Preface
The regulated migration of cells is essential for development and tissue homeostasis, and aberrant cell migration can lead to an impaired immune response and the progression of cancer. Primordial germ cells (PGCs), precursors to sperm and eggs, have to migrate across the embryo to reach somatic gonadal precursors (SGPs) and fulfill their function. Studies of model organisms have revealed that, despite important differences, several features of PGC migration are conserved. PGCs require both an intrinsic motility program and external guidance cues to survive and successfully migrate. Proper guidance involves both attractive and repulsive cues mediated by protein and lipid signalling.
Cell migration describes the directed movement of cells through the body. The basic features of cell migration have been deciphered by studies of cell culture systems as well as developing embryos1-5. Migrating cells exhibit directional polarity, with a leading edge at the front of the cell and lagging edge at the back. Movement is achieved by protrusion and adhesion of the leading edge of the cell and retraction of the lagging edge. These processes are regulated by transmembrane receptors that receive external chemoattractant signals, which are then translated to cytoskeletal changes by effector molecules such as phospholipids and small GTPases.
The study of how cells migrate is highly relevant to our understanding of both normal and pathological processes4, 5. Aberrant cell migration can cause developmental defects and impair the body's ability to respond to injury and disease. During embryonic development, gastrulation requires extensive coordinated cell migration as the embryo reorganizes to form the three germ layers (ectoderm, mesoderm and endoderm)6. Subsequently, the formation of organ systems, such as the vascular system and the nervous system, also requires highly regulated cell migration7-9. Following development, cell migration is also required to protect and heal mature organisms; for example, the migration of epidermal cells is required for wound healing, whereas the movement of lymphocytes towards sites of infection is part of the immune response. Furthermore, during metastasis cancerous cells travel to colonize new tissues, a process with dramatic effects on cancer treatment and on the survival of patients. It is clear that further understanding of the cellular and molecular mechanisms underlying cell migration has significant therapeutic importance.
In many animals the primordial germ cells (PGCs), precursors to sperm and eggs, arise far from the somatic cells of the developing gonad (somatic gonadal precursors (SGPs)) and therefore have to actively migrate across the embryo to reach their site of function10-13. This process provides a useful model system for the study of cell migration within the context of a developing organism. PGC migration must be finely regulated as it follows a complex path through a variety of developing tissues. In addition to the obvious effect of disrupted PGC migration on fertility, aberrant movement to ectopic sites in the body is one mechanism that could account for the incidence of extragonadal germ cell tumours in humans14, 15. Most of our understanding of PGC migration comes from the model genetic organisms Drosophila melanogaster, zebrafish and mice. In all of these, PGCs form early in development and can be readily identified by morphology, embryonic position and gene expression profile, facilitating their analysis by live and fixed imaging approaches. Such approaches, combined with genetic analysis, have begun to clarify the cellular behaviours and molecular mechanisms responsible for ensuring proper PGC migration.
The general events of PGC migration in model organisms have been well characterized10-13 (discussed below; FIG. 1). Although there are important differences in the specification and migration of PGCs in these organisms, there are also several shared principles emerging that both increase our understanding of how PGCs migrate and provide a conceptual framework for the study of other migrating cell types. In this Review, we begin with a brief summary of how PGCs are specified in three organisms that show pronounced PGC migration, D. melanogaster, zebrafish and mouse. We then focus specifically on the individual steps of PGC migration: How PGCs first acquire motility; how the path of PGC migration is determined and regulated, and how PGCs stop migrating once they reach their target. We also discuss the intriguing connections between PGC migration and survival, and conclude by highlighting emerging themes in studies of PGC migration.
Figure 1. Stages of primordial germ cell (PGC) migration.
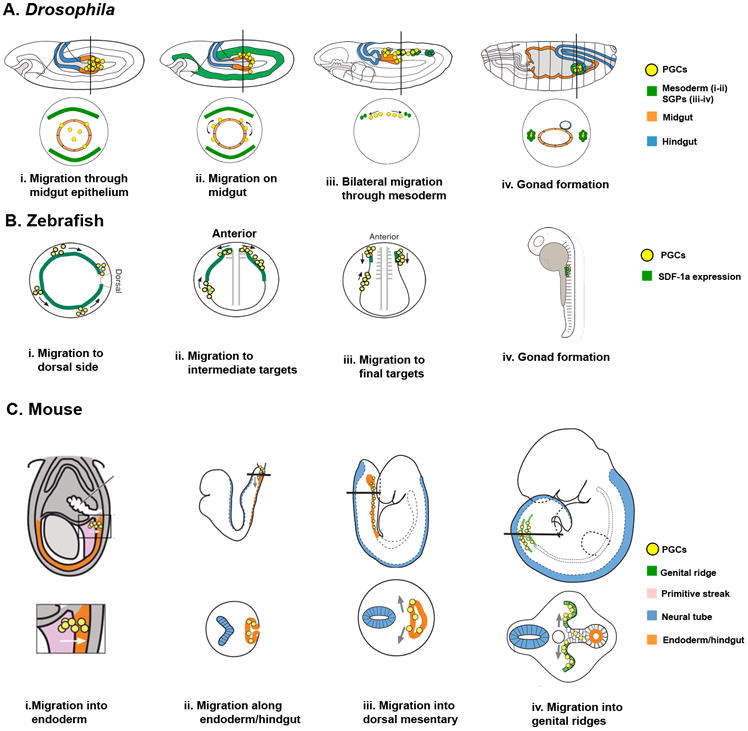
a | Drosophila melanogaster. i. After specification Primordial germ cells (PGCs) are carried into the embryo by the midgut primordium. PGCs polarize and migrate through the midgut epithelium at stages 9–10 (∼4.5h After Egg Laying (AEL)). ii. PGCs reorient on midgut towards the mesoderm at stage 10 (∼5h 10m AEL). iii. PGCs migrate bilaterally towards the somatic gonadal precursors (SGPs) at stage 11 (∼7h AEL). iv. PGCs associate with SGPs and coalesce to form the embryonic gonad. Lateral views (top) and transverse sections (bottom). b | Zebrafish. i. Following specification at four random locations, PGCs migrate dorsally (animal pole view; the animal pole refers to the portion of the blastula embryo that differentiates into mesoderm and ectoderm). ii. At gastrulation, 4.5 hours post-fertilization (hpf), PGCs follow expression of stromal derived factor 1a (SDF-1a). Somites 1-3 act as intermediate targets (lateral view, left side) at 10.5 hpf. iii. PGCs migrate towards the final target tissue at somites 8-10 (frontal view) at 13hpf. iv. At 24 hpf, PGCs coalesce with the somatic cells of the gonad (lateral view, left side). c | Mouse. i. PGCs, specified in proximal epiblast, migrate from the primitive streak to the endoderm (future hindgut) at embryonic day 7.5 (E7.5). Closeup shown on bottom. ii. At E8, PGCs migrate along the endoderm. iii, At E9.5, PGCs migrate bilaterally towards the dorsal body wall. iv. At E10.5, PGCs reach the genital ridges to form the embryonic gonad. Lateral views (top) and transverse sections (bottom). Adapted from Starz-Gaiano and Lehmann (2001) and Santos and Lehmann (2004)11, 114.
PGC specification
D. melanogaster, zebrafish and mice possess distinct strategies for forming PGCs. In particular, D. melanogaster and zebrafish require germ plasm, a specialized cytoplasm containing maternal RNAs and proteins. In the C. elegans embryo PGCs also form in germ plasm and much is known about their specification16. However, we chose not to cover C. elegans here because their PGCs do not show a pronounced migration and seem to reach the gonad by ingression during gastrulation17. There is no preformed germ plasm in mouse eggs; instead PGC specification requires cell-to-cell inductive signalling. Different types of PGC specification might relate to specific developmental constraints of a particular species, such as the timing of development and body plan11. However, there seem to be conserved molecular mechanisms for promoting PGC fate and maintenance, in particular transcriptional silencing of somatic gene expression.
In D. melanogaster, approximately 35 PGCs bud from the posterior of the embryo, adjacent to the forming somatic cells of the posterior midgut primordium (stages 4-5, which correspond to 1.5-3 hours after egg laying (AEL))11, 18. This process requires the activity of several germ plasm-specific RNAs and proteins. In particular, three germ plasm-localized RNAs, germ cell-less (gcl), nanos (nos) and polar granule component (pgc) have been implicated in the early events of germ cell specification, although only gcl seems to be directly required for PGC formation11. The precise mechanisms of gcl function remain unclear19, 20. pgc and nos function later in PGC development by regulating PGC gene expression and preserving their identity throughout development. Lack of pgc leads to improper expression of posterior somatic genes in PGCs, followed by disrupted PGC migration and death21-24. Loss of nos also leads to some inappropriate expression of somatic genes16, 25, 26. Later in development chromatin-based mechanisms of transcriptional repression seem to have important roles in maintaining PGC identity24, 27.
Zebrafish PGCs also form during early embryogenesis (3 hours post-fertilization (hpf)); however, zebrafish PGCs do not form at a single embryonic position. Instead, four PGC clusters, each containing approximately 4 cells, form at random locations in the early embryo28, 29. Relatively little is known about the mechanisms underlying germ cell specification in zebrafish. As in D. melanogaster, zebrafish PGCs require maternally supplied germ plasm and mRNAs such as nanos for their specification and maintenance28, 30, 31. Germ plasm assembly in zebrafish has recently been show to require Bucky ball, a novel, vertebrate-specific protein32, 33. Furthermore, a gcl homolog was recently identified in zebrafish and shown to have an expression pattern consistent with a role in PGC formation, although its function remains to be tested34.
In contrast to D. melanogaster and zebrafish, PGCs in the mouse are not specified by germ plasm but instead are induced during gastrulation by bone morphogenetic protein (BMP) signalling and yet unidentified signals from the extraembryonic ectoderm and visceral endoderm to underlying pluripotent epiblast cells (at embryonic day 6.5 (E6.5))35. This induction leads to transcriptional regulation of epiblast cells, mediated by the transcriptional repressor B-lymphocyte-induced maturation protein (BLIMP1, also known as PR-domain containing protein 1 (PRDM1)). BLIMP1 promotes expression of PGC-specific genes such as stella and represses expression of somatic cell genes, in particular members of the Hox gene family36-38. Correspondingly, PGCs lacking BLIMP1 do not properly differentiate or migrate. Recently, another transcriptional regulator, PRDM14, has been found to be important for PGC specification in mouse. Similar to BLIMP1, Prdm14 knockout mice fail to express PGC specific markers and are sterile due to a lack of proper PGC specification39, 40.
