughput screening, making them especially desirable platforms for therapeutic drug screening and toxicological studies. Their pluripotency allows them to be differentiated into diverse cell types. Although genetically quite stable, iPSCs are nonetheless cell lines and may still accumulate undesirable mutations during prolonged propagation in culture, possibly undermining their full potential for cell-based therapies. Thirdly, cancer-cell-derived iPSCs may be re-differentiated toward susceptible and resistant lineages, allowing us to study how specific oncogenic mutations impose the tumor phenotype on a particular cell lineage and/or developmental state. They may be used to study the interaction of specific oncogenic mutations with different cell types and their association with specific developmental states and cellular niches. The disadvantage is that the signaling pathways for re-creating developmental processes and fully differentiated cell types are still not well understood. Also, systemic processes cannot be fully modeled in vitro.
Cellular assays can be developed for large-throughput drug screening by converting cancer-cell-specific iPSCs into the cell types of interest. If directed differentiation can recapitulate tumor formation in vitro, drugs that can selectively eliminate the cancerous cells may be identified and also tested in a range of other cell types. Chemotherapy takes a huge toll on patients due to undesirable side effects. A differential cytotoxicity screen could develop drugs that are more specific to their targets cells. If it turns out that cancer-derived iPSCs are more resistant to differentiation compared to normal tissue-derived iPSCs, that inability to differentiate could be studied and further exploited for developing therapies that can enhance differentiation and aid tumor regression. The iPSC approach is particularly attractive for pharmacological screens. Efforts to harness the scalability and pluripotency of iPSCs have been shown for various neurological disorders36 and diabetic cardiomyopathy.37
Current genetic engineering technologies enable precise genome surgery in diseased cells. The feasibility of genetic manipulation in iPSCs has been demonstrated with several technologies, such as gene knockdown with RNA interference and gene correction using homologous recombination, combined with genome-editing tools like zinc-finger nucleases, TALENs, and the CRISPR/Cas9 systems.38 One can test the effects of genes in iPSCs by deliberately introducing mutations into an isogenic background to generate a specific genetic dysfunction or correct a particular genetic defect in the context of the patient genome. Genetically defective cells could be corrected in vitro and re-introduced into patients, which is an autologous transplantation approach that was shown to work in principle using a humanized mouse model of sickle cell anemia.39 Human iPSCs are a potential source of cells for tissue reconstruction in the long term. For example, engraftable hematopoietic precursor cells can be generated from iPSCs,40 potentially offering new cell sources for cell reconstitution for hematological cancer patients after treatment. More recently, AML patient-derived dermal fibroblasts were reprogrammed into normal iPSCs that did not carry any of the chromosomal aberrations present in the AML patients’ bone marrow cells, and could be differentiated into normal hematopoietic progenitors.41 For reconstitution of tissues lost during disease progression or treatment, allogeneic stem cell transplantation is often the only option. Use of iPSC-derived cells opens up the possibility of autologous transplantation or tissue banks that can cover a large swath of the population such as the iPS Cell Stock Project in Japan (http://www.cira.kyoto-u.ac.jp/e/research/stock.html).
Human iPSCs could also serve as a new source of hematopoietic cells for developing immunotherapies targeting cancers.42 For human iPSCs to be used in the implementation of such cell-based cancer therapies, their safety is of paramount concern. The use of nonintegrative reprogramming strategies has largely addressed concerns about the use of proto-oncogenes in reprogramming. However, transplantation of iPSCs into patients carries inherent risks due to the need for in vitro propagation and their propensity to form tumors. HSV-thymidine kinase, yeast cytosine deaminase, and inducible Caspase-9 suicide mechanisms have been engineered to eliminate potentially oncogenic or malfunctioning iPSC derivatives in vivo.43,44 Perhaps, multiple safety switches will have to be developed to make stem cell therapies safer for applications involving cell delivery in vivo.
