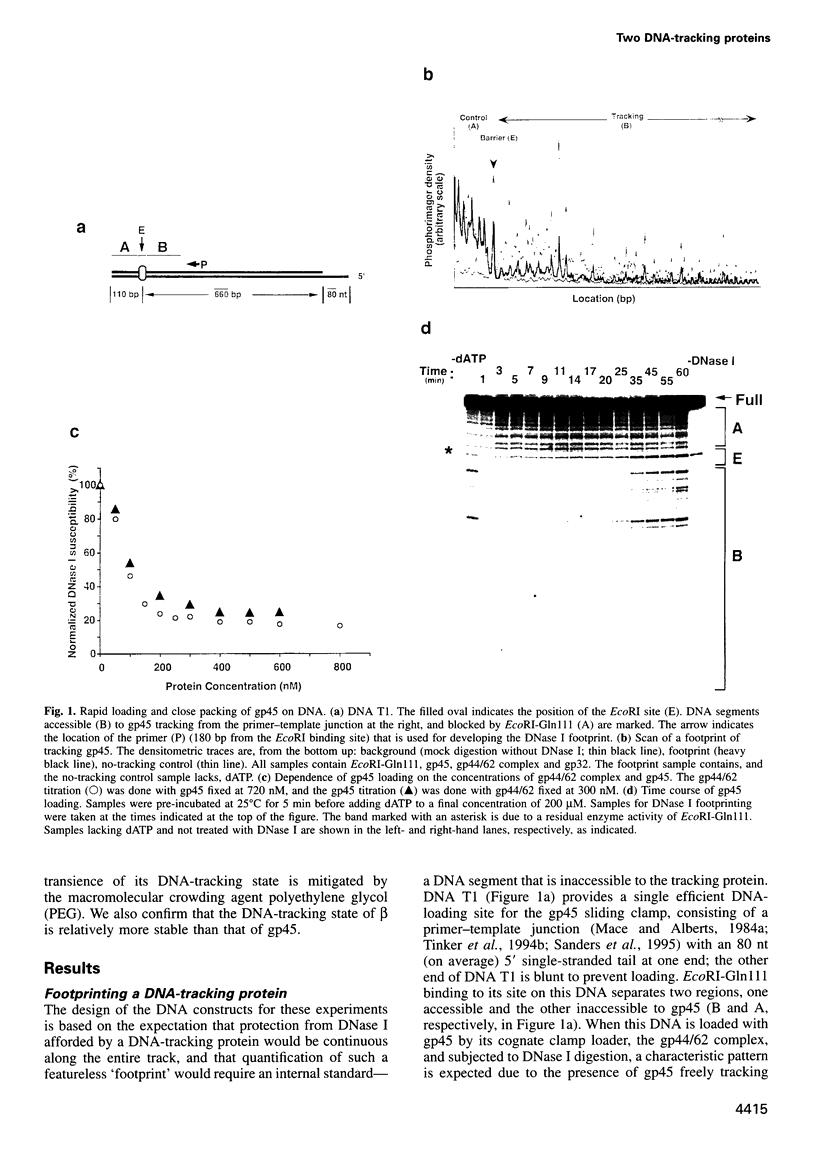
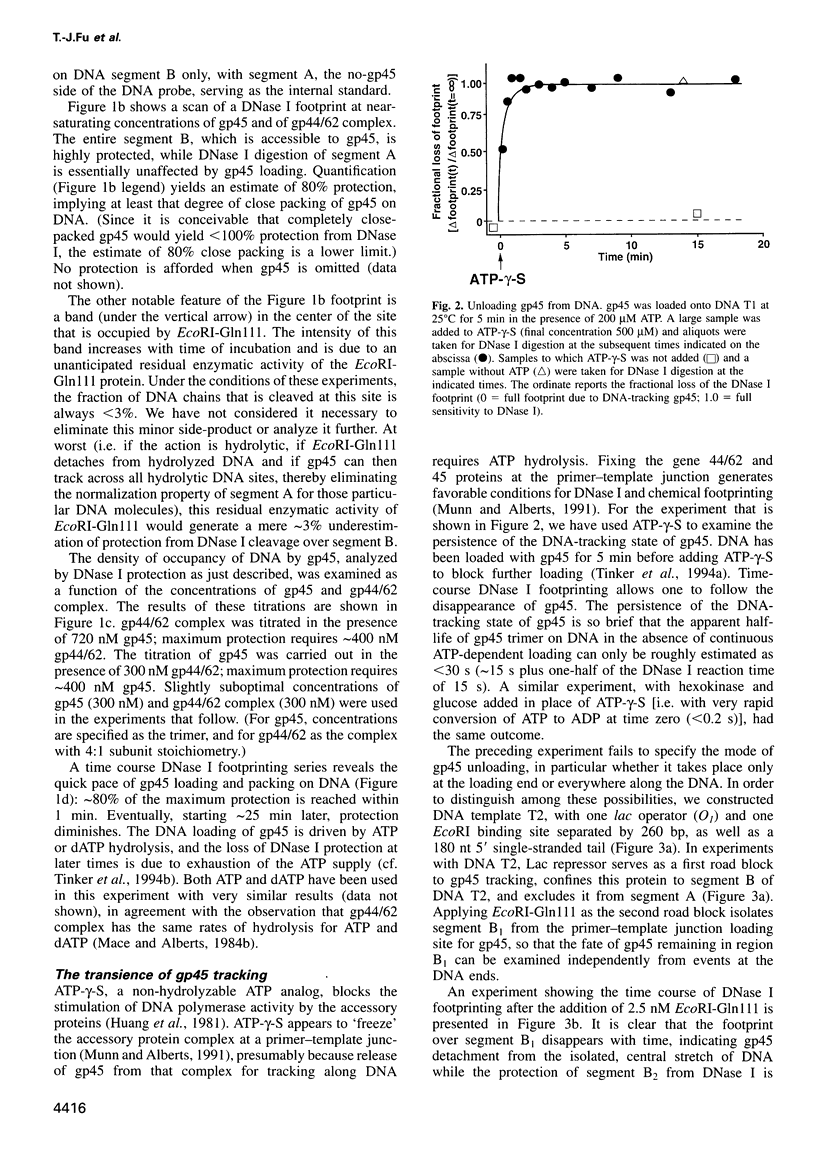
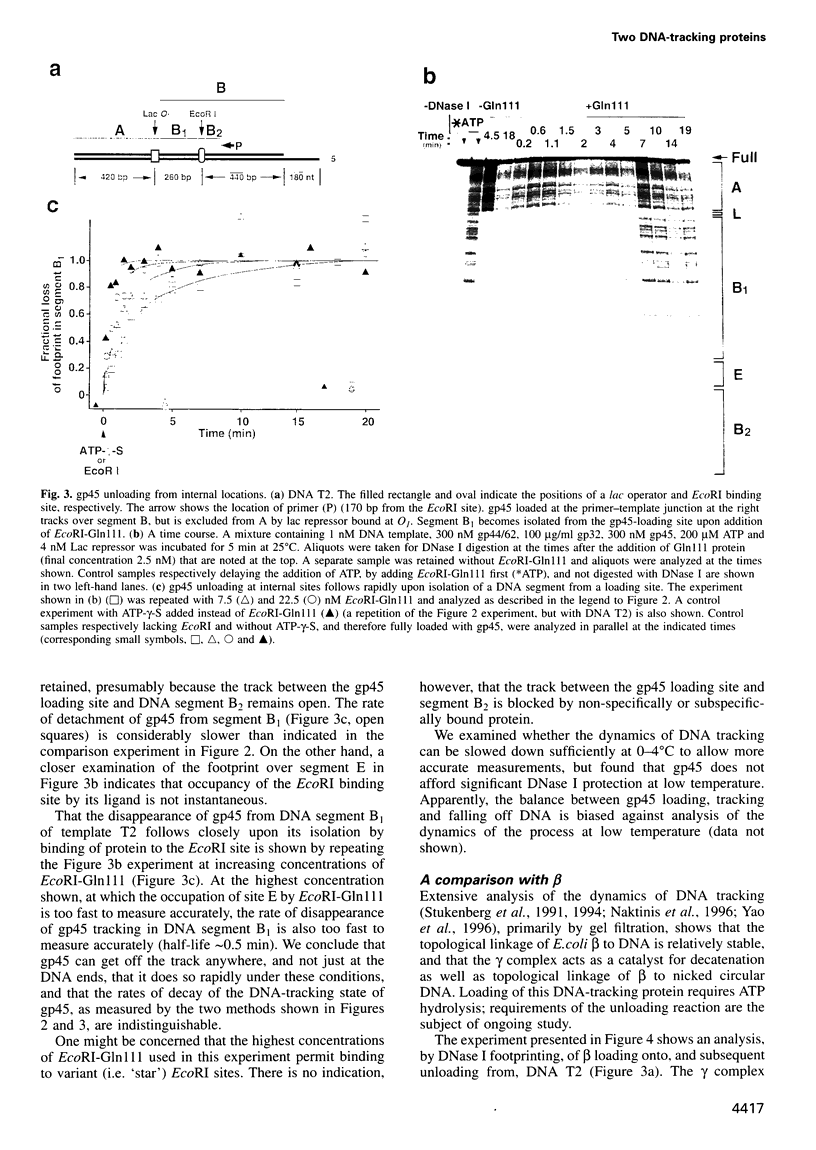
Abstract
Bacteriophage T4 gene 45 protein (gp45) and Escherichia coli beta are DNA-tracking sliding-clamp proteins that increase processivity by tethering their conjugate DNA polymerases to DNA. gp45 also activates T4 late transcription. DNA loading of gp45 and beta requires ATP or dATP hydrolysis; efficient loading at primer-template junctions is assisted by single-stranded DNA-binding proteins. The kinetics of gp45 loading and tracking have been examined by DNase I footprinting of linear DNA with one blunt end, one primer-template junction, and binding sites for proteins that block gp45 tracking. DNA loading of gp45 can also be interrupted by adding the non-hydrolyzable ATP analog ATP-gamma-S. At saturation, DNA is very closely packed with gp45 or beta. When gp45 loading is interrupted, or when a segment of the track is blocked off, the gp45 footprint dissipates within seconds, but the DNA-tracking state of beta is much more stable. The stability of the tracking state of gp45 is, however, increased by the macromolecular crowding agent polyethylene glycol. We suggest that labile gp45 catenation directly generates the coupling of late transcription to DNA replication during bacteriophage T4 multiplication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M. Prokaryotic DNA replication mechanisms. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):395–420. doi: 10.1098/rstb.1987.0068. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Yoder B. L. ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J Biol Chem. 1993 Sep 25;268(27):19923–19926. [PubMed] [Google Scholar]
- Cayley S., Lewis B. A., Guttman H. J., Record M. T., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991 Nov 20;222(2):281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- Elliott T., Geiduschek E. P. Defining a bacteriophage T4 late promoter: absence of a "-35" region. Cell. 1984 