Initiation of PGC migration
Following specification, PGCs must become motile and receive directional cues to begin migrating. This is achieved by distinct mechanisms involving signalling and cell polarity in D. melanogaster and transcriptional regulation in zebrafish.
D. melanogaster
Live imaging studies indicate that shortly after specification, D. melanogaster PGCs display migratory behaviours (stage 5, 2-2.5h AEL)41, 42. During gastrulation (stages 7-8, 3-3.5h AEL) the PGCs are carried by tissue movement into the forming posterior midgut (PMG) pocket of the embryo (stage 9, ∼4h AEL; FIG. 1a)42. In the lumen of the PMG, PGCs form a tight cluster with each other but make little contact with the surrounding somatic cells of the PMG (FIG. 2a)41-43. This PGC cluster takes on a characteristic radial organization with the leading edge of each cell facing outward toward the PMG. Subsequently, PGCs begin extending cellular protrusions toward the surrounding PMG cells and begin to lose adhesion to each other (Supplementary information S1 (movie))41. Active PGC migration begins as the cells disperse from the cluster and individually migrate through the PMG (stage 10, 4.5h AEL; FIG. 2a).
Figure 2. Initiation of primordial germ cell migration.
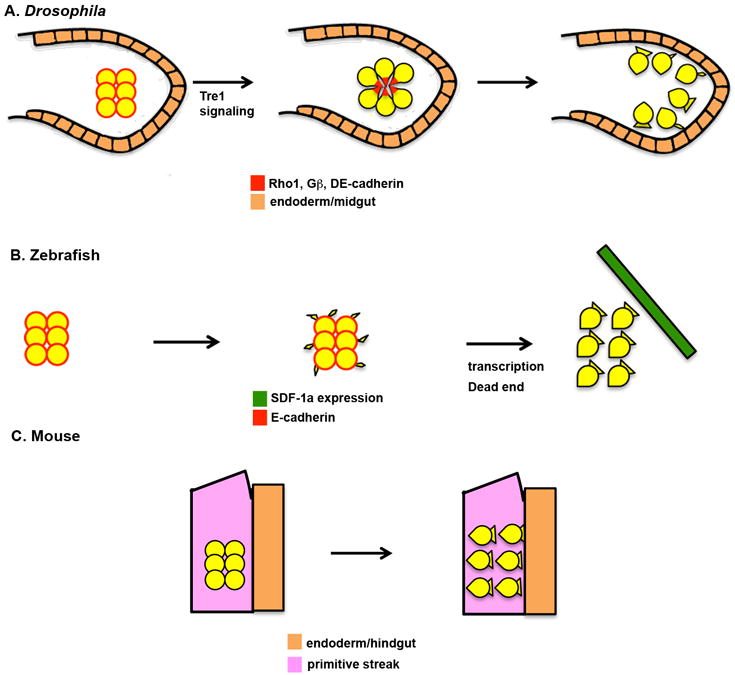
a | Drosophila melanogaster. I. At early stage 9 (∼4h After Egg Laying (AEL)), germ cells are tightly clustered in the midgut pocket. Primordial germ cells (PGCs) are not polarized at this stage and show little interaction with the midgut primordium. E-Cadherin, the small GTPase Rho1 and Gβ proteins are present uniformly at the cell periphery. Trapped in endoderm 1 (Tre1) signalling leads to the polarization of the PGCs, which take on a radial organization with the tails of the cells facing the inside of the cluster and the leading edges facing the midgut primordium. E-Cadherin, Rho1 and Gβ are redistributed to the tails of the cells. Next, the PGCs lose adhesion to each other and begin to extend cellular protrusions towards the epithelial cells of the midgut. b | Zebrafish. At specification, PGCs have a smooth, round morphology and do not posses migratory activity (3 hours post-fertilization (hpf)). PGCs begin to randomly extend small cellular protrusions in multiple directions at 3.5hpf. These protrusions disappear during mitosis. At 4.5hpf, PGCs become polarized, individualize and extend broad protrusions at the leading edge. This step is dependent on transcription and the Dead End protein, and is necessary for the cells to respond to stromal derived factor 1a (SDF-1a, also known as CXCL12a) chemokine signalling. c | Mouse. Following specification in the posterior primitive streak (embryonic day 7.5), PGCs have a smooth, round morphology. PGCs acquire a polarized morphology prior to initiating their migration into the endoderm. The molecular mechanisms regulating this polarization are not understood.
Recent studies have shown that the initiation of D. melanogaster PGC migration is regulated by the protein Trapped in endoderm 1 (Tre1), a G protein-coupled receptor (GPCR) of the rhodopsin family (TABLE 1)41. tre1 is expressed in germ cells and was initially identified as important to migration across the PMG epithelium by signalling through small G proteins and the GTPase Rho1 (discussed below)44. However, subsequent experiments have shown that Tre1 also acts earlier in regulating proper PGC polarization and dispersal41. Polarization of PGCs is concurrent with redistribution of the Gβ protein along with Rho1 and adherens junction components, such as D. melanogaster E-cadherin (DE-cad) and catenins, from the cell periphery to tails at the lagging edges of cells found within the cluster of PGCs (FIG. 2a, TABLE 1). Furthermore, reducing DE-cad levels in PGCs leads to premature PGC dispersal. However, this dispersal alone is not sufficient to promote PGC migration in the absence of Tre1, suggesting that Tre1 possesses additional functions in mediating the directed migration of PGCs, presumably though reception of an attractive signal (see section on migratory path of PGCs). As of yet, the link between Tre1 and the redistribution of Gβ, Rho1 and the adherens junction components remains unclear. Interestingly, Tre1 is closely related to the GPCR Moody, which is required within surface glia cells to regulate actin dynamics and cell polarization during the formation of the blood-brain barrier44-46. Therefore, the regulation of cell polarity might be a conserved function for this class of GPCRs.
Table 1. Genes regulating primordial germ cell migration (signalling and adhesion).
| Gene product (Molecular function) | Gene | Expression | Function | Refs |
|---|---|---|---|---|
| G-Protein Coupled Receptor Signalling | ||||
| Trapped in Epithelium 1 (GPCR) | tre1 (Dm) | Germ cells | dispersal, polarity, transepithelial migration | 41, 44 |
| Rho1 (GTPase) | Rho1 (Dm) | Uniform | dispersal, polarity, transepithelial migration | 41, 44 |
| SDF-1 (chemokine) | sdf-1a (Dr) | Target somatic tissues | Attractant | 75, 77, 78 |
| SDF-1 (Mm) | Genital ridges | Attractant | 86, 87 | |
| CXCR4 (GPCR) | cxcr4b (Dr) | Germ cells | Response to attractant | 75, 77, 78 |
| CXCR4 (Mm) | Germ cells | Response to attractant | 86, 87 | |
| CXCR7b (GPCR) | CXCR7b (Mm) | Somatic cells | Attractant turnover | 80 |
| G proteins (GPCR signalling) | Gβ13f, Gγ1 (Dm) | Germ cells | Transepithelial migration | 41 |
| Gαi (Dr) | Unknown | Response to attractant | 79 | |
| Other signalling pathways | ||||
| STAT (JAK-STAT signalling, Transcription Factor) | stat92E (Dm) | germ cells | Migration to mesoderm | 72, 73 |
| C-kit (Receptor tyrosine kinase) | C-kit (Mm) | germ cells | Migration and survival | 90, 101-103 |
| Steel (C-kit ligand) | Steel (Mm) | somatic cells surrounding germ cells | Migration and survival | 90, 101-103,54, 91 |
| Cell adhesion and polarity | ||||
| E-cadherin (adherens junction component) | shg (Dm) | Germ cells, gonad | Dispersal, polarity coalescence | 41, 96 |
| E-cad (Dr) | Germ cells | Initiation | 47 | |
| E-cad (Mm) | Germ cells | Migration | 92, 93 | |
| Integrin B1 (integrin) | Integrin B1 (Mm) | Germ cells, mesoderm | Migration to genital ridges | 94 |
Dr, Danio rerio; Dm, Drosophila melanogaster; E-cad, E-cadherin; GPCR, G Protein-Coupled Receptor; GTP, Guanosine-5′-Triphosphate; Mm, Mus musculus; SDF-1, Stromal Derived Factor 1; CXCR, Chemokine (CXC motif) Receptor; STAT, Signal Transducer and Activator of Transcription; JAK, Janus-Activated Kinase; shg, shotgun.
Zebrafish
In zebrafish, PGCs undergo multiple steps to acquire motility (FIG. 1b, 2b)47. Following their specification, zebrafish PGCs initially have a smooth, round morphology (at 3hpf). Approximately 30 minutes later, PGCs begin to randomly extend small cellular protrusions, but do not begin migrating and lose these protrusions as they undergo mitosis. One hour later (at 4.5hpf), PGCs extend broad protrusions and become polarized as the cells individualize and initiate directional migration, presumably in response to chemokine signalling from somatic cells (see section on migratory paths of PGCs)47.
Initiating migration requires de novo transcription in zebrafish PGCs. Cells treated with an RNA polymerase inhibitor are capable of randomly extending small cellular protrusion, but cannot extend broad protrusions or begin directional migration, presumably due to the requirement of zygotically transcribed gene products specific to this process47. Additionally, the activity of the RNA-binding protein Dead end (Dnd) is required for PGCs to start their migration (FIG. 2b, TABLE 2)47, 48. Knockdown of dnd by morpholino injection blocks polarization and migration of PGCs. Intriguingly, Dnd seems to function in part by regulating zebrafish E-cad during PGC individualization. Similar to D. melanogaster, zebrafish E-cad is normally downregulated as PGCs begin to polarize and disperse. However, this down-regulation does not occur in PGCs depleted of dnd, and cells remain in groups that maintain close cell-cell contacts48. The exact mechanism of how dnd regulates E-cad is unclear. This phenotype is also caused by E-cad overexpression in PGCs. More recent studies have shown that Dnd functions by counteracting the inhibitory function of the microRNAs, in particular miR-430, allowing the expression of PGC-specific proteins such as Nos and Tudor domain containing protein 7 (Tdrd7)49. How this relates to the mechanism of E-cad down-regulation and initiation of migration remains to be determined.