Finally, the use of three-dimensional organoids to engineer tissues that are histologically and functionally more faithful to their in vivo counterparts will bring about major advances in how we model human disorders, perform drug screens, and engineer replacement tissues and/or organs. Methods combining directed differentiation with organoid cultures are being developed for many complex tissues such as the liver, kidney, intestine, eye, and brain, to name a few.45 The advantages of human organoid cultures are multifold: more complete recreation of the cellular milieu, better spatial organization of cell types, improved tissue function, and no ethical concerns. For these reasons, human organoids might start to replace animal testing, where a whole-organism readout is not necessary. Human liver organoids would be particularly useful for toxicological studies, because the human liver often metabolizes drugs differently than animal livers.46 Incomplete understanding of developmental processes and the difficulty in delivering nutrients and gas exchange are realistic limitations of organoid cultures, but these are mainly tissue engineering issues that can be overcome with further research. Therefore, the bioengineering of new replacement organs for tissues resected during cancer treatment is no longer such a far-fetched idea.
Conclusion
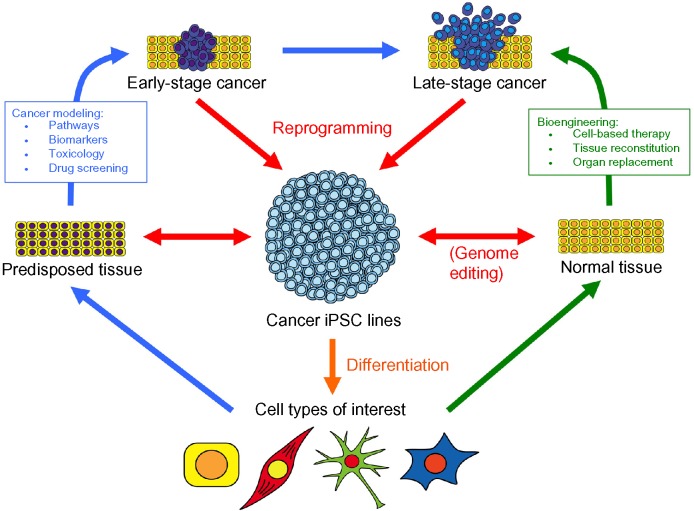
The wide-ranging use of iPSC technology is gathering momentum in the biomedical sciences. The development of patient-specific iPSCs from various somatic tissues is revolutionizing the way we approach human disease modeling, novel drug development, and autologous/allogeneic cell therapy for many disorders. In particular, cancer iPSCs offer a new paradigm in cancer modeling and tissue bioengineering (Fig. 1). Once current technological and biological challenges are fully overcome, cancer-derived iPSCs may enhance our understanding of tumorigenesis, the relationship between tumors and the surrounding tissue environment, and how epigenetic events contribute to various cancers. This knowledge will in turn enable the development of better drug screening platforms, more targeted and less toxic therapies, and effective tissue reconstitution. Efforts to harness the versatility of iPSCs for modeling human disease and for screening effective therapeutic drugs will undoubtedly accelerate the bench-to-bedside translation of new discoveries in cancer research in the future.
Figure 1.
Use of cancer-derived iPSCs in biomedical research. A variety of tissue sources may be used to develop human cancer iPSC lines (red arrows). Cancer iPSC lines can be differentiated into various cell types of interest (orange arrow) in order to investigate the features of cancer progression and drug screening (blue arrows) or develop cell-based therapies (green arrows).
Acknowledgments
I would like to thank Catherine Gillespie, Senior Editor at the Center for Cell and Gene Therapy, Baylor College of Medicine, for a critical reading of the manuscript.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
FUNDING: Author discloses no funding sources.