Jan;36(1):211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- Gogol E. P., Young M. C., Kubasek W. L., Jarvis T. C., von Hippel P. H. Cryoelectron microscopic visualization of functional subassemblies of the bacteriophage T4 DNA replication complex. J Mol Biol. 1992 Mar 20;224(2):395–412. doi: 10.1016/0022-2836(92)91003-8. [DOI] [PubMed] [Google Scholar]
- Hacker K. J., Alberts B. M. The rapid dissociation of the T4 DNA polymerase holoenzyme when stopped by a DNA hairpin helix. A model for polymerase release following the termination of each Okazaki fragment. J Biol Chem. 1994 Sep 30;269(39):24221–24228. [PubMed] [Google Scholar]
- Hacker K. J., Alberts B. M. The slow dissociation of the T4 DNA polymerase holoenzyme when stalled by nucleotide omission. An indication of a highly processive enzyme. J Biol Chem. 1994 Sep 30;269(39):24209–24220. [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Geiduschek E. P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992 May 29;256(5061):1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Williams K. P., Kassavetis G. A., Geiduschek E. P. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990 May 4;248(4955):573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E., Alberts B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981 Apr 25;256(8):4087–4094. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., Hockensmith J. W., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12717–12729. [PubMed] [Google Scholar]
- Jarvis T. C., Paul L. S., von Hippel P. H. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem. 1989 Jul 25;264(21):12709–12716. [PubMed] [Google Scholar]
- Jarvis T. C., Ring D. M., Daube S. S., von Hippel P. H. "Macromolecular crowding": thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem. 1990 Sep 5;265(25):15160–15167. [PubMed] [Google Scholar]
- Johanson K. O., McHenry C. S. Purification and characterization of the beta subunit of the DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem. 1980 Nov 25;255(22):10984–10990. [PubMed] [Google Scholar]