Table 2. Genes regulating primordial germ cell migration (lipid biology and survival).
| Gene product (Molecular Function) | Gene | Expression | Function | Refs |
|---|---|---|---|---|
| Lipid Biology | ||||
| Hmgcr (isoprenoid and cholesterol synthesis) | clb (Dm) | Mesoderm, gonad | Attraction to mesoderm | 68 |
| Hmgcr2 (Dr) | Uniform | Migration | 82 | |
| Hmgcr (Mm) | Uniform | Migration, Survival | 95 | |
| Farnesyl-diphosphate synthase (isoprenoid synthesis) | fpps (Dm) | Mesoderm, CNS, foregut, midgut | Attraction to mesoderm | 70 |
| Geranylgeranyl diphosphate synthase (isoprenoid synthesis) | qm (Dm) | Mesoderm, CNS, midgut | Attraction to mesoderm | 70 |
| Geranylgeranyl transferase (prenylation) | β-ggt1 (Dm) | Unknown | Attraction to mesoderm | 70 |
| ggt1 (Dr) | Uniform | Migration | 82 | |
| Multidrug resistance 49 (ABC transporter) | mdr49 (Dm) | Mesoderm | Attraction to mesoderm | 71 |
| Prenyl protease type 1 (prenyl processing) | ste24a,b,c (Dm) | Unknown | Attraction to mesoderm | 71 |
| Isoprenylcysteine carboxylmethyltransferase (prenyl processing) | ste14 (Dm) | Unknown | Attraction to mesoderm | 71 |
| Lipid phosphate phosphatase 3 (phospholipid hydrolysis, lipid uptake) | wunen, wunen2 (Dm) | Epidermis, CNS germ cells, midgut | Repulsion and survival | 58, 60,61, 63,62 |
| Cell survival | ||||
| Dead End (RNA regulation) | dnd (Dr) | Germ cells | Initiation, survival | 47, 48 |
| Dnd1 (Mm) | Germ cells | Survival | 53, 107 | |
| Bax | Bax (Mm) | germ cells | germ cell survival | 104 |
| Outsider (monocarboxylate transporter) | out (Dm) | unknown | germ cell programmed cell death | 99, 100 |
| p53 | p53 (Dm) | mesoderm, gut, germ cells | germ cell programmed cell death | 100 |
Bax, Bcl-2-associated X protein; clb, columbus; Dr, Danio rerio; Dm, Drosophila melanogaster; dnd; dead end; fpps, farnesyl-diphosphate synthase; ggt, geranylgeranyl transferase; Hmgcr, Hydroxy-3-methylglutaryl coenzyme A reductase; out, outsider; qm, quemao; ste, sterile
Mouse
In contrast to D. melanogaster and zebrafish, little is known about how PGC migration is initiated in the mouse. Mouse PGCs are initially identifiable in the posterior primitive streak (at E7.5; FIG. 1c, 2c)50. Soon thereafter, cells begin to exhibit polarized morphology and extend cytoplasmic protrusions as they initiate migration through the primitive streak into the adjacent posterior embryonic endoderm, extraembryonic endoderm and allantois50.
This initiation of mouse PGC migration was initially thought to be regulated by the interferon induced transmembrane protein 1 (IFITM1)51. IFITM proteins are cell surface proteins implicated in diverse cellular processes, including cell adhesion. Knockdown of IFITM1 by RNA interference in the primitive streak leads to failure of PGC migration into the endoderm, suggesting that IFITM1 expression functions to repel PGCs from the mesoderm into the endoderm. However, a more recent study in which the IFITM gene family was deleted from the embryo showed no defects in PGC migration, leaving the mechanism of migration initiation in mouse an open question52.
Although mouse possesses a Dnd homolog, it seems to be primarily required for PGC survival53. The C-kit receptor tyrosine kinase (RTK) and its ligand Steel are required for general PGC motility at E7.5 (see section on migratory paths of PGCs), although it seems that PGCs are still capable of initiating migration when this pathway is disrupted54.
Migratory paths of PGCs
Following initiation of a motility program and directional migration, PGC migration must be carefully regulated to promote the migration of PGCs through the developing embryo towards the SGPs. These migratory paths are regulated by a combination of attractive and repulsive signals (BOX 1), specifically tailored to individual steps of the migration process.
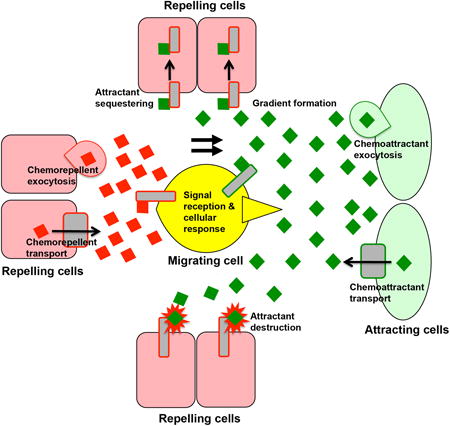
Box 1. Principals of attracting and repelling migrating cells.
Distant cells promote chemoattraction by secreting attractant molecules (see the figure). Possible mechanisms of secretion include exocytosis or the use of transmembrane transporters, such as members of the ABC protein family. Chemoattractive signals are received by transmembrane proteins, such as G-protein coupled receptors (GPCRs), expressed on the surface of migrating cells. Migrating cells are thought to read and decipher gradients of chemoattractant concentration, leading to polarized cell protrusions (in form of blebbing or lamellipodia formation) and directional migration towards the highest levels of chemoattractant. This is accomplished by localized polarization of migrating cells and cytoskeletal rearrangements brought about by downstream signalling effectors and small GTPases. For example, the GPCR Chemokine (CXC motif) Receptor 4 (CXCR4) mediates attraction of many cell types, including zebrafish and mouse primordial germ cells (PGCs) towards the chemoattractant Stromal Derived Factor 1 (SDF-1). Migrating cells can be repelled directly by the expression of a chemorepellent, which is detected by the migrating cell and avoided. The semaphorin family of proteins is one example of a diffusible protein that repels axons during nervous system development. However, this type of repulsion has not been seen in PGC migration. Alternatively, migrating cells can also be repelled indirectly by the sequestering or destruction of an attractant signal by another population of cells. This mechanism might promote the formation of finely tuned gradients of chemoattractants in space and developmental time. For example, a chemoattractant phospholipid is thought to be degraded by the proteins Wunen and Wunen2 during Drosophila melanogaster PGC migration. Similarly, the CXCR7b protein functions to sequester SDF-1 by endocytic uptake during zebrafish PGC migration. The combined effects of these diverse regulators of cell migration lead to the precise migratory paths observed during embryonic development. Figures adapted from Santos and Lehmann (2004) after drawings by Michelle Starz-Gaiano11.
D. melanogaster
D. melanogaster PGC migration lasts for approximately four hours and can be subdivided into three distinct steps: First, transepithelial migration across the midgut, second, reorientation to the dorsal side of the midgut and third, bilateral migration into the mesoderm toward the SGPs11, 13. Prior to active migration, PGCs are found in the pocket of the PMG primordium. As the PGCs begin to disperse, they extend protrusions towards the somatic cells of the midgut. During migration across the midgut epithelium, PGCs are polarized and actin is enriched at both the leading edge and tail41, 42. The rearrangement of the epithelial cells of the PMG also seems important, as ultrastructural and confocal analysis have shown transient deformations and intercellular gaps between these cells as the PGCs pass through42, 43. Supporting this idea, mutations that transform the PMG epithelium into more rigid hindgut epithelium, such as serpent and huckebein, prevent PGC migration42, 55, 56.
Migration across the PMG also depends on Tre1 (FIG. 3a, TABLE 1)41, 44. tre1 mutants display a complete defect in transepithelial migration, with the PGCs found clustered within the fully developed PMG late in development. Expression of a dominant negative form of the small GTPase Rho1 in PGCs leads to a similar defect in PGC migration, suggesting that Tre1 signalling leads to the activation of this cytoskeletal regulator44. Tre1 presumably functions in transepithelial migration by mediating an attractive response to an extracellular ligand. GPCR signalling has a widely appreciated role in other cell types of mediating the cellular response to attractive chemokines, often through redistribution of phosphoinositides and cytoskeletal reorganization1. The identity and location of the Tre1 ligand remains to be determined and should provide insight into how this pathway regulates PGC migration. Fatty acids act as ligands for the GPCR GPR84, the closest mammalian homolog of Tre1, during leukocyte migration. This is intriguing given the multiple roles of lipids in regulation of PGC migration (see below)57.
Figure 3. Molecular regulation of PGC migration paths.
a | Drosophila melanogaster. i. The G-protein coupled receptor (GPCR) Trapped in endoderm 1 (Tre1) regulates transepithelial migration of primordial germ cells (PGCs) through the midgut. Tre1 might regulate Rho1, triggering cytoskeletal changes necessary for migration. ii. Wunen and Wunen2 (Wun and Wun2) regulate migration into the mesoderm. Wun and Wun2 are expressed at sites that PGCs avoid, such as the ventral midgut, and in PGCs. Data suggest that Wun and Wun2 hydrolyze an extracellular phospholipid that functions as a PGC attractant and survival factor. iii. PGCs are attracted to somatic gonad precursors (SGPs) by the 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgcr) pathway, which adds a geranyl-geranyl (GG) group to a putative chemoattractant. Multidrug resistance 49 (Mdr49), an ABC transporter, is required for chemoattractant secretion. b | Zebrafish. PGCs expressing the GPCR Chemokine (CXC motif) Receptor 4b (CXCR4b) migrate towards the CXCR4b ligand, stromal derived factor 1a (SDF-1a), secreted by somatic cells. Another somatically-expressed GPCR, CXCR7b, promotes internalization and degradation of SDF-1a, which might lead to proper gradient formation and precise targeting of PGCs. Following PGC migration to an intermediate target, a new group of distant somatic cells begins expressing SDF-1a, directing PGCs to new targets. c | Mouse. PGC migration to the genital ridges is controlled by the GPCR CXCR4 and its ligand SDF-1. SDF-1 is expressed by the somatic cells of the genital ridge and PGCs express CXCR4. Integrin β1 is also required for this step. PGC motility and survival requires the receptor tyrosine kinase c-Kit and its ligand Steel. Steel is expressed by somatic cells surrounding PGCs throughout migration.
