Abstract
The enzyme dihydrofolate reductase (DHFR, E) from Escherichia coli is a paradigm for the role of protein dynamics in enzyme catalysis. Previous studies have shown that the enzyme progresses through the kinetic cycle by modulating the dynamic conformational landscape in the presence of substrate dihydrofolate (DHF), product tetrahydrofolate (THF), and cofactor (NADPH or NADP+). This study focuses on the quantitative description of the relationship between protein fluctuations and product release, the rate-limiting step of DHFR catalysis. NMR relaxation dispersion measurements of millisecond time scale motions for the E:THF:NADP+ and E:THF:NADPH complexes of wild-type and the Leu28Phe (L28F) point mutant reveal conformational exchange between an occluded ground state and a low population of a closed state. The backbone structures of the occluded ground states of the wild-type and mutant proteins are very similar, but the rates of exchange with the closed excited states are very different. Integrated analysis of relaxation dispersion data and THF dissociation rates measured by stopped-flow spectroscopy shows that product release can occur by two pathways. The intrinsic pathway consists of spontaneous product dissociation and occurs for all THF-bound complexes of DHFR. The allosteric pathway features cofactor-assisted product release from the closed excited state and is utilized only in the E:THF:NADPH complexes. The L28F mutation alters the partitioning between the pathways and results in increased flux through the intrinsic pathway relative to the wild-type enzyme. This repartitioning could represent a general mechanism to explain changes in product release rates in other E. coli DHFR mutants.
Introduction
An enzyme enhances the rate of a chemical reaction compared to the uncatalyzed reaction in solution.1 However, the rate-limiting step does not necessarily correspond to the actual chemical step since, in contrast to uncatalyzed reactions in solution, enzyme catalysis requires substrate binding and product unbinding steps, which commonly determine the overall catalytic rate.2 There is much evidence that protein dynamics play an important role in ligand binding and unbinding events.3−9 Nuclear magnetic resonance (NMR) experiments are crucial for dissecting protein dynamics on different time scales.10 Fast picosecond–nanosecond time scale dynamics reflect rapid angular fluctuations of the backbone and side chains, while loop conformational changes usually occur on a slower microsecond–millisecond time scale.11 Although prior research suggests an important role for dynamics in catalysis, a quantitative description remains elusive.
The enzyme dihydrofolate reductase (DHFR) from Escherichia coli has been widely used as a model system to investigate the role of protein dynamics in enzyme catalysis.5,12−20 DHFR catalyzes the nicotinamide adenine dinucleotide phosphate (NADPH, cofactor)-dependent reduction of dihydrofolate (DHF, substrate) to tetrahydrofolate (THF, product), with concurrent oxidation of the cofactor to form NADP+. Under physiological substrate and cofactor concentrations, DHFR catalysis progresses through a preferred catalytic cycle of five different intermediates (Figure 1A).21
Figure 1.
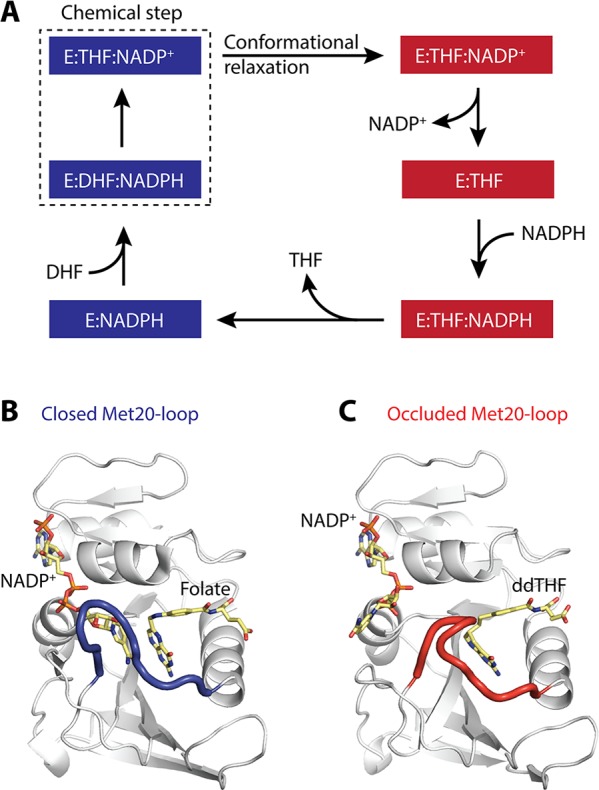
(A) E. coli DHFR kinetic cycle adapted from Fierke et al.21 Closed states of the enzyme are highlighted in blue, occluded states in red. An extra step (within the hatched box) has been added between the Michaelis complex (E:DHF:NADPH) and the first product complex (E:THF:NADP+) to represent relaxation of the closed excited state of the product ternary complex, formed immediately following hydride transfer, into its occluded ground state conformation.61 (B) Closed conformation of the Met20 loop (blue) with bound ligands, the nicotinamide moiety occupies the active site. (C) Occluded conformation of the Met20 loop (red) with bound ligands, the nicotinamide moiety hangs outside of the active site.
Extensive X-ray and NMR characterization identified structural differences between these intermediates in the active site region.22,23 In particular, the active site loop (Met20 loop) takes up either a closed (Figure 1B) or an occluded conformation (Figure 1C) during catalysis. Boehr et al. characterized the microsecond–millisecond time scale motions for each of these five intermediates using 15N NMR relaxation dispersion experiments.5 It was found that each intermediate samples a small population of a minor state that resembles the next and/or previous intermediate in the catalytic cycle and, further, that the exchange time scales are similar to the corresponding ligand exchange kinetics.5,21
In order to establish the mechanistic relationship between protein dynamics and product dissociation kinetics, we chose to study a mutant protein in which Leu 28 is changed to Phe; this mutation was chosen because of its large effect on the product dissociation rate (10–20-fold increase compared to wild-type product dissociation rates).24 Comprehensive NMR relaxation dispersion data sets were acquired with both 15N and 1H probes for the ternary product complexes, E:THF:NADP+ and E:THF:NADPH, of wild-type (WT) and L28F mutant DHFR, incorporating non-uniform sampling (NUS) and scan-interleaving in constant-time CPMG pulse sequences.25−28
Quantitative analysis of the relaxation dispersion data, together with stopped-flow kinetic data measured under NMR conditions, provides new mechanistic insights and shows that the THF product is released via two parallel pathways, an intrinsic pathway and an allosteric pathway. Product release from WT DHFR occurs primarily by the allosteric pathway, whereas the intrinsic pathway is dominant in L28F DHFR. Changes in partitioning between the two pathways may be a common response of E. coli DHFR to mutations. Our results show that new insights into the role of dynamics and mechanism of ligand exchange in proteins can be obtained through a combined approach of NMR and stopped-flow spectroscopy.
Methods
General Procedures
β-Nicotinamide adenine dinucleotide phosphate reduced tetrasodium salt hydrate (NADPH), β-nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP+), d-glucose-6-phosphate solution, glucose-6-phosphate dehydrogenase from Leuconostoc mesenteroides, methotrexate, folic acid and (6R)-5,10-dideazatetrahydrofolate (ddTHF, also known as lometrexol hydrate) were purchased from Sigma-Aldrich. (6S)-5,6,7,8-tetrahydrofolic acid (THF) was obtained from Schircks Laboratories. The L28F DHFR was generated by site-directed mutagenesis using the QuikChange Multi kit (Agilent) as described elsewhere.29 Plasmid construction, protein expression, and purification of WT and L28F DHFRs were performed as described previously.29,30 All experiments, including kinetic measurements, were performed in NMR buffer (70 mM KPi, 25 mM KCl, 1 mM DTT, pH 7.6) unless otherwise specified.