COMPETING INTERESTS: Author discloses no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the study: JJK. Wrote the first draft of the manuscript: JJK. Made critical revisions and approved final version: JJK. The author reviewed and approved of the final manuscript.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A. 2010;107:14152–7. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 6.Maekawa M, Yamaguchi K, Nakamura T, et al. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–9. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 7.Maekawa M, Yamanaka S. Glis1, a unique pro-reprogramming factor, may facilitate clinical applications of iPSC technology. Cell Cycle. 2011;10:3613–4. doi: 10.4161/cc.10.21.17834. [DOI] [PubMed] [Google Scholar]
- 8.Schlaeger TM, Daheron L, Brickler TR, et al. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kareta MS, Gorges LL, Hafeez S, et al. Inhibition of pluripotency networks by the rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. doi: 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krizhanovsky V, Lowe SW. Stem cells: the promises and perils of p53. Nature. 2009;460:1085–6. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao J, Marumoto T, Yamaguchi S, et al. Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Mol Ther. 2013;21:1242–50. doi: 10.1038/mt.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banito A, Gil J. Induced pluripotent stem cells and senescence: learning the biology to improve the technology. EMBO Rep. 2010;11:353–9. doi: 10.1038/embor.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banito A, Rashid ST, Acosta JC, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–9. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos-Mejia V, Fraga MF, Menendez P. iPSCs from cancer cells: challenges and opportunities. Trends Mol Med. 2012;18:245–7. doi: 10.1016/j.molmed.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–10. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi N, Ishii H, Nagai K, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2010;107:40–5. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carette JE, Pruszak J, Varadarajan M, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–42. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieu J, Zhang Z, Zhou W, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corominas-Faja B, Cufi S, Oliveras-Ferraros C, et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013;12:3109–24. doi: 10.4161/cc.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stricker SH, Feber A, Engstrom PG, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–69. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32(18):e1–21. doi: 10.1038/onc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Cheng H, Gao S, et al. Reprogramming of MLL-AF9 leukemia cells into pluripotent stem cells. Leukemia. 2014;28:1071–80. doi: 10.1038/leu.2013.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu K, Yu J, Suknuntha K, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–19. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumano K, Arai S, Hosoi M, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–42. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- 26.Gandre-Babbe S, Paluru P, Aribeana C, et al. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood. 2013;121:4925–9. doi: 10.1182/blood-2013-01-478412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Hoffman JP, Alpaugh RK, et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–99. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotini AG, Chang CJ, Boussaad I, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol. 2015;33(6):646–55. doi: 10.1038/nbt.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee DF, Su J, Kim HS, et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–54. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore JB, Loeb DM, Hong KU, et al. Epigenetic reprogramming and re-differentiation of a Ewing sarcoma cell line. Front Cell Dev Biol. 2015;3:15. doi: 10.3389/fcell.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochedlinger K, Blelloch R, Brennan C, et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–85. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Ron-Bigger S, Bar-Nur O, Isaac S, Bocker M, Lyko F, Eden A. Aberrant epigenetic silencing of tumor suppressor genes is reversed by direct reprogramming. Stem Cells. 2010;28:1349–54. doi: 10.1002/stem.468. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi K, Semi K, Yamamoto T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–77. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Broxmeyer HE. Will iPS cells enhance therapeutic applicability of cord blood cells and banking? Cell Stem Cell. 2010;6:21–24. doi: 10.1016/j.stem.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Xu XH, Zhong Z. Disease modeling and drug screening for neurological diseases using human induced pluripotent stem cells. Acta Pharmacol Sin. 2013;34:755–64. doi: 10.1038/aps.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drawnel FM, Boccardo S, Prummer M, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014;9:810–21. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 38.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna J, Wernig M, Markoulaki S, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–3. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 40.Doulatov S, Vo LT, Chou SS, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–70. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salci KR, Lee JH, Laronde S, et al. Cellular reprogramming allows generation of autologous hematopoietic progenitors from AML patients that are devoid of patient-specific genomic aberrations. Stem Cells. 2015;33(6):1839–49. doi: 10.1002/stem.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachamitr P, Hackett S, Fairchild PJ. Induced pluripotent stem cells: challenges and opportunities for cancer immunotherapy. Front Immunol. 2014;5:176. doi: 10.3389/fimmu.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Hong SG, Winkler T, et al. Development of an inducible caspase-9 safety switch for pluripotent stem cell—based therapies. Mol Ther Methods Clin Dev. 2014;1:14053. doi: 10.1038/mtm.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B, Watts KL, Gori JL, et al. Safeguarding nonhuman primate iPS cells with suicide genes. Mol Ther. 2011;19:1667–75. doi: 10.1038/mt.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 46.Graham MJ, Lake BG. Induction of drug metabolism: species differences and toxicological relevance. Toxicology. 2008;254:184–91. doi: 10.1016/j.tox.2008.09.002. [DOI] [PubMed] [Google Scholar]