Following their migration into the mesoderm, PGCs move along the midgut into the posterior mesoderm (FIG. 1a). Once in the mesoderm, PGCs sort bilaterally and migrate toward the SGPs, which are specified in the lateral mesoderm (Supplementary information S2 (movie); stage 11, 7h AEL). These steps of migration are regulated by two related proteins with redundant functions, Wunen and Wunen2 (Wun and Wun2; FIG. 3a, TABLE 2). While loss of either of these gene products has a mild effect on PGC migration, removal of both from the somatic cells of the embryo leads to a dramatic disruption of PGC migration, with PGCs found scattered throughout the embryo late in development58-60. These genes are expressed in somatic cells in areas of the embryo that PGCs normally avoid, such as the ventral midgut, central nervous system (CNS) and epidermis. Conversely, overexpression of wun or wun2 in the mesoderm is sufficient to repel PGCs58, 60. Taken together, these data indicate that Wun and Wun2 are necessary and sufficient to repel PGCs. Forced exposure of PGCs to Wun or Wun2 leads to nonapoptotic cell death (discussed below)61, 62.
Wun and Wun2 encode lipid phosphate phosphatases (LPPs), which are transmembrane ectoenzymes that hydrolyze extracellular phospholipids58, 60. Wun and Wun2 activity specifically mediates hydrolysis and uptake of phosphatidic acid (PA) and lysophosphatidic acid (LPA) when transfected into insect Hi5 cells63. These phospholipids have been shown in other systems and cell types to function in intercellular signalling and promote cell migration63-65. However, the in vivo substrate for Wun and Wun2 activity during PGC migration has not been identified. The current model for Wun and Wun2 regulation of PGC migration is that they hydrolyze a phospholipid attractant molecule, thereby destroying its attractant function. The localized expression patterns of Wun and Wun2 create an inverse gradient of phospholipid attractant, and PGCs migrate towards the highest concentration of phospholipid61, 63, 66.
Interestingly, Wun and Wun2 activity is also required within PGCs for proper migration and survival. Loss of Wun and Wun2 expression within PGCs leads to a failure of migration and extensive PGC death shortly after transepithelial migration61, 63. These data, in addition to in vitro studies demonstrating that Wun and Wun2 activity promotes the uptake of hydrolyzed lipid into cells, has led to the model that somatic cells and PGCs compete for the same extracellular phospholipid. PGCs require this substrate for their migration and survival, while somatic cells locally deplete the lipid and therefore create an environment that is not permissive for PGCs13, 66. A recent study by Steinhauer et al demonstrates a role for lysophospholipid acyltransferases in D. melanogaster PGC migration. Genetic interactions suggest a shared pathway with Wun and Wun267.
During their final step of migration D. melanogaster PGCs move into the lateral mesoderm to meet with the SGPs. Molecularly, this step is regulated by the 3-hydroxy-3-methylglutaryl coenzyme A reductase (Hmgcr) enzymatic pathway (FIG. 3a. TABLE 2). Mutation in Hmgcr (also known as columbus) leads to defects in PGC migration to the lateral mesoderm and SGPs68. Conversely, ectopic expression of Hmgcr in the nervous system or ectoderm is sufficient to attract PGCs. In situ analysis determined that Hmgcr is expressed in the lateral mesoderm and enriched in the SGPs, consistent with a role in the production of a PGC attractant.
Hmgcr is responsible for the synthesis of mevalonate, an essential intermediate in the metabolic pathway that produces cholesterol69. However, analysis of the D. melanogaster genome has determined that the fly lacks enzymes required for this process, suggesting cholesterol-independent roles for Hmgcr70. Indeed, the formation of isoprenoids, an alternative branch of the Hmgcr pathway, is required for PGC migration. Isoprenylation describes a post-translational lipid modification that involves the covalent attachment of farnesyl or geranyl-geranyl groups to the carboxyl terminus of a protein. Mutations in other enzymes of the isoprenylation pathway lead to a PGC migration defect similar to Hmgcr mutants, strongly suggesting that isoprenylation of a PGC attractant occurs in the SGPs (TABLE 2)70.
Recent insight into the mechanism of Hmgcr function in PGC migration has come from studies demonstrating that the multidrug resistance 49 (mdr49) gene product is important for this process (TABLE 2). mdr49 encodes an ATP-binding cassette (ABC) transporter, a family of proteins that regulate the export of farnesyl-modified mating factors in yeast. mdr49 is expressed in the mesoderm, and mdr49 mutants have defects in PGC migration71. mdr49 genetically interacts with Hmgcr, supporting a model in which these gene products function in a pathway to produce and export a PGC attractant. This study also suggested that D. melanogaster homologs of yeast Sterile24 (Ste24), a prenyl protease type 1 and Sterile14 (Ste14), an isoprenylcysteine carboxylmethyltransferase, other components of the isoprenylation pathway, regulate PGC migration71. Importantly, this study utilized an adapted in vitro transwell migration assay to demonstrate that expression of Hmgcr and mdr49 in cultured cells is sufficient to attract PGCs independently of other embryonic cues. The exact identity of the PGC attractant regulated by these gene products remains to be discovered and should provide insight to the exact mechanisms of this final step of PGC migration.
A role for the Janus-activated kinase (JAK)–signal transducer and activator of transcription (STAT) signalling pathway in PGC migration through the mesoderm has also been indicated (TABLE 1). Two Unpaired family members (Upd and Upd3), which are ligands for this pathway, as well as the STAT92E transcription factor are expressed in PGCs72, 73. Mutations removing upd ligands, stat92E or the JAK–STAT receptor domeless (dome) all had defects in migration of PGCs to the SGPs72, 73. Furthermore, constitutive activation of the Torso RTK seems to activate STAT and lead to premature PGC migration during gastrulation72. This pathway seems to function by promoting the migratory behaviour of PGCs, such as the formation of cellular protrusions, but does not have an instructive role by providing a directional cue73.
Zebrafish
Zebrafish PGCs make their way to the gonad by a complex migration path through six distinct migration steps using intermediate targets throughout the embryo (6 hpf-24 hpf)74: migration to the dorsal side of the embryo, exclusion from the dorsal midline, alignment with the anterior and lateral mesoderm, the formation of two lateral PGC clusters at somite 1-3, anterior migration of trailing PGCs to join the main PGC clusters and posterior positioning of PGC clusters at somite 8 (Supplementary information S3 (movie)). During migration, PGCs alternate between migratory ‘run’ phases as they move between targets and stationary ‘tumble’ phases in which they lose their polarity at intermediate targets75. Although cells move as a cluster at each step, careful analysis has revealed that cells move individually. Unlike many migratory cells, zebrafish PGCs do not exhibit increased actin polymerization within the advancing cellular protrusion76. Instead, Myosin-dependent contractility at the cell cortex generates local hydrostatic pressure or ruptures in the cortex that lead to membrane detachment from the cytoskeleton and flow of cytoplasm that expands directed cellular protrusions (known as membrane blebbing). The conservation of these cell behaviours with migrating PGCs in other organisms awaits further study.
The main molecules guiding zebrafish PGC migration are Stromal Derived Factor 1 (SDF-1, also known as Chemokine (CXC motif) Ligand 12 (CXCL12)) and its receptor, the GPCR Chemokine (CXC motif) Receptor 4b (CXCR4b), which is expressed in PGCs (FIG. 3b, TABLE 1)77, 78. The migratory path of PGCs is tightly correlated with the dynamic somatic expression of SDF-1, which marks intermediate and final targets of migration75. Furthermore, expression of SDF-1 is sufficient to attract PGC to ectopic positions in the embryo. Loss of either SDF-1 or CXCR4b does not disrupt migratory activity of PGCs but instead leads to random migration through the embryo. Downstream of CXCR4b, the G protein Gαi is required for PGC migration (TABLE 1)79. Further downstream factors that regulate the cellular response to chemokine signalling remain to be identified.
A recent study has shed light on how the proper distribution of SDF-1 in the embryo is regulated80, 81. A second SDF-1 receptor, CXCR7b, which is also required for proper PGC migration, functions mainly in somatic tissues and is uniformly distributed throughout the embryo (FIG. 3b, TABLE 1). Cells expressing CXCR7b show enhanced internalization of SDF-1, and knockdown of CXCR7b suggests that it is required to establish a gradient of SDF-1a activity. In contrast to CXCR4b, which is localized at the membrane, CXCR7b is localized in intracellular structures that colocalize with both SDF-1 and a lysosomal marker. This observation suggests that CXCR7b functions by mediating continuous clearing of the ligand from somatic tissues, providing a mechanism for achieving fast SDF-1 turnover and precise spatial and temporal control of its activity and resultant PGC migration. Consistent with this model, PGC migration defects are suppressed by simultaneously reducing SDF-1 and CXCR7b levels.
The Hmgcr pathway also has a role in zebrafish PGC migration (TABLE 2)82. As in D. melanogaster, the isoprenoid branch of the pathway seems to be required, as inhibiting either Hmgcr or geranylgeranyl transferase disrupts PGC migration. Recent data suggest a role for the Hmgcr pathway in the geranylation of the Gγ subunits required directly for GPCR signalling in zebrafish PGCs83. Additional experiments are required to determine whether geranylation also affects the guidance of germ cells by the soma similar to D. melanogaster.
Mouse
The initial step in mouse PGC migration is the movement of cells from the posterior primitive streak to the endoderm (E7.5)50. Following subsequent migration within the hindgut during its anterior extension (E8-E9.5), mouse PGCs follow a path remarkably similar to D. melanogaster, in which they migrate through hindgut tissue to the mesoderm, followed by bilateral migration to the gonadal ridges and gonad formation (Supplementary information S4 (movie); E10.5-11.5)84. As in D. melanogaster, the gut seems to have an important role in the regulation of this process. Removal of the SRY (sex determining region Y)-box 17 (Sox17) transcription factor prevents proper expansion of hindgut endoderm. In these mutants, PGCs fail to migrate properly to the genital ridges and instead scatter in the extraembryonic endoderm85.