Enzyme Kinetics
The THF dissociation rate constants for the binary and ternary complexes were measured at 300 K on an Applied Photophysics stopped-flow spectrophotometer using the competition method.21,31 Premixed saturated E:THF:NADP(H) or E:THF complex ([DHFR] = 4 μM, [THF] = 32 μM, [NADP(H)] = 160 μM) was combined with a large excess of methotrexate (1 mM). When the concentration of the competing ligand (MTX) is large enough, the observed rate constant for this reaction is equal to the THF dissociation rate constant. For each experiment, 50 independent runs were averaged and fitted to a general equation for single exponential decay. Errors were estimated using the bootstrap method.32
X-ray Crystallography
The WT E:ddTHF:NADP+ and L28F E:ddTHF:NADP+ complexes were crystallized from solutions containing 1 mM (WT or L28F) DHFR, 3 mM ddTHF, 3 mM NADP+, and 10 mM imidazole (pH7). Crystallization trials were set up using the Rigaku Crystalmation robot at the Joint Center for Structural Genomics (JCSG). Crystals for data collection were grown by sitting drop vapor diffusion using a well solution containing 0.1 M tris(hydroxymethyl) aminomethane pH7.0, 20% w/v PEG3350, and 0.2 M calcium acetate. Crystals were grown at 277 K and were observed within 3 days. The WT E:ddTHF:NADP+ and L28F E:ddTHF:NADP+ crystals were cryoprotected by soaking in a well solution supplemented with 30% PEG 400 or 30% ethylene glycol, respectively.
Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline BL11–1. Data collection and processing statistics are detailed in Table S1. Data sets were indexed, integrated, and scaled using the HKL-2000 package.33 The structures were solved by molecular replacement using PHASER34 with a previously published DHFR structure (PDB code 1RX6) as a search model and further refined using phenix.refine(35) combined with manual building cycles in Coot.36 Cα main-chain coordinate RMSDs between the WT and L28F crystal structures were calculated using Superpose from the CCP4 suite of programs.37,38
NMR Spectroscopy
Isotopically labeled DHFR was overexpressed and purified as described previously.30 Samples for relaxation dispersion CPMG NMR experiments contained 1 mM 15N, 2H-labeled WT, or L28F (∼80% deuteration), 18 mM THF and/or 10 mM NADP(H), 5 mM ascorbic acid, 1 mM dithiothreitol (DTT), 25 mM KCl, and 10% D2O in 70 mM KPi pH 7.6. For backbone assignments, 13C, 15N-labeled proteins were used instead. DHFR ligands are extremely sensitive to oxygen and/or light. When NADPH was present, a recycling system consisting of 10 mM glucose-6-phosphate and 20 units/mL glucose-6-phosphate dehydrogenase was added to regenerate NADPH from oxidized cofactor. Buffers were thoroughly degassed through freeze–pump–thaw cycles prior to addition of ascorbic acid as oxygen scavenger. All samples were prepared under argon atmosphere in a glovebox, placed into amber NMR tubes, and flame-sealed to prevent reoxygenation of the buffer.
NMR spectra were acquired on Bruker Avance spectrometers operating at 500, 750, or 800 MHz. Backbone resonance assignments for the L28F E:THF, L28F E:THF:NADP+, and L28F E:FOL:NADP+ complexes were made at 300 K using standard triple resonance experiments at 750 or 800 MHz.39−41 Assignments for L28F E:NADPH were made at 283 K due to lower sample stability for this complex and were transferred to an HSQC spectrum measured at 300 K for the calculation of equilibrium chemical shift differences Δδ. The triple resonance data were processed using NMRpipe42 and analyzed using CCPN.43 The HSQC spectrum of the L28F E:THF:NADPH complex is virtually identical to that of L28F E:THF:NADP+, apart from small shifts in the cross peaks of residues 96–98, and assignments were therefore transferred from L28F E:THF:NADP+. Relaxation compensated, constant time Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion NMR experiments44,45 for amide 15N and 1H were performed for the L28F E:THF, L28F E:THF:NADP+, L28F E:THF:NADPH, WT E:THF:NADP+, and WT E:THF:NADPH complexes at 500 and 800 MHz using 60% NUS in the indirect dimension and 16 scans per sampling point. For the 1H dispersion experiments, we placed the CPMG period after the t1 evolution and replaced the nonselective 1H 180° pulse in the rcINEPT period by a REBURP pulse that selectively refocuses amide proton magnetization. Partially (∼80%) deuterated protein was used to remove artifacts from 3J(HN–Hα) couplings.45 The total relaxation time for all CPMG experiments was 40 ms. Relaxation dispersion data were processed and reconstructed using MDDNMR,26−28 NMRpipe,42 and FuDA (http://pound.med.utoronto.ca) and were fitted to the Bloch–McConnell equations11 for two-site exchange using the program GLOVE.46 Errors were set to 2% and 4% for the 800 and 500 MHz data points, respectively, unless the estimated error based on three repeat points was larger.
Global exchange rates and minor state populations were determined by simultaneously fitting a subset of the 15N and 1H dispersion curves [for residues 14, 23, 26, 29, 30, 32, 33, 57, 59, 116, 117, 120, 121, 124 (excluded for the WT E:THF:NADP+ data), 148 and 149] that were well-defined and could be fitted to a two-site exchange model. For each of these residues, the χ2 value for the global fit was <2 times larger than for the individual fit. Next, all remaining dispersion curves were force fitted using the rates and populations for each complex determined from the global fits. It was clear from this procedure that the dispersion data for several residues reflect additional exchange processes and cannot be fit by a two-site exchange model; these residues were excluded from further analysis. Uncertainties in the parameters were estimated using Monte Carlo simulations. Signs for Δω were determined by comparing the 1H–15N HSQC and HMQC spectra.47
Results
Product Dissociation Rates Under NMR Conditions
Previous kinetic measurements on WT and L28F DHFR were performed at 298 K in MTEN buffer (50 mM 2-(N-morpholino)-ethanesulfonic acid, 25 mM tris(hydroxymethyl)-aminomethane, 25 mM ethanolamine, and 100 mM sodium chloride).21,24 In order to minimize differences between NMR and kinetics measurements, the THF dissociation rate constants for the WT and L28F E:THF:NADPH, E:THF:NADP+, and E:THF complexes were measured by stopped-flow spectroscopy using 15N, 2H-labeled protein in the same buffer and at the same temperature (300 K) as relaxation dispersion CPMG NMR experiments (Table 1). Under these conditions, the THF dissociation rate constants for the WT E:THF:NADP+ and WT E:THF complexes are the same within experimental error, while the dissociation rate constant for the WT E:THF:NADPH complex is increased ∼2.5-fold. The THF dissociation rate constants for the L28F E:THF:NADP+ and L28F E:THF complexes are also identical at 36 s–1, while the dissociation rate constant for the L28F E:THF:NADPH complex is increased 1.3-fold. The THF dissociation rate constants for the E:THF:NADP+ and E:THF complexes are faster by a factor of 10 in L28F, while the THF dissociation rate constant for the E:THF:NADPH complex is increased 5-fold.
Table 1. Dissociation Rates of THF from WT and L28F Enzyme (E) Complexesa.
| enzyme species | koff (s–1), pH 7.6, 300 K | koff (s–1), pH 6.0, 298 K | koff (s–1), pH 9.0, 298 K |
|---|---|---|---|
| WT E:THF:NADPH | 9.0 ± 0.3 | 12 ± 2b | 18 ± 2b |
| WT E:THF:NADP+ | 3.8 ± 0.2 | 2.4 ± 0.2b | 5.7 ± 0.7b |
| WT E:THF | 3.7 ± 0.3 | 1.4 ± 0.2b | 2.4 ± 0.3b |
| L28F E:THF:NADPH | 48 ± 2 | 80 ± 5c | NA |
| L28F E:THF:NADP+ | 36 ± 1 | 34 ± 3c | NA |
| L28F E:THF | 36 ± 2 | 40 ± 2c | NA |
Structures of WT and L28F E:THF:NADP+
X-ray crystal structures of WT ecDHFR and L28F ecDHFR in complex with NADP+ and ddTHF (a THF analog with greater long-term stability) were determined in order to establish whether protein structural differences can account for the increased product release rates that occur with the L28F single point mutation. Both WT and mutant enzyme complexes crystallized under identical buffer conditions in space group P212121. The new structure of the WT E:ddTHF:NADP+ complex is at 1.5 Å resolution and is similar to the 1.9 Å resolution structure of the same complex reported previously (PDB accession code 1RX4).22 The structure of the L28F E:ddTHF:NADP+ complex was determined at 1.2 Å resolution. The backbone conformations of the WT and L28F structures (Figure 2A) and the structure of the bound NADP+ (Figure 2B) are very similar, with an average RMSD of 0.29 Å at the Cα atoms (Figure S1). The Met20 loop adopts the occluded conformation in both structures and sterically prevents the nicotinamide moiety of the oxidized cofactor from binding in the active site. The ribosyl-nicotinamide moiety projects into the solvent, where it packs against residues 134–138 in a neighboring molecule in the crystal lattice. In the L28F complex, the carboxamide occupies two sites, each of ∼50% occupancy, which are related by a 180° rotation about the axis of the nicotinamide ring. The two conformers are stabilized through alternate intermolecular packing interactions in the crystal lattice. There are no steric barriers to nicotinamide ring rotation in the DHFR monomer, and any conformational averaging in solution would therefore occur on a much faster time scale than that of CPMG dispersion.