- Kaboord B. F., Benkovic S. J. Accessory proteins function as matchmakers in the assembly of the T4 DNA polymerase holoenzyme. Curr Biol. 1995 Feb 1;5(2):149–157. doi: 10.1016/s0960-9822(95)00036-4. [DOI] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- Kelman Z., O'Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995 Sep 25;23(18):3613–3620. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X. P., Onrust R., O'Donnell M., Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992 May 1;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Krishna T. S., Kong X. P., Gary S., Burgers P. M., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994 Dec 30;79(7):1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993 Dec 20;234(4):915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- Mace D. C., Alberts B. M. Characterization of the stimulatory effect of T4 gene 45 protein and the gene 44/62 protein complex on DNA synthesis by T4 DNA polymerase. J Mol Biol. 1984 Aug 5;177(2):313–327. doi: 10.1016/0022-2836(84)90459-5. [DOI] [PubMed] [Google Scholar]
- Mace D. C., Alberts B. M. T4 DNA polymerase. Rates and processivity on single-stranded DNA templates. J Mol Biol. 1984 Aug 5;177(2):295–311. doi: 10.1016/0022-2836(84)90458-3. [DOI] [PubMed] [Google Scholar]
- Munn M. M., Alberts B. M. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem. 1991 Oct 25;266(30):20034–20044. [PubMed] [Google Scholar]
- Naktinis V., Turner J., O'Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996 Jan 12;84(1):137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- Onrust R., Finkelstein J., Naktinis V., Turner J., Fang L., O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J Biol Chem. 1995 Jun 2;270(22):13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., Daube S. S., Jarvis T. C., von Hippel P. H. Using macromolecular crowding agents to identify weak interactions within DNA replication complexes. Methods Enzymol. 1995;262:466–476. doi: 10.1016/0076-6879(95)62038-9. [DOI] [PubMed] [Google Scholar]