Similar to zebrafish, SDF-1 and CXCR4 (mammals only posses one CXCR4 protein) function as an attractant system for mouse PGCs (FIG. 3c, TABLE 1)86, 87. This signalling pathway seems to be dispensable for migration out of the endoderm but is specifically required for later stages of PGC migration to the genital ridge. SDF-1 is expressed at the genital ridges and in the surrounding mesenchyme, while CXCR4 is expressed within the PGCs. Removal of either SDF-1 or CXCR4 leads to very few PGCs reaching the genital ridge, while ectopic expression of SDF-1 causes PGCs to migrate to new locations86, 87.
The c-Kit RTK and its ligand Steel have long been appreciated for their roles in PGC proliferation, migration and survival. During the initial characterizations of mice mutant for Kit and Steel, some PGCs were found outside of the gonad 88,89. Further studies also suggested that the Kit–Steel interaction is required for PGCs to move along the endoderm of the hindgut 90. Recent studies have clarified the specific migratory role of these factors (FIG. 3c, TABLE 1)54, 91. Rather than providing a directional cue, Steel and c-Kit are thought to regulate general PGC motility, as removal of Steel function leads to PGCs that migrate in the proper direction, but at a greatly reduced rate54. This phenotype is reminiscent of disruption in JAK–STAT signalling in D. melanogaster (see above)73. Consistent with their roles in PGC motility, PGCs express c-Kit protein, while surrounding somatic cells expressing Steel throughout all stages of their migration.
In addition to signalling, there is also evidence for adhesion molecules having a role in mouse PGC migration. E-cad is expressed in PGCs as they migrate out of the hindgut, and disrupting E-cad function causes problems with PGC-PGC adhesion and causes PGCs to be left outside the gonad (TABLE 1)92, 93. PGCs also express integrin β1, which is required for proper PGC migration out of the hindgut into the genital ridges(FIG. 3c, TABLE 1)94. Previously another member of the IFITM family, IFITM3, was thought to regulate PGC migration out of the hindgut, based on gene knockdown using RNAi51. However, as with IFITM1, data from a targeted knockout 52.
Finally, there is recent evidence of a role for the Hmgcr pathway in mouse PGC migration (TABLE 2)95. Inhibition of Hmgcr in a tissue culture system impairs germ cell migration. Interestingly, in the mouse cholesterol synthesis seems to be involved in this process, as both cholesterol and isoprenoids are required to rescue this phenotype. Additionally, cholesterol was found to be enriched in the genital ridges, further suggesting a potential role in PGC migration. The in vivo role of the Hmgcr pathway in mouse PGC migration remains to be clarified and will benefit from a targeted knockout approach and further gene expression analysis.
Stopping PGC migration
At the end of their migration, PGCs presumably lose their motile properties as they associate with somatic cells to form the gonad. Evidence for this model comes from D. melanogaster, in which PGCs round up and become non-motile, cluster together and form tight contacts with each other and the somatic cells of the gonad42. An important outstanding question concerns the mechanisms by which PGCs cease migrating once they reach the gonad. Evidence from D. melanogaster and zebrafish supports the simple model that PGCs stop directional migration at the site of highest attractant expression. In D. melanogaster, SGPs express high levels of Hmgcr at the site where PGCs stop migrating, and ectopic expression of Hmgcr in other tissues leads to both attraction of PGCs and migration arrest at those tissues68. Similarly in zebrafish, regions of high SDF-1a seem to dictate where PGCs stop75. This is evident not only at the somatic gonad, but also at intermediate targets sites, where PGCs temporarily lose their directional migration until a new region of somatic cells begins to express SDF-1a and initiates further migration.
In addition to a loss of directional migration caused by PGCs reaching the site of highest attractant expression, it seems likely that the inherent motile behaviour of PGCs needs to be suppressed for proper gonad formation. Although the molecular mechanisms of suppressing the motility of germ cells are unclear, it is likely that cell-cell contacts between PGCs and somatic cells are important for this process. Supporting this model, both DE-cad and Fear of intimacy (Foi), a Zn transporter, are required for gonad coalescence and compaction in D. melanogaster96-98. However, initial PGC–soma interactions are unperturbed in these mutants, suggesting that additional factors mediate this process. Further genetic and imaging approaches, are needed to determining how PGCs stop migrating.
Once PGCs reach the gonad, stop migrating, and associate with somatic cells of the gonad, they begin to fulfill their functions as germ cells by acquiring sex-specific morphologies. Currently there does not seem to be any evidence for sex-specific differences during germ cell migration in any organism. A subset of germ cells in the gonad acquire the ability to function as germline stem cells, which undergo meiosis to produce sperm and eggs and promote the next generation of embryonic development and PGC migration.
PGC migration and survival
A continuous theme through studies of PGC migration is the tight linkage between proper migration and PGC survival. Evidence from D. melanogaster suggests that not all PGCs specified at early embryogenesis successfully migrate to the gonads99. The elimination of PGCs that mismigrate seems to be a priority in each organism, presumably due to the importance of preventing deleterious effects of PGC trans-differentiation at ectopic locations in the body. Supporting this hypothesis, in humans ectopic PGCs often correlate with the locations of where germ cell tumours arise14. Therefore, understanding the mechanisms of PGC death and its relationship to migration has medical relevance.
As mentioned above, D. melanogaster wun and wun2 are closely associated with PGC survival (TABLE 2). Loss of wun2 activity in PGCs or high expression of wun or wun2 in somatic cells both lead to PGC death61-63. Interestingly, this death does not seem to require apoptosis. However, programmed cell death also has a role in removing PGCs that mismigrate. This process depends on the monocarboxylate transporter Outsiders as well as the p53 tumour suppressor gene (TABLE 2)99, 100. Mutation of either of these genes leads to excess PGCs that are found outside of the gonads.
In vertebrates, many of the same genes required for PGC migration also have roles in survival. Both zebrafish and mouse Dnd are required to prevent PGC death in the late stages of embryogenesis (TABLE 2)48, 53. In the mouse, Steel is required for PGC survival and loss of Steel from the midline during late stages of development leads to the death of any PGCs remaining (TABLE 1)91, 101-103. Downstream of Steel–Kit signalling, this elimination of ectopic PGCs is dependent on the gene Bax (TABLE 2)104. Bax is a member of the Bcl2 protein family, and upon activation promotes the release of pro-apoptotic factors from mitochondria, caspase activation and the progression of apoptosis105, 106.
We propose three non-exclusive models for why ectopic PGCs die in the embryo. First, these PGCs might lack an essential growth factor, such as SDF-1 or the Wun and Wun2-regulated phospholipid. Second, the differentiation program of PGCs might require an interaction with somatic gonad cells, and PGCs cells might die in the absence of proper differentiation. Third, ectopic PGCs might transdifferentiate and begin to exhibit somatic characteristics, and subsequently die due to disrupted cellular function. A failure to die in this later case can lead to drastic consequences, as in mice harbouring the Ter mutation in Dnd, which leads to germline teratomas53. The incidence of these tumours is increased in Bax mutants, highlighting the importance of eliminating ectopic or dysfunctional PGCs107.
Cell adhesion during PGC migration
Another important theme in the study of PGC migration is a role for cell-cell adhesion. Most apparently, there are multiple roles found for E-cad, specifically in the initiation of D. melanogastor and zebrafish PGC migration, as well as the cessation of D. melanogastor PGC migration (FIG. 2a-b, TABLE 1)41, 48, 96. E-cad is a crucial component of adherens junctions, which function at cellular junctions to link cells together108. These data suggest that cell adhesion represents an important mechanism for both starting and stopping PGC migration. Less is known about the types of cell-cell interactions that regulate the migratory paths of PGCs. In all three organisms, PGCs must migrate through a variety of tissue types such as epithelial endoderm and mesoderm. In mouse, integrins are important for PGC migration, presumably through an interaction with other cells or the extracellular matrix (FIG. 3c, TABLE 1). Supporting this idea, mouse PGCs might use fibronectin as a substrate for their migration109. Integrins seem dispensible for D. melanogastor PGC migration, although the motility of PGCs cultured in vitro increases on laminin-coated surfaces42, 110. In a recent study Erez Raz and colleagues provide evidence that zebrafish PGCs use the retrograde flow of actin-rich structures for the generation of E-cadherin mediated forces that provide traction between the germ cells and the surrounding tissue111. Future studies concerning PGC interactions with other cells and/or ECM provide an exciting avenue for further research.
Conclusions and future perspectives
PGC migration in D. melanogaster, zebrafish and mouse involves significant differences in the rate at which the cells move and the distances they need to travel13. For example, mouse PGCs must migrate a greater distance through a larger embryo over a longer developmental time than D. melanogastor PGCs. These differences might help explain divergent strategies for achieving proper PGC migration in these organisms. Despite these differences, there are several striking similarities and general themes linking this process. For example, in each of these organisms PGCs possess an inherent motility, often mediated by RTK signalling, but require external factors to impart directionality, such as chemokines that signal through GPCRs. The loss of PGC-PGC adhesion, often mediated by the regulation of the adhesion molecule E-cad, is also closely correlated with the acquisition of directional migration41, 47. However, the causal relationship between loss of adhesion and initiation of migration remains to be clearly demonstrated. Future genetic studies should lead to the identification of further intrinsic and extrinsic factors guiding the initiation, migratory paths and termination of PGC migration.
It is clear that GPCRs represent a class of molecules crucial for PGC migration. These seven-pass transmembrane proteins have been shown to have important roles in many types of migrating cells, generally through the reception of extracellular attractive signals1. In D. melanogaster, GPCR signalling seems to be limited to the earlier steps of dispersal and transepithelial migration mediated by Tre141, 44. In zebrafish GPCR signalling by CXCR4 seems to be the main pathway regulating PGC migration75, 77, 78, while in mouse it has a role in the final steps of migration86, 87. The discovery of a novel role for CXCR7b in regulating chemoattractant distribution in zebrafish highlights that there is still much to learn about the roles of GPCRs and dynamic regulation of ligand distribution during PGC migration80.