Figure 2.
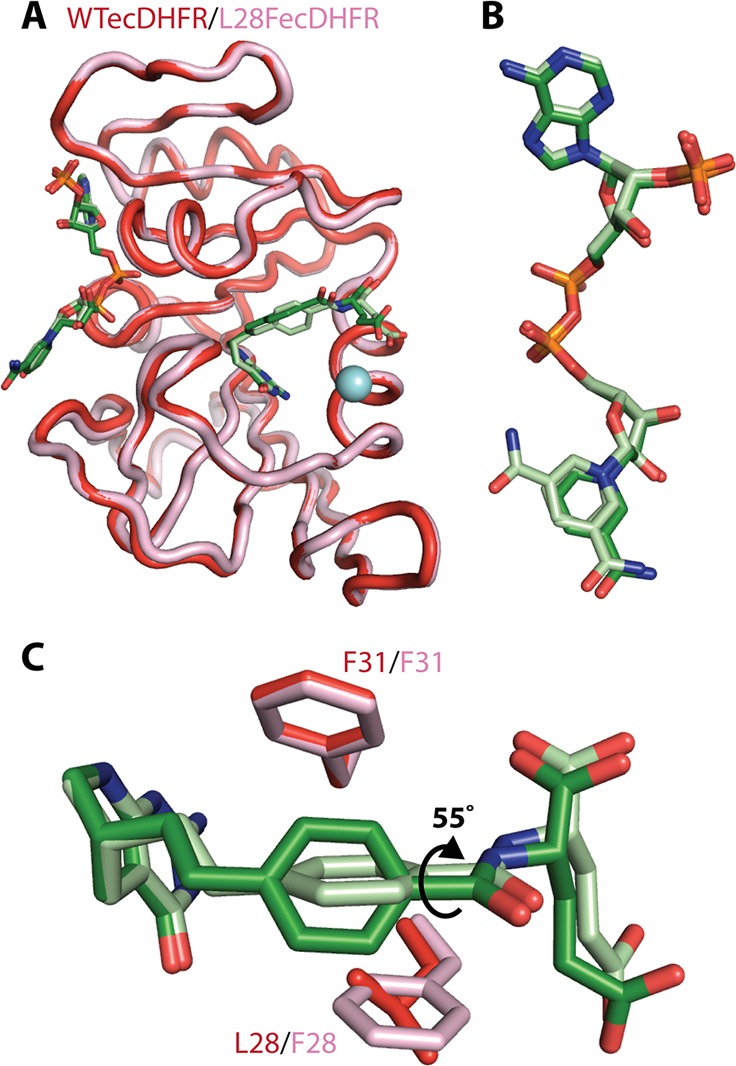
(A) Overlay of the WT E:ddTHF:NADP+ (red) and L28F E:ddTHF:NADP+ (pink) crystal structures. The backbone is shown as a cartoon and the ddTHF and NADP+ ligands as sticks. The site of the L28F mutation is shown as a blue sphere. (B) Conformation of NADP+ in WT E:ddTHF:NADP+ (dark green) and L28F E:ddTHF:NADP+(pale green) complexes. In both structures, the nicotinamide ring projects into the solvent and packs against a neighboring molecule in the crystal lattice. The carboxamide moiety is found in two orientations (related by 180° flips about the axis of the nicotinamide ring) in the L28F E:ddTHF:NADP+ structure. (C) The benzene ring of the of the para-ethyl benzoyl glutamic acid tail (PEBA) of ddTHF is rotated 55° around its C1′-C4′ axis in L28F E:ddTHF:NADP+ (pale green, pink) relative to its orientation in the WT complex (dark colors), altering its contacts with F31 and aliphatic side chains lining the binding pocket.
Only one of these conformers is observed in the WT E:ddTHF:NADP+ crystal structure. As might be expected from its close proximity to the site of mutation, the conformation of the product analogue ddTHF, bound within the active site, differs between the WT and L28F E:ddTHF:NADP+ structures. The benzene ring of the benzoyl glutamic acid tail of ddTHF is rotated 55° around its C1′-C4′ axis in the L28F E:ddTHF:NADP+ crystal structure (light colors in Figure 2C) compared to the WT E:ddTHF:NADP+ crystal structure (dark colors in Figure 2C). This reorientation of the benzoyl ring establishes staggered coplanar packing with the aromatic side chain of F28, while tilting it away from the edge-to-face contact made with the ring of F31 in the WT protein. The benzoyl ring also rotates away from the side chains of I50, L54, and I94, creating a substantial cavity in the binding pocket of the L28F mutant complex.
Relaxation Dispersion Experiments
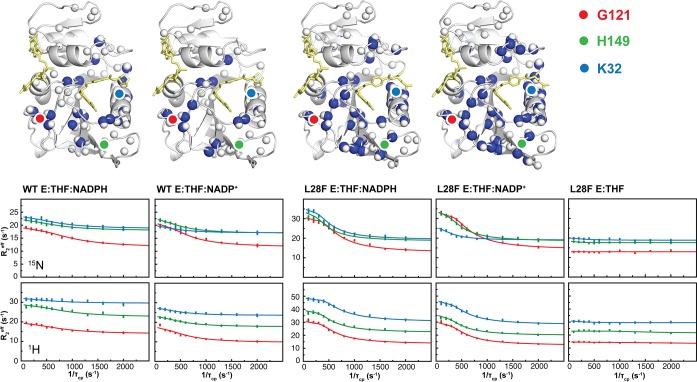
In order to determine the effect of the L28F mutation on the μs–ms time scale dynamics of DHFR, we measured amide 15N and 1H R2 relaxation dispersion for the WT E:THF:NADPH, WT E:THF:NADP+, L28F E:THF, L28F E:THF:NADPH, and L28F E:THF:NADP+ complexes using relaxation-compensated, constant time CPMG experiments.44,45 CPMG relaxation dispersion experiments monitor exchange on a μs–ms time scale between the ground state and one or more higher-energy conformational substates. The effective R2 relaxation rates measured as a function of the CPMG pulsing interval were fitted to exchange models using the Bloch-McConnell equations.11 For two-site exchange between a major state (A) and a minor state (B), fitting yields an exchange rate constant (kex = ka + kb), populations (pa, pb), and chemical shift differences between the ground and excited states (Δω = ωa – ωb). The majority of the dispersion curves for the ternary WT E:THF:NADP+, WT E:THF:NADPH, L28F E:THF:NADP+, and L28F E:THF:NADPH complexes could be fitted to a global two-site exchange process (Figure 3, Table 2, and Figures S2–S5).
Figure 3.
R2 relaxation dispersion data for the WT E:THF:NADPH, WT E:THF:NADP+, L28F E:THF:NADPH, L28F E:THF:NADP+, and L28F E:THF complexes. Amide 1H and 15N probes that undergo two-site exchange are represented as white (1H) and blue (15N) spheres on the structures of each of the various complexes. The location of G121 (red) and H149 (green), which are markers of the closed to occluded transition, and K32 (blue) in the substrate/product binding pocket are indicated on the structures. Representative dispersion profiles, measured at 800 MHz, are shown for the amide 1H and 15N probes of K32 (blue), G121 (red), and H149 (green) for each of the complexes.