- Reddy M. K., Weitzel S. E., von Hippel P. H. Assembly of a functional replication complex without ATP hydrolysis: a direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders G. M., Kassavetis G. A., Geiduschek E. P. Rules governing the efficiency and polarity of loading a tracking clamp protein onto DNA: determinants of enhancement in bacteriophage T4 late transcription. EMBO J. 1995 Aug 15;14(16):3966–3976. doi: 10.1002/j.1460-2075.1995.tb00068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders G. M., Kassavetis G. A., Geiduschek E. P. Use of a macromolecular crowding agent to dissect interactions and define functions in transcriptional activation by a DNA-tracking protein: bacteriophage T4 gene 45 protein and late transcription. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7703–7707. doi: 10.1073/pnas.91.16.7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Smart machines at the DNA replication fork. Cell. 1994 Sep 9;78(5):725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Stukenberg P. T., Studwell-Vaughan P. S., O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991 Jun 15;266(17):11328–11334. [PubMed] [Google Scholar]
- Stukenberg P. T., Turner J., O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994 Sep 9;78(5):877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- Tinker R. L., Kassavetis G. A., Geiduschek E. P. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J. 1994 Nov 15;13(22):5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker R. L., Williams K. P., Kassavetis G. A., Geiduschek E. P. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994 Apr 22;77(2):225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11816–11821. [PubMed] [Google Scholar]
- Yao N., Turner J., Kelman Z., Stukenberg P. T., Dean F., Shechter D., Pan Z. Q., Hurwitz J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996 Jan;1(1):101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Minton A. P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]