The sequestration of SDF-1 by CXCR7b in zebrafish is also reminiscent of the proposed function of Wun and Wun2 in destroying a phospholipid chemoattractant66. In both cases, these gene products function by promoting the proper distribution of a chemoattractant and prevent migration of PGCs to improper places in the embryo. Further studies of how chemoattractant gradients are established and maintained by these molecules will allow better understanding into this intriguing mechanism of regulating cell migration (BOX 1).
Another important theme of PGC migration is the importance of lipids and lipid modifications to this process. The Hmgcr enzymatic pathway has been linked to PGC migration in D. melanogaster, zebrafish and mouse68, 70, 71, 82, 95. This pathway is responsible for adding lipid moieties to proteins, which can regulate signalling properties and might be important in the generation of a chemoattractant. Furthermore, Wun and Wun2 function by hydrolyzing phospholipids and have also been shown to promote the uptake of the dephosphorylated lipids58, 60, 61, 63. The identities of both the Hmgcr-modified chemoattractant and the phospholipid hydrolyzed by Wun and Wun2 remain unknown and are crucial next steps in our understanding of PGC migration. Furthermore, interplay between these two pathways should be examined.
The migration of PGCs differs from many other well characterized types of cell migration such as fibroblasts. PGC migration most closely resembles amoeboid migration as well as the migration of immune cells. This is characterized by individually migrating cells with a broad leading edge, highly dynamic morphology and low adhesiveness. This type of cell migration may be optimized for cell movement through diverse external environments composed of various tissues. In particular, PGCs share many features with both migrating leukocytes and certain types of metastatic cells112, 113. Intriguingly, the SDF-1–CXCR4 pathway as well as phospholipid signalling through S1P and its receptors are important to the migration of these cell types. Therefore, continued studies of PGC migration should help uncover other mechanisms that have relevance to human health and disease. These future studies should focus on utilizing creative genetic approaches, recent advances in imaging techniques, and development of new in vivo and in vitro assays to further promote our understanding of the mechanisms guiding PGC migration.
Supplementary Material
Supplementary information S1 (movie) | in vivo timelapse of primordial germ cell migration through the midgut in Drosophila melanogaster. Wild type stage 9-10 (∼4-5.5h after egg laying) D. melanogaster embryo with primordial germ cells (PGCs) labelled by an Enhanced Green Fluorescent Protein–Moesin fusion protein. PGCs are initially clustered and highly polarized, but lose adhesion with each other and begin to disperse and migrate through the surrounding posterior midgut cells. Images were taken every 2 min with a 63× objective. The playback rate is three frames per second. The embryo is oriented with its anterior to the left and dorsal side up. Movie is reproduced, with permission, from Kunwar, P.S. et al. J Cell Biol 183, 157-168 (2008).
Supplementary information S2 (movie) | in vivo timelapse of bilateral migration of primordial germ cells into the mesoderm in Drosophila melanogaster. Wild-type stage 10-11 (∼5.5-7h after egg laying) D. melanogaster embryo with primordial germ cells (PGCs) labelled by an Enhanced Green Fluorescent Protein–Moesin fusion protein. PGCs migrate laterally from the midline of the embryo. Images were taken every 2 min with a 40× objective. The playback rate is six frames per second. The embryo is oriented with its anterior to the top and dorsal side up. Movie is reproduced, with permission, from Sano, H., Renault, A.D. & Lehmann, R. J Cell Biol 171, 675-683 (2005).
Supplementary information S3 (movie) | in vivo timelapse of primordial germ cell migration in zebrafish. Wild type zebrafish embryo with primordial germ cells PGCs labelled by farnesylated (membrane-bound) GFP and the nuclei of all cells labelled by H2B-mCherry. 1s of playback represents approximately 25 minutes of development. The embryo is shown from a dorsal view with the anterior at the upper right of the frame. Courtesy of Azadeh Paksa and Erez Raz, University of Münster, and Nadine Peyriéras, CNRS-DEPSN.
Supplementary information S4 (movie) | in vivo timelapse of primordial germ cell migration in mouse. Wild type organ culture at embryonic day 9.5 with primordial germ cells (PGCs) labelled by Oct4ΔPE∷GFP+. Transverse slices from the hindgut regions of mouse embryos were cultured and filmed as described in reference 1. PGCs migrate out of the dorsal aspect of the hindgut, separate into two bilateral streams and migrate into the dorsal ridges. The movie covers about 12h of migration beginning at E9.5. The culture is oriented with its dorsal side to the left of the frame. Courtesy of Brian Dudley and Kathleen Molyneaux, Department of Genetics, Case Western University.
Online summary.
Primordial germ cell (PGC) migration provides a useful system for studying a group of individually migrating cells in vivo.
PGC migration in all species follows similar steps: initiation of polarity and directed migration, regulated migration by attractive and repulsive cues, and termination of migration at the site of gonad formation.
PGCs frequently utilize G protein-coupled receptor signalling to reach their targets tissues, a mechanism found in many types of migrating cells.
Lipids have an essential role in regulating PGC migration and seem to work both directly as chemoattractants and by modifying and activating protein chemoattractants.
Cell adhesion molecules, in particular cadherins, have important roles in several steps of PGC migration, such as initiation of migration, migrating through somatic tissues, and cessation of migration and gonad coalescence.
The migration of PGCs is closely linked with their survival, and PGCs that do not properly migrate to the gonad are usually eliminated through cell death. However, mechanisms of PGC death might differ between species.
Acknowledgments
We thank members of the Lehmann laboratory for comments on the manuscript, and A. Paksa, N. Peyriéras and E. Raz for the zebrafish movies; and B. Dudley and K. Molyneaux for sharing the mouse movie. B. Richardson is supported by NIH/NRSA fellowship F32HD062160. Our work on PGC migration is supported by NIH grant RO1HD041900. R. Lehmann is an HHMI investigator.
Glossary terms
- Gastrulation
A crucial step in animal development in which the layout of the embryo is dramatically restructured by cell migration to form the three germ layers, ectoderm, mesoderm and endoderm
- Primordium
An organ or tissue at its earliest stages of development
- Extraembryonic ectoderm
A cell layer in mouse that lies outside of the embryo and eventually differentiates to form the chorion
- Visceral endoderm
An extraembryonic cell layer that covers the early mouse embryo and has important signalling functions during development
- Epiblast
The inner layer of the developing vertebrate embryo that gives rise to the fetus
- Posterior midgut (PMG) pocket
A luminal structure in the developing Drosophila melanogaster embryo formed by the midgut primordium during gastrulation
- Adherens junction
A protein complex found at cell-cell junctions in epithelial tissues, composed of catenins, cadherins and actin filaments
- Primitive streak
The site of gastrulation in many vertebrates, including mouse, where precursors to the mesoderm and endoderm ingress into the embryo
- Allantois
An extraembryonic membrane formed near the hindgut of mammalian embryos that is important collecting embryonic waste and for development of the umbilical cord and placenta
- Somite
Mesodermal structure found on either side of the neural tube in vertebrate embryos that eventually give rise to muscle, skin and vertebrae and are often used to stage embryos
- Genital ridge
Mesodermal precursor to the somatic gonads in vertebrate embryos (also known as gonadal ridge)
- Apoptosis
A process of programmed cell death characterized by DNA fragmentation and loss of membrane integrity
Biographies
Brian Richardson received his Ph.D. from the Weill Graduate School of Medical Sciences at Cornell University, under the mentorship of Mary Baylies. He joined Ruth Lehmann's lab in 2008 to study the role of phospholipid signalling in Drosophila melanogaster primordial germ cell migration.
Ruth Lehmann, a HHMI Investigator, is the director of the Skirball Institute and the Laura and Isaac Perlmutter Professor of Cell Biology at NYU School of Medicine. Before moving to NYU in 1996 she was a member of the Whitehead Institute and an Associate Professor of Biology at MIT. She received her PhD. in 1985 with Christiane Nüsslein-Volhard at the MPI in Tübingen, Germany, studying the role of maternal genes in establishing the embryonic axes in Drosophila melanogaster. Her laboratory studies RNA regulation and germ line development in D. melanogaster.