Table 2. Fitted Two-Site Exchange Parameters.
| kex (s–1) | pb (%) | ka (s–1) | kb (s–1) | |
|---|---|---|---|---|
| WT E:THF:NADPH | 1890 ± 80 | 0.69 ± 0.02 | 1880 ± 80 | 13.0 ± 0.3 |
| WT E:THF:NADP+ | 1420 ± 70 | 1.25 ± 0.07 | 1400 ± 70 | 17.7 ± 0.4 |
| L28F E:THF:NADPH | 770 ± 20 | 2.75 ± 0.04 | 740 ± 20 | 21.0 ± 0.3 |
| L28F E:THF:NADP+ | 910 ± 20 | 2.67 ± 0.04 | 890 ± 20 | 24.3 ± 0.3 |
However, the dispersion profiles for several residues could not be fitted to a two-state process; these residues experience multistate exchange, the description of which is beyond the scope of the present work. The 15N and 1H amide probes undergoing two-site exchange are located in the active site loops and the product-binding site (blue and white spheres, respectively, in Figure 3). In contrast, the backbone 15N and 1H probes located in the product-binding site and active site loops of the binary L28F E:THF complex do not exhibit relaxation dispersion (Figure 3, right).
To gain insights into the structural features of the minor states of the various complexes, we determined the dynamic chemical shift differences Δω between the major and minor state for each probe. Fitting of the dispersion curves with the Bloch–McConnell equations yields only absolute values for ΔωN and ΔωH. The sign of ΔωN for the WT and L28F E:THF:NADPH and E:THF:NADP+ complexes was determined by comparing their 1H–15N HSQC and HMQC spectra.47 Reliable results could only be obtained for a small fraction of residues, especially for the WT complexes (Tables S2, S4, S6, and S8). The dynamic chemical shift differences ΔωN for a subset of residues that have been identified as markers for the occluded-to-closed transition48 (N23, L24, I94, G95, I115, D116, E118, G121, H124, and H149, identified by labels or orange data points in Figures 4 and 5) and markers for binding of the nicotinamide ring in the active site (A7 and G15) correlate well with the equilibrium chemical shift differences (ΔδN) measured between the occluded E:THF:NADPH or E:THF:NADP+ complexes and the closed E:NADPH and E:FOL:NADP+ complexes, respectively. For the L28F E:THF:NADPH complex, signs of ΔωN were available for 3 out of 10 marker residues and absolute values of the chemical shifts were used for the other 7 residues.
Figure 4.
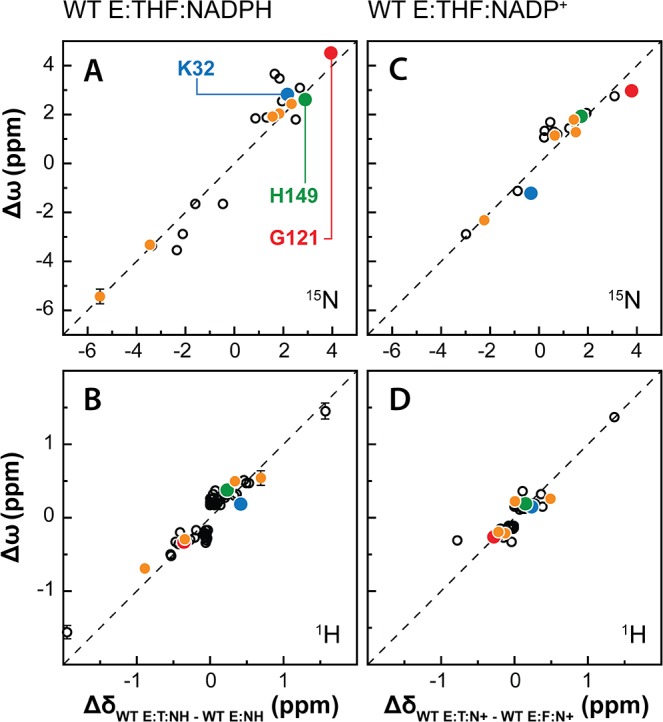
Correlation between the dynamic chemical shift differences (Δω) and equilibrium chemical shift differences (Δδ) for WT DHFR complexes. (A) Plot of ΔωN for E:THF:NADPH versus ΔδN, the difference in 15N chemical shift between the WT E:THF:NADPH and E:NADPH complexes. (B) Plot of ΔωH for E:THF:NADPH versus ΔδH, the difference in 1H chemical shift between the WT E:THF:NADPH and E:NADPH complexes. (C) Plot of ΔωN for E:THF:NADP+ versus ΔδN, the difference in 15N chemical shift between the WT E:THF:NADPH and E:FOL:NADP+ complexes. (D) Plot of ΔωH for E:THF:NADP+ versus ΔδH, the difference in 1H chemical shift between the WT E:THF:NADPH and E:FOL:NADP+ complexes. For each of the panels, signs were transferred from the equilibrium chemical shift values (Δδ) to the dynamic chemical shift values (Δω). The hatched lines represent 1:1 correlations. Data points for residues that act as markers of the occluded to closed transition are indicated in orange, while the data points for K32, G121, and H149 (highlighted on the structures in Figure 3) are colored blue, red, and green, respectively.
Figure 5.
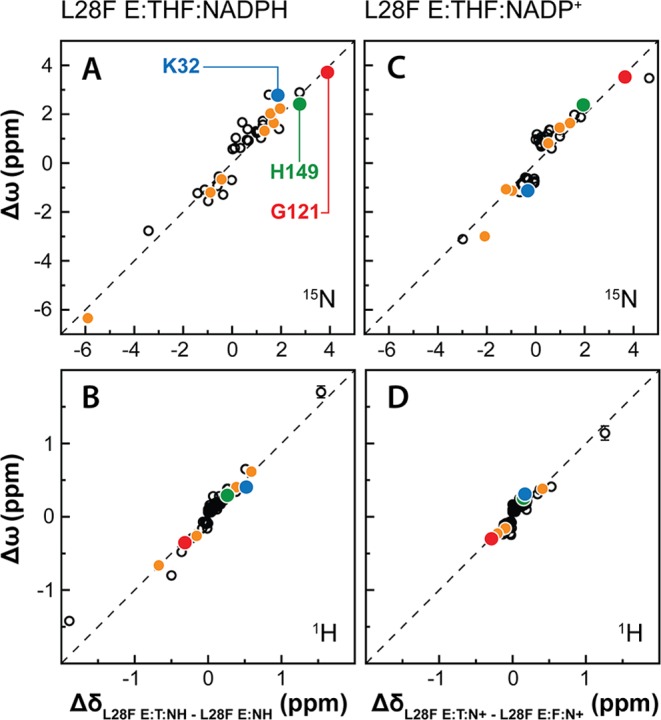
Correlation between the dynamic chemical shift differences (Δω) and equilibrium chemical shift differences (Δδ) for L28F DHFR complexes. (A) Plot of ΔωN for E:THF:NADPH versus ΔδN, the difference in 15N chemical shift between the L28F E:THF:NADPH and E:NADPH complexes. (B) Plot of ΔωH for E:THF:NADPH versus ΔδH, the difference in 1H chemical shift between the L28F E:THF:NADPH and E:NADPH complexes. (C) Plot of ΔωN for E:THF:NADP+ versus ΔδN, the difference in 15N chemical shift between the L28F E:THF:NADPH and E:FOL:NADP+ complexes. (D) Plot of ΔωH for E:THF:NADP+ versus ΔδH, the difference in 1H chemical shift between the L28F E:THF:NADPH and E:FOL:NADP+ complexes. For each of the panels, signs were transferred from the equilibrium chemical shift values (Δδ) to the dynamic chemical shift values (Δω). The hatched lines represent 1:1 correlations. Data points for residues that act as markers of the occluded to closed transition are indicated in orange, while the data points for K32, G121, and H149 (highlighted on the structures in Figure 3) are colored blue, red, and green, respectively.
The ΔωN and ΔδN are strongly correlated (Figure S6); a linear least-squares fit yields slope = 1.00 and R2 = 0.95, providing strong evidence that the process that gives rise to relaxation dispersion involves exchange between the occluded ground state and a closed excited state. Linear correlations were also observed between ΔωN and the equilibrium chemical shift difference ΔδN for the L28F E:THF:NADP+ complex and for each of the WT complexes (Figure S6), showing that each complex undergoes exchange between an occluded ground state, with the nicotinamide ring projecting into solvent, and a small population of a closed excited state in which the nicotinamide ring occupies the active site.