References
- 1.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–6. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 3.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–84. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webb DJ, Zhang H, Horwitz AF. Cell migration: an overview. Methods Mol Biol. 2005;294:3–11. [PubMed] [Google Scholar]
- 5.Franz CM, Jones GE, Ridley AJ. Cell migration in development and disease. Dev Cell. 2002;2:153–8. doi: 10.1016/s1534-5807(02)00120-x. [DOI] [PubMed] [Google Scholar]
- 6.Keller R. Cell migration during gastrulation. Curr Opin Cell Biol. 2005;17:533–41. doi: 10.1016/j.ceb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro C, Petit V, Affolter M. Signaling systems, guided cell migration, and organogenesis: insights from genetic studies in Drosophila. Dev Biol. 2003;260:1–8. doi: 10.1016/s0012-1606(03)00211-2. [DOI] [PubMed] [Google Scholar]
- 8.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–39. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 9.Rossant J, Howard L. Signaling pathways in vascular development. Annu Rev Cell Dev Biol. 2002;18:541–73. doi: 10.1146/annurev.cellbio.18.012502.105825. [DOI] [PubMed] [Google Scholar]
- 10.Raz E. Guidance of primordial germ cell migration. Curr Opin Cell Biol. 2004;16:169–73. doi: 10.1016/j.ceb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Santos AC, Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr Biol. 2004;14:R578–89. doi: 10.1016/j.cub.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Molyneaux K, Wylie C. Primordial germ cell migration. Int J Dev Biol. 2004;48:537–44. doi: 10.1387/ijdb.041833km. [DOI] [PubMed] [Google Scholar]
- 13.Kunwar PS, Siekhaus DE, Lehmann R. In vivo migration: a germ cell perspective. Annu Rev Cell Dev Biol. 2006;22:237–65. doi: 10.1146/annurev.cellbio.22.010305.103337. [DOI] [PubMed] [Google Scholar]
- 14.Gobel U, et al. Germ-cell tumors in childhood and adolescence. GPOH MAKEI and the MAHO study groups. Ann Oncol. 2000;11:263–71. doi: 10.1023/a:1008360523160. [DOI] [PubMed] [Google Scholar]
- 15.Schneider DT, et al. Multipoint imprinting analysis indicates a common precursor cell for gonadal and nongonadal pediatric germ cell tumors. Cancer Res. 2001;61:7268–76. [PubMed] [Google Scholar]
- 16.Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–27. doi: 10.1242/dev.022434. [DOI] [PubMed] [Google Scholar]
- 17.Nance J, Priess JR. Cell polarity and gastrulation in C. elegans. Development. 2002;129:387–97. doi: 10.1242/dev.129.2.387. [DOI] [PubMed] [Google Scholar]
- 18.Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell. 2008;132:559–62. doi: 10.1016/j.cell.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Leatherman JL, Levin L, Boero J, Jongens TA. germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr Biol. 2002;12:1681–5. doi: 10.1016/s0960-9822(02)01182-x. [DOI] [PubMed] [Google Scholar]
- 20.Jongens TA, Ackerman LD, Swedlow JR, Jan LY, Jan YN. Germ cell-less encodes a cell type-specific nuclear pore-associated protein and functions early in the germ-cell specification pathway of Drosophila. Genes Dev. 1994;8:2123–36. doi: 10.1101/gad.8.18.2123. [DOI] [PubMed] [Google Scholar]
- 21.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–3. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–9. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 23.Deshpande G, Calhoun G, Schedl P. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development. 2004;131:1247–57. doi: 10.1242/dev.01004. [DOI] [PubMed] [Google Scholar]
- 24.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–65. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 25.Asaoka M, Sano H, Obara Y, Kobayashi S. Maternal Nanos regulates zygotic gene expression in germline progenitors of Drosophila melanogaster. Mech Dev. 1998;78:153–8. doi: 10.1016/s0925-4773(98)00164-6. [DOI] [PubMed] [Google Scholar]
- 26.Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 1999;99:271–81. doi: 10.1016/s0092-8674(00)81658-x. [DOI] [PubMed] [Google Scholar]
- 27.Schaner CE, Deshpande G, Schedl PD, Kelly WG. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 2003;5:747–57. doi: 10.1016/s1534-5807(03)00327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–65. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 29.Raz E. Primordial germ-cell development: the zebrafish perspective. Nat Rev Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 30.Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–88. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–85. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bontems F, et al. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19:414–22. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 33.Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, et al. Germ cell-less expression in zebrafish embryos. Dev Growth Differ. 2006;48:333–8. doi: 10.1111/j.1440-169X.2006.00868.x. [DOI] [PubMed] [Google Scholar]
- 35.Saga Y. Mouse germ cell development during embryogenesis. Curr Opin Genet Dev. 2008;18:337–41. doi: 10.1016/j.gde.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Saitou M, Payer B, O'Carroll D, Ohinata Y, Surani MA. Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle. 2005;4:1736–40. doi: 10.4161/cc.4.12.2209. [DOI] [PubMed] [Google Scholar]
- 37.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–13. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 38.Vincent SD, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–25. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 39.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–22. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 40.Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7:3514–8. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- 41.Kunwar PS, et al. Tre1 GPCR initiates germ cell transepithelial migration by regulating Drosophila melanogaster E-cadherin. J Cell Biol. 2008;183:157–68. doi: 10.1083/jcb.200807049. Demonstrates a link between G protein-coupled receptor signalling and PGC adhesion and polarity, mediated by E-Cadherin, at the initiation of migration in D. melanogaster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaglarz MK, Howard KR. The active migration of Drosophila primordial germ cells. Development. 1995;121:3495–503. doi: 10.1242/dev.121.11.3495. [DOI] [PubMed] [Google Scholar]
- 43.Callaini G, Riparbelli MG, Dallai R. Pole cell migration through the gut wall of the Drosophila embryo: analysis of cell interactions. Dev Biol. 1995;170:365–75. doi: 10.1006/dbio.1995.1222. [DOI] [PubMed] [Google Scholar]
- 44.Kunwar PS, Starz-Gaiano M, Bainton RJ, Heberlein U, Lehmann R. Tre1, a G protein-coupled receptor, directs transepithelial migration of Drosophila germ cells. PLoS Biol. 2003;1:E80. doi: 10.1371/journal.pbio.0000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bainton RJ, et al. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123:145–56. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 46.Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in drosophila. Cell. 2005;123:133–44. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 47.Blaser H, et al. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–38. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- 48.Weidinger G, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–34. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 49.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–86. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–8. doi: 10.1016/s0925-4773(99)00271-3. Describes the first in-depth analysis of PGC migration in mouse using live imaging. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PP. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell. 2005;9:745–56. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 52.Lange UC, et al. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol Cell Biol. 2008;28:4688–96. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Youngren KK, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–4. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development. 2009;136:1295–303. doi: 10.1242/dev.030619. Clarifies the role of the Steel–c-Kit signalling pathway in regulating PGC migration in the mouse using a PGC-specific reporter line in live embryos. It determines that Steel expression continuously surrounds PGCs throughout their migration and promotes general motility, but does not provide directional information. [DOI] [PubMed] [Google Scholar]
- 55.Warrior R. Primordial germ cell migration and the assembly of the Drosophila embryonic gonad. Dev Biol. 1994;166:180–94. doi: 10.1006/dbio.1994.1306. [DOI] [PubMed] [Google Scholar]
- 56.Moore LA, Broihier HT, Van Doren M, Lunsford LB, Lehmann R. Identification of genes controlling germ cell migration and embryonic gonad formation in Drosophila. Development. 1998;125:667–78. doi: 10.1242/dev.125.4.667. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Wu X, Simonavicius N, Tian H, Ling L. Medium-chain fatty acids as ligands for orphan G protein-coupled receptor GPR84. J Biol Chem. 2006;281:34457–64. doi: 10.1074/jbc.M608019200. [DOI] [PubMed] [Google Scholar]
- 58.Zhang N, Zhang J, Purcell KJ, Cheng Y, Howard K. The Drosophila protein Wunen repels migrating germ cells. Nature. 1997;385:64–7. doi: 10.1038/385064a0. [DOI] [PubMed] [Google Scholar]
- 59.Zhang N, Zhang J, Cheng Y, Howard K. Identification and genetic analysis of wunen, a gene guiding Drosophila melanogaster germ cell migration. Genetics. 1996;143:1231–41. doi: 10.1093/genetics/143.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starz-Gaiano M, Cho NK, Forbes A, Lehmann R. Spatially restricted activity of a Drosophila lipid phosphatase guides migrating germ cells. Development. 2001;128:983–91. doi: 10.1242/dev.128.6.983. [DOI] [PubMed] [Google Scholar]
- 61.Hanyu-Nakamura K, Kobayashi S, Nakamura A. Germ cell-autonomous Wunen2 is required for germline development in Drosophila embryos. Development. 2004;131:4545–53. doi: 10.1242/dev.01321. [DOI] [PubMed] [Google Scholar]
- 62.Sano H, Renault AD, Lehmann R. Control of lateral migration and germ cell elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J Cell Biol. 2005;171:675–83. doi: 10.1083/jcb.200506038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renault AD, Sigal YJ, Morris AJ, Lehmann R. Soma-germ line competition for lipid phosphate uptake regulates germ cell migration and survival. Science. 2004;305:1963–6. doi: 10.1126/science.1102421. Along with reference 61, it demonstrates that D. melanogaster PGCs and somatic cells compete for a phospholipid that is required for both PGC migration and survival. [DOI] [PubMed] [Google Scholar]
- 64.Burnett C, Howard K. Fly and mammalian lipid phosphate phosphatase isoforms differ in activity both in vitro and in vivo. EMBO Rep. 2003;4:793–9. doi: 10.1038/sj.embor.embor900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burnett C, Makridou P, Hewlett L, Howard K. Lipid phosphate phosphatases dimerise, but this interaction is not required for in vivo activity. BMC Biochem. 2004;5:2. doi: 10.1186/1471-2091-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Renault AD, Lehmann R. Follow the fatty brick road: lipid signaling in cell migration. Curr Opin Genet Dev. 2006;16:348–54. doi: 10.1016/j.gde.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 67.Steinhauer J, et al. Drosophila Lysophospholipid Acyltransferases Are Specifically Required for Germ Cell Development. Mol Biol Cell. 2009 doi: 10.1091/mbc.E09-05-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Doren M, Broihier HT, Moore LA, Lehmann R. HMG-CoA reductase guides migrating primordial germ cells. Nature. 1998;396:466–9. doi: 10.1038/24871. [DOI] [PubMed] [Google Scholar]