Having established that the exchange process involves fluctuations between occluded and closed states, we extended the ΔωN/ΔδN correlations to include all residues with dispersion curves that fit a two-site exchange process (Figures 4 and 5). Signs were transferred from the equilibrium chemical shifts (Δδ) to the dynamic chemical shifts (Δω) derived from the fits of the relaxation dispersion profiles. Linear correlations are observed between ΔωN (ΔωH) and the equilibrium chemical shift difference ΔδN (ΔδH) for each of the WT and L28F complexes, showing that the effects of the occluded to closed conformational fluctuations are propagated throughout much of the protein. The occluded-closed conformational exchange processes in the product ternary complexes, E:THF:NADPH and E:THF:NADP+, are comparable to the fluctuations closed-occluded processes previously characterized for the WT E:FOL:NADP+ complex.5,14
Discussion
The large amount of data available for E. coli DHFR from kinetics studies,21,24 X-ray crystallography,22 and NMR spectroscopy5,12−14,23 makes this enzyme a paradigm for understanding the role of protein dynamics in mediating ligand flux and catalysis. Despite the identification of the closed-occluded transitions during catalysis (Figure 1), a quantitative description of how conformational fluctuations regulate ligand flux has remained elusive. Here, we set out to develop a kinetic scheme for product release in DHFR, and potentially other enzymes, using a combination of stopped-flow and NMR relaxation dispersion experiments.
Ligand Exchange Rates Are Sensitive to Buffer Conditions
Steady-state and pre-steady-state kinetic data for WT and L28F DHFR have previously been measured in MTEN buffer at 298 K.21,24 However, the buffer conditions for the NMR relaxation dispersion experiments in the present and previous work5,16,49,50 are different and might therefore influence the kinetic parameters. The THF dissociation rates measured under NMR conditions (Table 1) differ slightly from previously recorded rates in MTEN buffer. For example, the THF dissociation rate for the WT E:THF:NADPH complex is 9.0 s–1 under NMR conditions (pH 7.6, 300 K) compared to 12 s–1 at pH 6 in MTEN buffer at 298 K.21 Similarly, the THF dissociation rate for the L28F E:THF:NADPH complex is reduced from 80 s–1 in MTEN buffer to 48 s–1 in NMR buffer.24 Thus, it is essential to measure the kinetic parameters under identical conditions if pre-steady-state kinetics and relaxation dispersion experiments are to be analyzed in an integrated fashion. Indeed, it has been shown recently that the buffer composition can have a substantial effect on the ms time scale fluctuations of enzymes.51
Differences in temperature, pH, or the incorporation of heavy isotopes52 for NMR experiments could contribute to the observed differences in THF dissociation kinetics. However, it is likely that the major factor is the difference in the nature and concentration of the cation between MTEN buffer (Na+, 100 mM) and NMR buffer (K+, 156 mM). Cations are known to bind to DHFR and inhibit product release,53,54 and the extent of inhibition increases with the cation radius and concentration. Thus, both the larger radius of potassium compared to sodium and the higher cation concentration will contribute to reduction of the THF dissociation rate for the WT and L28F E:THF:NADPH complexes in NMR buffer compared to MTEN buffer.
Conformational Fluctuations in the WT E:THF:NADPH Complex
Previous 15N NMR relaxation dispersion studies of WT E. coli DHFR suggested that millisecond time scale conformational fluctuations play an important role in governing progression of the enzyme through its catalytic cycle.5 In particular, the close correspondence between the rate (12–18 s–1) of conformational fluctuations in the active site of the E:THF:NADPH complex and the kinetics of THF release (12.5 s–1)21 provided circumstantial evidence that protein motions may play a direct role in controlling product release, the rate-determining step in the catalytic cycle. In order to obtain a more detailed and quantitative description of the product release mechanism, we acquired new and extended relaxation dispersion data for the E:THF:NADPH and E:THF:NADP+ complexes of WT DHFR and a mutant (L28F) with a higher rate of product release.
The E:THF:NADPH complex is of limited stability, with a sample lifetime of ∼1.5 days under strictly anaerobic conditions and in the presence of an enzymatic NADPH recycling system. By implementing scan interleaving and non-uniform sampling in the constant time CPMG pulse programs,25−28 we were able to acquire more scans (16 versus 8 per sampling time-point) and obtain higher quality 15N dispersion data than in the earlier experiments.5 In addition, the measurements were extended to include amide 1H dispersion. Since many amides that have no 15N dispersion do exhibit 1H dispersion (Figures S2–S5), the 1H CPMG experiments are a valuable complement to 15N dispersion measurements and provide probes at many additional sites in the polypeptide chain. For both the WT E:THF:NADPH and E:THF:NADP+ complexes, we were able to fit the amide 15N and 1H dispersion profiles for most of the residues in the active site loops and product binding site to a two-state global process. A linear correlation was observed between the ΔωN and ΔωH values for the WT E:THF:NADPH complex and the equilibrium chemical shift differences ΔδN and ΔδH between the occluded WT E:THF:NADPH and closed WT E:NADPH complexes (Figure 4), showing that the E:THF:NADPH complex fluctuates between its occluded ground state and a weakly populated closed conformation. Previous 15N relaxation dispersion experiments provided evidence, based on ΔωN values for G15, L8, and G95, for entry of the nicotinamide moiety of NADPH into the active site in the closed excited state.49 Additional evidence comes from the large |ΔωH| of A7 (1.56 ppm) observed in the present experiments; the A7 amide 1H resonance is shifted downfield by 1–2 ppm by formation of a hydrogen bond to the carboxamide moiety of the nicotinamide when the ribosyl-nicotinamide moiety occupies the active site.23 While 15N dispersion was observed for residues in the active site loop in previous studies,5 the structure of the excited state formed by E:THF:NADPH could not be discerned given the limited amount and quality of dispersion data available. However, the extensive, high-quality 15N and 1H dispersion data provided by the current work shows clearly that the active site loops in the higher energy substate of the E:THF:NADPH complex adopt the closed conformation. A similar linear correlation is observed between ΔωN and ΔωH values for the WT E:THF:NADP+ complex and the equilibrium chemical shift differences ΔδN and ΔδH between the occluded WT E:THF:NADP+ and closed WT E:FOL:NADP+ complexes (Figure 4). This correlation shows that WT E:THF:NADP+ also transiently samples a closed conformational substate, in accord with previous results,5 with the oxidized nicotinamide ring of NADP+ occupying the active site (|ΔωH| of A7 = 1.36 ppm).
Simultaneous fitting of the current 15N and 1H R2 dispersion data yields values for kex and pb for the WT E:THF:NADPH and E:THF:NADP+ complexes that differ somewhat from those reported earlier.5,49 Visual inspection shows that the new and old sets of 15N dispersion profiles are very similar, although the newer data are of much higher quality due to the use of interleaved scans and non-uniform sampling. We suspect that the differences arise because the old data was of poorer quality (more noise) and because it is difficult to obtain robust fits of kex and pb using 15N dispersion data alone when, as in the present case, exchange is relatively fast and the dispersion curves are rather featureless. By simultaneously fitting 15N and 1H dispersion profiles at two magnetic fields, as in the present work, the exchange rate and excited-state population can be determined with a high degree of confidence both because of the greatly increased amount of data and because many of the 1H dispersion curves do not increase monotonically but have features that allow more robust extraction of exchange parameters. The new data for E:THF:NADPH indicate a smaller excited state population (pb) than was obtained in the previous analysis, together with an increased rate of egress of the nicotinamide ring from the binding pocket, and show that occupation of the active site by the reduced nicotinamide ring of NADPH is disfavored relative to that of NADP+.