- 69.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–30. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 70.Santos AC, Lehmann R. Isoprenoids control germ cell migration downstream of HMGCoA reductase. Dev Cell. 2004;693:283. doi: 10.1016/s1534-5807(04)00023-1. Identifies the ABC transporter Mdr49 as important for germ cell migration in D. melanogaster, presumably through export of an Hmgcr-modified chemoattractant, and proves the ability of Mdr49 to promote PGC attraction using sorted PGCs in an in vitro chemotaxis assay. [DOI] [PubMed] [Google Scholar]
- 71.Ricardo S, Lehmann R. An ABC transporter controls export of a Drosophila germ cell attractant. Science. 2009;323:943–6. doi: 10.1126/science.1166239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Xia F, Li WX. Coactivation of STAT and Ras is required for germ cell proliferation and invasive migration in Drosophila. Dev Cell. 2003;5:787–98. doi: 10.1016/s1534-5807(03)00328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown S, Zeidler MP, Hombria JE. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev Dyn. 2006;235:958–66. doi: 10.1002/dvdy.20709. [DOI] [PubMed] [Google Scholar]
- 74.Weidinger G, Wolke U, Koprunner M, Klinger M, Raz E. Identification of tissues and patterning events required for distinct steps in early migration of zebrafish primordial germ cells. Development. 1999;126:5295–307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]
- 75.Reichman-Fried M, Minina S, Raz E. Autonomous modes of behavior in primordial germ cell migration. Dev Cell. 2004;6:589–96. doi: 10.1016/s1534-5807(04)00074-7. [DOI] [PubMed] [Google Scholar]
- 76.Blaser H, et al. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell. 2006;11:613–27. doi: 10.1016/j.devcel.2006.09.023. Provides a detailed cellular analysis of PGC migration in zebrafish and determines that PGCs migrate by extending bleb-like protrusions that are regulated by myosin contraction in response to an increase in local calcium contration. [DOI] [PubMed] [Google Scholar]
- 77.Knaut H, Werz C, Geisler R, Nusslein-Volhard C. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–82. doi: 10.1038/nature01338. Along with references 78, 86 and 87, identifies the CXCR4/SDF-1 signalling pathway as essential for PGC migration in zebrafish and mouse. [DOI] [PubMed] [Google Scholar]
- 78.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–59. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 79.Dumstrei K, Mennecke R, Raz E. Signaling pathways controlling primordial germ cell migration in zebrafish. J Cell Sci. 2004;117:4787–95. doi: 10.1242/jcs.01362. [DOI] [PubMed] [Google Scholar]
- 80.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–73. doi: 10.1016/j.cell.2007.12.034. Identifies a role for CXCR7b, a G protein-coupled receptor, in sequestering SDF-1 in somatic cells and promoting proper distribution of this chemokine and PGC migration. [DOI] [PubMed] [Google Scholar]
- 81.Mahabaleshwar H, Boldajipour B, Raz E. Killing the messenger: The role of CXCR7 in regulating primordial germ cell migration. Cell Adh Migr. 2008;2:69–70. doi: 10.4161/cam.2.2.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thorpe JL, Doitsidou M, Ho SY, Raz E, Farber SA. Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev Cell. 2004;6:295–302. doi: 10.1016/s1534-5807(04)00032-2. [DOI] [PubMed] [Google Scholar]
- 83.Mulligan T, Blaser H, Raz E, Farber SA. Prenylation-deficient G protein gamma subunits disrupt GPCR signaling in the zebrafish. Cell Signal. 2009 doi: 10.1016/j.cellsig.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev Biol. 2001;240:488–98. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- 85.Hara K, et al. Evidence for crucial role of hindgut expansion in directing proper migration of primordial germ cells in mouse early embryogenesis. Dev Biol. 2009;330:427–39. doi: 10.1016/j.ydbio.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 86.Ara T, et al. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proc Natl Acad Sci U S A. 2003;100:5319–23. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Molyneaux KA, et al. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–86. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- 88.McCoshen JA, McCallion DJ. A study of the primordial germ cells during their migratory phase in Steel mutant mice. Experientia. 1975;31:589–90. doi: 10.1007/BF01932475. [DOI] [PubMed] [Google Scholar]
- 89.Buehr M, McLaren A, Bartley A, Darling S. Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev Dyn. 1993;198:182–9. doi: 10.1002/aja.1001980304. [DOI] [PubMed] [Google Scholar]
- 90.Mahakali Zama A, Hudson FP, 3rd, Bedell MA. Analysis of hypomorphic KitlSl mutants suggests different requirements for KITL in proliferation and migration of mouse primordial germ cells. Biol Reprod. 2005;73:639–47. doi: 10.1095/biolreprod.105.042846. [DOI] [PubMed] [Google Scholar]
- 91.Runyan C, et al. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development. 2006;133:4861–9. doi: 10.1242/dev.02688. [DOI] [PubMed] [Google Scholar]
- 92.Di Carlo A, De Felici M. A role for E-cadherin in mouse primordial germ cell development. Dev Biol. 2000;226:209–19. doi: 10.1006/dbio.2000.9861. [DOI] [PubMed] [Google Scholar]
- 93.Bendel-Stenzel MR, Gomperts M, Anderson R, Heasman J, Wylie C. The role of cadherins during primordial germ cell migration and early gonad formation in the mouse. Mech Dev. 2000;91:143–52. doi: 10.1016/s0925-4773(99)00287-7. [DOI] [PubMed] [Google Scholar]
- 94.Anderson R, et al. Mouse primordial germ cells lacking beta1 integrins enter the germline but fail to migrate normally to the gonads. Development. 1999;126:1655–64. doi: 10.1242/dev.126.8.1655. [DOI] [PubMed] [Google Scholar]
- 95.Ding J, et al. Inhibition of HMG CoA reductase reveals an unexpected role for cholesterol during PGC migration in the mouse. BMC Dev Biol. 2008;8:120. doi: 10.1186/1471-213X-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jenkins AB, McCaffery JM, Van Doren M. Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development. 2003;130:4417–26. doi: 10.1242/dev.00639. [DOI] [PubMed] [Google Scholar]
- 97.Van Doren M, et al. fear of intimacy encodes a novel transmembrane protein required for gonad morphogenesis in Drosophila. Development. 2003;130:2355–64. doi: 10.1242/dev.00454. [DOI] [PubMed] [Google Scholar]
- 98.Mathews WR, Ong D, Milutinovich AB, Van Doren M. Zinc transport activity of Fear of Intimacy is essential for proper gonad morphogenesis and DE-cadherin expression. Development. 2006;133:1143–53. doi: 10.1242/dev.02256. [DOI] [PubMed] [Google Scholar]
- 99.Coffman CR. Cell migration and programmed cell death of Drosophila germ cells. Ann N Y Acad Sci. 2003;995:117–26. doi: 10.1111/j.1749-6632.2003.tb03215.x. [DOI] [PubMed] [Google Scholar]
- 100.Yamada Y, Davis KD, Coffman CR. Programmed cell death of primordial germ cells in Drosophila is regulated by p53 and the Outsiders monocarboxylate transporter. Development. 2008;135:207–16. doi: 10.1242/dev.010389. [DOI] [PubMed] [Google Scholar]
- 101.Matsui Y, et al. Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature. 1991;353:750–2. doi: 10.1038/353750a0. [DOI] [PubMed] [Google Scholar]
- 102.Godin I, et al. Effects of the steel gene product on mouse primordial germ cells in culture. Nature. 1991;352:807–9. doi: 10.1038/352807a0. [DOI] [PubMed] [Google Scholar]
- 103.Dolci S, et al. Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature. 1991;352:809–11. doi: 10.1038/352809a0. [DOI] [PubMed] [Google Scholar]
- 104.Stallock J, Molyneaux K, Schaible K, Knudson CM, Wylie C. The pro-apoptotic gene Bax is required for the death of ectopic primordial germ cells during their migration in the mouse embryo. Development. 2003;130:6589–97. doi: 10.1242/dev.00898. [DOI] [PubMed] [Google Scholar]
- 105.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burlacu A. Regulation of apoptosis by Bcl-2 family proteins. J Cell Mol Med. 2003;7:249–57. doi: 10.1111/j.1582-4934.2003.tb00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cook MS, Coveney D, Batchvarov I, Nadeau JH, Capel B. BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev Biol. 2009;328:377–83. doi: 10.1016/j.ydbio.2009.01.041. Examines the formation of teratomas in mice with disrupted Dnd function caused by the Ter mutation and finds that apoptosis mediated by Bax is responsible for both germ cell loss and protection from tumour formation in these mutants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–9. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alvarez-Buylla A, Merchant-Larios H. Mouse primordial germ cells use fibronectin as a substrate for migration. Exp Cell Res. 1986;165:362–8. doi: 10.1016/0014-4827(86)90590-2. [DOI] [PubMed] [Google Scholar]
- 110.Devenport D, Brown NH. Morphogenesis in the absence of integrins: mutation of both Drosophila beta subunits prevents midgut migration. Development. 2004;131:5405–15. doi: 10.1242/dev.01427. [DOI] [PubMed] [Google Scholar]
- 111.Kardash E, et al. Single-cell motility in vivo: a role for Rac, RhoA and cell-cell adhesion. Nat Cell Biol in press. 2009 doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- 112.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 113.Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 114.Starz-Gaiano M, Lehmann R. Moving towards the next generation. Mech Dev. 2001;105:5–18. doi: 10.1016/s0925-4773(01)00392-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information S1 (movie) | in vivo timelapse of primordial germ cell migration through the midgut in Drosophila melanogaster. Wild type stage 9-10 (∼4-5.5h after egg laying) D. melanogaster embryo with primordial germ cells (PGCs) labelled by an Enhanced Green Fluorescent Protein–Moesin fusion protein. PGCs are initially clustered and highly polarized, but lose adhesion with each other and begin to disperse and migrate through the surrounding posterior midgut cells. Images were taken every 2 min with a 63× objective. The playback rate is three frames per second. The embryo is oriented with its anterior to the left and dorsal side up. Movie is reproduced, with permission, from Kunwar, P.S. et al. J Cell Biol 183, 157-168 (2008).
Supplementary information S2 (movie) | in vivo timelapse of bilateral migration of primordial germ cells into the mesoderm in Drosophila melanogaster. Wild-type stage 10-11 (∼5.5-7h after egg laying) D. melanogaster embryo with primordial germ cells (PGCs) labelled by an Enhanced Green Fluorescent Protein–Moesin fusion protein. PGCs migrate laterally from the midline of the embryo. Images were taken every 2 min with a 40× objective. The playback rate is six frames per second. The embryo is oriented with its anterior to the top and dorsal side up. Movie is reproduced, with permission, from Sano, H., Renault, A.D. & Lehmann, R. J Cell Biol 171, 675-683 (2005).
Supplementary information S3 (movie) | in vivo timelapse of primordial germ cell migration in zebrafish. Wild type zebrafish embryo with primordial germ cells PGCs labelled by farnesylated (membrane-bound) GFP and the nuclei of all cells labelled by H2B-mCherry. 1s of playback represents approximately 25 minutes of development. The embryo is shown from a dorsal view with the anterior at the upper right of the frame. Courtesy of Azadeh Paksa and Erez Raz, University of Münster, and Nadine Peyriéras, CNRS-DEPSN.
Supplementary information S4 (movie) | in vivo timelapse of primordial germ cell migration in mouse. Wild type organ culture at embryonic day 9.5 with primordial germ cells (PGCs) labelled by Oct4ΔPE∷GFP+. Transverse slices from the hindgut regions of mouse embryos were cultured and filmed as described in reference 1. PGCs migrate out of the dorsal aspect of the hindgut, separate into two bilateral streams and migrate into the dorsal ridges. The movie covers about 12h of migration beginning at E9.5. The culture is oriented with its dorsal side to the left of the frame. Courtesy of Brian Dudley and Kathleen Molyneaux, Department of Genetics, Case Western University.