The L28F Mutation Alters μs–ms Time Scale Backbone Dynamics
Relaxation dispersion data for the E:THF:NADPH and E:THF:NADP+ complexes of L28F DHFR show that the mutation changes the μs–ms time scale backbone dynamics (Table 2, Figure 3) without altering the occluded conformation of the ground states as shown by X-ray crystallography (Figure 2A). Fitting of the dispersion curves for the L28F complexes yielded approximately 2-fold slower exchange rates and 2–4-fold higher excited-state populations compared to the corresponding WT complexes (Table 2), indicating that the excited conformational substate is thermodynamically more favorable in the L28F mutant than in WT DHFR. Linear correlations are observed between ΔωN and ΔωH for active site amides of L28F E:THF:NADPH and L28F E:THF:NADP+ and the equilibrium shift differences, ΔδN and ΔδH, between the occluded E:THF:NADPH and E:THF:NADP+ complexes and the closed E:NADPH and E:FOL:NADP+ complexes of L28F (Figure 5). Thus, as in WT DHFR, the E:THF:NADP(H) ternary complexes of the L28F mutant sample a small population of a closed conformational substate. The large values of |ΔωH| for the A7 amide (1.42 and 1.06 ppm for the L28F E:THF:NADPH and E:THF:NADP+ complexes, respectively) show that the nicotinamide moiety of the cofactor transiently enters the active site in the closed conformational substate. Further confirmation comes from the signs of ΔωN and ΔδN for G15, which are positive (upfield shift; ΔωN = 1.39 ppm, ΔδN = 0.63 ppm) when the reduced nicotinamide ring occupies the active site and are negative (ΔωN = −3.11 ppm, ΔδN = −2.98 ppm) when the oxidized nicotinamide ring occupies the active site (Tables S6 and S8). The L28F mutation results in a ∼50% increase in the rate at which the nicotinamide moiety of the cofactor enters the active site, while decreasing the rate at which it leaves (Table 2). For each of the WT and L28F complexes, the rates of entry of the oxidized and reduced nicotinamide rings into the active site pocket are comparable (Table 2), suggesting that the barrier for insertion is determined primarily by protein conformational changes.49
Product Remains Bound in the Excited State
The relaxation dispersion experiments show clearly that the WT and L28F E:THF:NADPH product release complexes transiently sample a weakly populated conformational substate that closely resembles the closed ground-state conformation of the respective binary E:NADPH complexes. Several lines of evidence suggest strongly that the THF product remains physically bound to the enzyme in this excited conformational substate. For WT E:THF:NADPH, the population of the E:NADPH binary complex at equilibrium in the presence of 18 mM THF was estimated from the binding kinetics in MTEN buffer21 to be 0.03–0.05% (at pH 6–9), which is very much smaller than the 0.7% minor population observed in the relaxation dispersion experiments. Second, it has been shown in previous relaxation dispersion studies that the exchange rate and excited-state population are independent of THF concentration, which would not be the case if the exchange process involved a physical dissociation event.5 Finally, simulations using the kinetic scheme of Figure 6 with rate constants measured in the present work show that the population of the WT binary E:NADPH complex that would be formed by dissociation of THF is only 0.02% (Table S10). For L28F, the THF on-rate is not available, but the general similarity between the behavior of WT and L28F makes it highly probable that the product also remains fully bound in the minor conformational substate of the L28F E:THF:NADPH complex.
Figure 6.
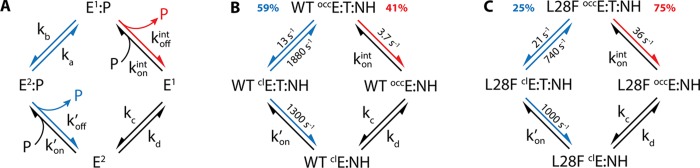
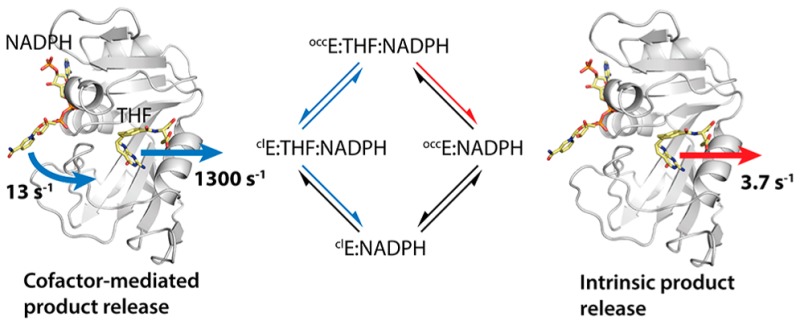
(A) Generalized kinetic scheme for product release. E1 and E2 are two different protein conformational states. E1 is the major conformational state (ground state) when product is bound, and E2 is the major conformational state (ground state) in the product-dissociated state. The red arrows highlight the intrinsic product release pathway, in which product dissociates spontaneously from the E1 state at rate koffint. The blue arrows indicate an allosteric pathway, in which the enzyme undergoes a conformational change prior to product release to populate the E2 state, which facilitates product dissociation (koff >koffint). (B) Kinetic scheme for product release from the WT E:THF:NADPH complex. The measured and calculated rate constants in NMR buffer are shown (Tables 1–3). Product release occurs predominantly through the allosteric pathway (60%) instead of the intrinsic pathway (40%). (C) Kinetic scheme for product release from the L28F E:THF:NADPH complex. The measured and calculated rate constants are shown (Tables 1–3). The L28F mutation alters the partitioning between the allosteric and intrinsic pathway from 60/40 to 25/75 for the WT and mutant enzymes, respectively.
Product Release Occurs via Allosteric and Intrinsic Pathways
Stopped-flow measurements of the THF dissociation rate for the WT and L28F E:THF:NADPH complexes show that the reduced (but not the oxidized) cofactor assists product release. Substituting NADPH for NADP+, the THF dissociation rate increases from 3.8 to 9.0 s–1 for the WT enzyme and from 36 to 48 s–1 for the L28F enzyme (Table 1). In contrast, the rate of THF dissociation is the same from the binary E:THF and the ternary E:THF:NADP+ complexes of each enzyme. Relaxation dispersion measurements for the WT E:THF:NADPH complex indicate transient entry of the nicotinamide moiety into the active site, resulting in a closed state with a population of 0.7% (Table 2). Since relaxation dispersion is also observed for several probes in the product binding site (Figure 3), it is evident that entry of the nicotinamide ring into the active site leads to changes in the product binding site. We have hypothesized previously that this transiently populated closed state may function to facilitate product release.5 Binding of the adenosine moiety of NADPH, at a site distant from the active site, causes a change in the enzyme dynamics and a shift in the conformational ensemble from 100% occluded in the E:THF complex to 99.3% occluded/0.7% closed in E:THF:NADPH. In the occluded ground state of the E:THF:NADPH complex, the nicotinamide ring of the cofactor is outside the active site where it projects into the solvent and cannot promote product release. In the minor conformational substate, the ring transiently enters the pocket and binds in a site that is distinct from the product binding site. Both the nicotinamide and product can occupy the active site simultaneously, however the resulting steric strain enhances the probability of THF release. Our results are in accord with current views that link allostery to a population shift in the conformational ensemble55−57 and also exemplify the recently introduced anchor and driver concept for allosteric effectors.58 The adenosine moiety of NADPH acts as an anchor, binding the cofactor to the enzyme in both the occluded ground state and the higher energy, closed conformational substate. By binding transiently in the active site, the nicotinamide ring acts as a driver by creating a repulsive interaction caused by steric clash with the pterin ring of THF. However, the allosteric pathway is not solely responsible for product release from the WT E:THF:NADPH complex.
Residues in the Met20 loop and product binding site of the WT E:THF complex exhibit no relaxation dispersion, showing that μs–ms time scale backbone fluctuations that might promote product release do not occur in the binary product complex.49 Nevertheless, THF can spontaneously dissociate from the occluded WT E:THF binary complex at a rate of 3.7 s–1 (Table 1), indicating the existence of an intrinsic pathway for product dissociation. It is likely that THF can also dissociate directly from the ternary occluded WT E:THF:NADPH complex by this intrinsic pathway. We therefore describe THF dissociation from the WT E:THF:NADPH complex as occurring via two parallel pathways, an allosteric pathway and an intrinsic pathway (Figure 6A). The overall rate of dissociation is then given by
| 1 |
where koffobs, koff, and koffint are the observed, allosteric, and intrinsic THF dissociation rates, respectively. The observed THF dissociation rate (koff) is the rate measured by stopped-flow competition experiments (Table 1). The intrinsic THF dissociation rate (koffint) is assumed to be identical to the measured rate for the binary WT E:THF complex in absence of cofactor. The allosteric THF dissociation rate (koff), calculated from koffobs, and koff, is described by the standard equation for the effective dissociation rate in a two-step dissociation process:59
| eq 2 |
where kb and ka are the rates of formation and relaxation of the transiently populated closed state obtained from globally fitting the relaxation dispersion data (Table 2), and koff′ is the rate for product release from this state. In Table 3, we summarize the rates for the individual steps in the kinetic scheme of Figure 6A. To highlight the pathway preference, we also calculated the partitioning ratio (Table 3), which is the ratio of the allosteric and the intrinsic product release rates. If the partitioning ratio is larger than 1, then the allosteric pathway is preferred. The partitioning ratio of 1.43 for product release from the WT E:THF:NADPH complex in NMR buffer shows that the allosteric pathway is dominant with approximately 60% of the total product release flux taking this route versus 40% going through the intrinsic pathway (Figure 6B). A full simulation of the flux through the two pathways is shown in Figure S7. The flux that goes through each pathway will change depending on the experimental conditions; for example, the allosteric pathway becomes more dominant (90% flux) in MTEN buffer at pH 6.0 (Table 3).21
Table 3. Rate Constants Involved in the Proposed Kinetic Schemea.
| koff′ (s–1) | koffallos (s–1) | koffint (s–1) | partitioning ratio | allosteric pathway flux (%) | |
|---|---|---|---|---|---|
| WT E:THF:NADPH | 1300 ± 200 | 5.3 ± 0.4 | 3.7 ± 0.3 | 1.4 ± 0.2 | 59 ± 5 |
| WT E:THF:NADP+ | 8 ± 29 | 0.1 ± 0.4 | 3.7 ± 0.3 | 0.0 ± 0.1 | 3 ± 9 |
| L28F E:THF:NADPH | 1000 ± 400 | 12 ± 3 | 36 ± 2 | 0.33 ± 0.08 | 25 ± 6 |
| L28F E:THF:NADP+ | 0 ± 0 | 0 ± 2 | 36 ± 2 | 0 ± 0 | 0 ± 0 |
| WT E:THF:NADPHb | ND | 10.6 | 1.4 | 7.6 | 88 |
| L28F E:THF:NADPHc | ND | 40 | 40 | 1.0 | 50 |
Unlike NADPH, binding of NADP+ to the E:THF complex does not promote THF release (Table 2), despite the fact that the oxidized nicotinamide ring enters the active site pocket in the transient excited state formed in the E:THF:NADP+ complex. It seems likely that the active site is able to accommodate both the puckered pterin ring of THF and the planar nicotinamide ring of the oxidized cofactor with minimal steric strain. In contrast, puckering of the nicotinamide ring of NADPH appears to cause steric clash with the puckered pterin ring of THF.14 Indeed, DHFR binds NADPH in its preferred conformation with the glycosidic bond nearly perpendicular to the nicotinamide ring, maximizing the puckering of the latter.60
Mutations Modulate the Partitioning Ratio
In absence of the reduced cofactor, the L28F mutation causes a large increase in the intrinsic THF dissociation rate from the binary E:THF complex, from 3.7 s–1 for WT to 36 s–1 for the mutant (Table 1). The L28F mutation is located in the product-binding site (Figure 2A) where it changes the conformation of bound THF by reorienting the benzoyl ring and altering the packing interactions of the glutamyl tail (Figure 2C). In the WT E:ddTHF:NADP+ X-ray structure, the benzoyl ring is packed tightly against the methyl side chains of I50, L54, and I94 and against the edge of the F31 aromatic ring. The L28F mutation causes the benzoyl ring to rotate away from these side chains, leaving a substantial cavity in the binding pocket, which likely destabilizes the interaction and contributes toward the increase in the intrinsic rate for dissociation of THF from the mutant protein. Like the WT E:THF complex,5 relaxation dispersion profiles for amide probes in the L28F E:THF active site loop and product binding site are flat (Figure 3), indicating the absence of an occluded to closed transition that might aid in product release. Thus, as for the WT complexes, we can assume that dissociation of THF from the L28F E:THF:NADPH complex also occurs via allosteric and intrinsic pathways, with dissociation by the latter pathway occurring at the same rate as dissociation from the binary L28F E:THF complex. In contrast to WT E:THF:NADPH, the flux of THF release from the ternary L28F E:THF:NADPH complex favors the intrinsic pathway (75%) over the allosteric pathway (25%) (Figure 6C; Table 3). The THF dissociation rate for the ternary L28F E:THF:NADP+ complex is identical to the rate for the binary L28F E:THF complex. We can therefore conclude that, just as for the WT enzyme, product release from the L28F E:THF:NADP+ complex is not assisted by oxidized cofactor and occurs solely via the intrinsic pathway.
Other DHFR mutant proteins can be treated similarly. For example, the G121V mutation reduces the product dissociation rate 7-fold.29 Like WT DHFR, the G121V E:THF:NADPH and G121V E:THF:NADP+ complexes assume an occluded conformation of the Met20 loop.23 However, unlike the WT protein, 15N relaxation–dispersion experiments show that the active site residues in the G121V complexes do not undergo μs–ms time scale conformational fluctuations to sample a closed excited-state conformation in which the nicotinamide ring transiently enters the active site pocket.50 Since the allosteric pathway is therefore not available, product release from the G121V DHFR occurs entirely via the intrinsic pathway and at the same rate (1.9 s–1) for both the E:THF:NADPH and E:THF:NADP+ complexes.29
Conclusions
Based on the integrated application of NMR relaxation dispersion and stopped-flow kinetics experiments, we propose a kinetic scheme for product release in E. coli DHFR that involves two parallel pathways, an intrinsic pathway, where the product THF dissociates spontaneously from the enzyme, and an allosteric pathway, which utilizes NADPH (but not NADP+) as an allosteric effector to enhance product release. Binding of cofactor to the E:THF product binary complex induces μs–ms time scale fluctuations in the active site and a population shift in the DHFR conformational ensemble. The rate enhancement for product release occurs through the transient formation of a small population of a closed excited state where the nicotinamide ring of the cofactor enters the active site, detected in relaxation dispersion experiments. Only the reduced cofactor is able to promote dissociation via the allosteric pathway, even though the ternary complexes containing both reduced and oxidized cofactors populate closed excited states. The ring pucker of the reduced NADPH cofactor appears to be the determining feature that increases the rate of product release through steric crowding of the pterin ring of the product THF. This mechanism can also explain the effects of DHFR mutations (and the effects of different solution conditions) on the rate of catalysis, as a general change in the partitioning between the intrinsic and allosteric pathways.
Acknowledgments
This work was supported by the US National Institutes of Health (NIH) grant GM75995 and the Skaggs Institute of Chemical Biology (P.E.W.). D.O. was supported by a Collen-Francqui fellowship from the Belgian American Educational Foundation (BAEF). We acknowledge Maria Yamout for assistance with protein purification, Gerard Kroon for assistance with NMR experiments, and Phillip Aoto for helpful discussions. We thank the Joint Center for Structural Genomics (JCSG) for use of its automated crystallization facility. The JCSG in funded under the NIH NIGMS Protein Structure Initiative (U54 GM094586). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Supporting Information Available
Tables of kinetic and relaxation data, crystallographic data, and Figures S1–S7. Coordinates and structure factors of WT ecDHFR:ddTHF:NADP+ and L28F ecDHFR:ddTHF:NADP+ have been deposited in the PDB with accession numbers 5CCC and 5CC9, respectively. Chemical shift assignments for the L28F complexes have been deposited in the BMRB with accession codes 26583, 26584, and 26585. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b05707.
The authors declare no competing financial interest.
Supplementary Material
References
- Fersht A.Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; W.H. Freeman: New York, 1999. [Google Scholar]
- Cleland W. W. Acc. Chem. Res. 1975, 8, 145. 10.1021/ar50089a001. [DOI] [Google Scholar]
- Rozovsky S.; McDermott A. E. J. Mol. Biol. 2001, 310, 259. 10.1006/jmbi.2001.4672. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; Dyson H. J.; Wright P. E. Chem. Rev. 2006, 106, 3055. 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- Boehr D. D.; McElheny D.; Dyson H. J.; Wright P. E. Science 2006, 313, 1638. 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. A.; Lei M.; Thai V.; Kerns S. J.; Karplus M.; Kern D. Nature 2007, 450, 913. 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. A.; Thai V.; Lei M.; Ott M.; Wolf-Watz M.; Fenn T.; Pozharski E.; Wilson M. A.; Petsko G. A.; Karplus M.; Hubner C. G.; Kern D. Nature 2007, 450, 838. 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Khajehpour M.; Wu L.; Liu S.; Zhadin N.; Zhang Z. Y.; Callender R. Biochemistry 2007, 46, 4370. 10.1021/bi602335x. [DOI] [PubMed] [Google Scholar]
- Whittier S. K.; Hengge A. C.; Loria J. P. Science 2013, 341, 899. 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. G. III J. Magn. Reson. 2014, 241, 3. 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. G. III Chem. Rev. 2004, 104, 3623. 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Osborne M. J.; Schnell J.; Benkovic S. J.; Dyson H. J.; Wright P. E. Biochemistry 2001, 40, 9846. 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- Schnell J. R.; Dyson H. J.; Wright P. E. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 119. 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- McElheny D.; Schnell J. R.; Lansing J. C.; Dyson H. J.; Wright P. E. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 5032. 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S.; Benkovic S. J. Annu. Rev. Biochem. 2006, 75, 519. 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- Bhabha G.; Lee J.; Ekiert D. C.; Gam J.; Wilson I. A.; Dyson H. J.; Benkovic S. J.; Wright P. E. Science 2011, 332, 234. 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk L. Y. P.; Javier Ruiz-Pernia J.; Dawson W. M.; Roca M.; Loveridge E. J.; Glowacki D. R.; Harvey J. N.; Mulholland A. J.; Tunon I.; Moliner V.; Allemann R. K. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 16344. 10.1073/pnas.1312437110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K.; Kohen A. Curr. Opin. Chem. Biol. 2014, 21, 19. 10.1016/j.cbpa.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoian P.; Liu C. T.; Hammes-Schiffer S.; Benkovic S. Acc. Chem. Res. 2015, 48, 482. 10.1021/ar500390e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk L. Y. P.; Loveridge E. J.; Allemann R. K. Phys. Chem. Chem. Phys. 2015, 10.1039/C5CP00794A. [DOI] [PubMed] [Google Scholar]
- Fierke C. A.; Johnson K. A.; Benkovic S. J. Biochemistry 1987, 26, 4085. 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- Sawaya M. R.; Kraut J. Biochemistry 1997, 36, 586. 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- Venkitakrishnan R. P.; Zaborowski E.; McElheny D.; Benkovic S. J.; Dyson H. J.; Wright P. E. Biochemistry 2004, 43, 16046. 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- Wagner C. R.; Thillet J.; Benkovic S. J. Biochemistry 1992, 31, 7834. 10.1021/bi00149a013. [DOI] [PubMed] [Google Scholar]
- Orekhov V. Y.; Korzhnev D. M.; Diercks T.; Kessler H.; Arseniev A. S. J. Biomol. NMR 1999, 14, 345. 10.1023/A:1008356809071. [DOI] [Google Scholar]
- Aoto P. C.; Fenwick R. B.; Kroon G. J. A.; Wright P. E. J. Magn. Reson. 2014, 246, 31. 10.1016/j.jmr.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orekhov V. Y.; Jaravine V. A. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 271. 10.1016/j.pnmrs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Kazimierczuk K.; Orekhov V. Y. Angew. Chem., Int. Ed. 2011, 50, 5556. 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- Cameron C. E.; Benkovic S. J. Biochemistry 1997, 36, 15792. 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- Bhabha G.; Tuttle L.; Martinez-Yamout M. A.; Wright P. E. FEBS Lett. 2011, 585, 3528. 10.1016/j.febslet.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsall B.; Burgen A. S.; Roberts G. C. Biochemistry 1980, 19, 3723. 10.1021/bi00557a013. [DOI] [PubMed] [Google Scholar]
- Efron B.An Introduction to the Bootstrap; Chapman and Hall: New York, 1993; Vol. 57. [Google Scholar]
- Otwinowski Z.; Minor W.. Methods in Enzymology: Macromolecular Crystallography Part A; Carter C. W. Jr; Sweet R. M., Eds.; Academic Press: New York, NY, 1997; pp 307–326. [DOI] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. J. Appl. Crystallogr. 2007, 40, 658. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2256. 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994, 50, 760. 10.1107/S0907444994003112 [DOI] [PubMed] [Google Scholar]
- Wittekind M.; Mueller L. J. Magn. Reson., Ser. B 1993, 101, 201. 10.1006/jmrb.1993.1033. [DOI] [Google Scholar]
- Grzesiek S.; Bax A. J. Am. Chem. Soc. 1992, 114, 6291. 10.1021/ja00042a003. [DOI] [Google Scholar]
- Muhandiram D. R.; Kay L. E. J. Magn. Reson., Ser. B 1994, 103, 203. 10.1006/jmrb.1994.1032. [DOI] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. J. Biomol. NMR 1995, 6, 277. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. Proteins: Struct., Funct., Genet. 2005, 59, 687. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Loria J. P.; Rance M.; Palmer A. G. III J. Am. Chem. Soc. 1999, 121, 2331. 10.1021/ja983961a. [DOI] [Google Scholar]
- Ishima R.; Torchia D. A. J. Biomol. NMR 2003, 25, 243. 10.1023/A:1022851228405. [DOI] [PubMed] [Google Scholar]
- Sugase K.; Konuma T.; Lansing J. C.; Wright P. E. J. Biomol. NMR 2013, 56, 275. 10.1007/s10858-013-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrynnikov N. R.; Dahlquist F. W.; Kay L. E. J. J. Am. Chem. Soc. 2002, 124, 12352. 10.1021/ja0207089. [DOI] [PubMed] [Google Scholar]
- Osborne M. J.; Venkitakrishnan R. P.; Dyson H. J.; Wright P. E. Protein Sci. 2003, 12, 2230. 10.1110/ps.03219603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D. D.; McElheny D.; Dyson H. J.; Wright P. E. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1373. 10.1073/pnas.0914163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D. D.; Schnell J. R.; McElheny D.; Bae S. H.; Duggan B. M.; Benkovic S. J.; Dyson H. J.; Wright P. E. Biochemistry 2013, 52, 4605. 10.1021/bi400563c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.; Khirich G.; Loria J. P. Biochemistry 2013, 52, 6548. 10.1021/bi400973e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Singh P.; Czekster C. M.; Kohen A.; Schramm V. L. J. Am. Chem. Soc. 2014, 136, 8333. 10.1021/ja501936d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccanari D.; Phillips A.; Smith S.; Sinski D.; Burchall J. Biochemistry 1975, 14, 5267. 10.1021/bi00695a006. [DOI] [PubMed] [Google Scholar]
- Ohmae E.; Miyashita Y.; Tate S. I.; Gekko K.; Kitazawa S.; Kitahara R.; Kuwajima K. Biochim. Biophys. Acta, Proteins Proteomics 2013, 1834, 2782. 10.1016/j.bbapap.2013.09.024. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Karplus M. Protein Sci. 2008, 17, 1295. 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilser V. J. Science 2010, 327, 653. 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh H. N.; Wrabl J. O.; Li J.; Hilser V. J. Nature 2014, 508, 331. 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R.; Tsai C.-J.; Liu J. J. J. Am. Chem. Soc. 2014, 136, 17692. 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikl T. R.; Boehr D. D. Proteins: Struct., Funct., Genet. 2012, 80, 2369. 10.1002/prot.24123. [DOI] [PubMed] [Google Scholar]
- Wu Y. D.; Houk K. N. J. Am. Chem. Soc. 1991, 113, 2353. 10.1021/ja00007a002. [DOI] [Google Scholar]
- Boehr D. D.; Dyson H. J.; Wright P. E. Biochemistry 2008, 47, 9227. 10.1021/bi801102e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.