Abstract
Familial Mediterranean fever (FMF) is an inherited disorder characterized by recurrent episodes of fever accompanied by sterile peritonitis, arthritis, and pleuritis. Many mutations in the MEFV gene have been identified as causing FMF. However, accompanying epidemiological information remains quite scarce except in some Mediterranean countries, and the degree of penetrance has been a subject of controversy. Here, I established a genetic epidemiology of full FMF mutations using two population exome studies. Of 57 mutations associated with FMF, 22 were detected in a total of 9007 individuals from two exome sequences. Exome-based epidemiology revealed the carrier rates of FMF in 28 populations in 19 countries by individual mutation and showed strong population specificity for the MEFV mutations. Unexpectedly high carrier rates suggested that some mutations are benign variants with no pathological significance and highlighted the need for caution in analyzing MEFV mutations. Similar approach could be used to uncover the incomplete or no penetrance of mutations in most inherited disorders.
Keywords: Allele frequency, epidemiology, exome, Familial Mediterranean fever, genetic diagnosis
Introduction
Familial Mediterranean fever (FMF; MIM# 249100) is an inherited disorder characterized by recurrent short episodes of fever, sterile peritonitis, arthritis, and pleurisy (Sohar et al. 1967; Livneh et al. 1997; Ben-Chetrit and Levy 1998). The febrile attacks are accompanied by a strong acute phase response, and the most severe complication is the development of renal amyloidosis (Sohar et al. 1967; Livneh et al. 1997; Ben-Chetrit and Levy 1998). FMF occurs most commonly among people from the Mediterranean basin (such as non-Ashkenazi Jews (Lazarin et al. 2013), Arabs (Majeed et al. 2005), Armenians (Cazeneuve et al. 1999), Greece (Konstantopoulos et al. 2003), and Turks (Dundar et al. 2011; Neocleous et al. 2015), and also in other countries (Sohar et al. 1967; Ben-Chetrit and Levy 1998).
The causative gene for FMF, MEFV, was first identified by two independent groups in 1997 (The French International FMF consortium 1997; The International FMF Consortium 1997). The MEFV gene is located on chromosome 16p13.3 and encodes a 781-amino acid protein (10 exons) known as pyrin (The French International FMF consortium 1997; The International FMF Consortium 1997). Previous studies on FMF patients and animal models suggest that MEFV mutations lead to gain of pyrin function, resulting in increased IL-1β secretion by monocytes and a prolonged inflammatory response when stimulated with lipopolysaccharide (Chae et al. 2011; Omenetti et al. 2014).
To date, over 50 MEFV mutations have been identified in FMF patients (Touitou 2001; Giancane et al. 2015). The five founder mutations, E148Q, M680I, M694I, M694V, and V726A, were reported to account for approximate 70% of cases of FMF from the Mediterranean origin and the nonfounder mutations would constitute the remaining proportion (Touitou 2001) although the pathogenic role of some mutations including E148Q remains debatable (Tchernitchko et al. 2003, 2006; Giancane et al. 2015). Draft guidelines for the genetic diagnosis of FMF were also prepared based on the current practice and data from the literature (Shinar et al. 2012). However, despite the recent advances in FMF studies, the epidemiological information is still insufficient and inconclusive, except in some Mediterranean countries. In addition, the degree of penetrance of many mutations has not been a subject of research for many years. The aim in this study was to establish the global epidemiology of autosomal recessive FMF (MIM# 249100) mutations using the population exome sequences and to evaluate the penetrance of each MEFV mutation.
Methods
Analysis of genetic variants using two representative exome projects
Genetic pipelines from 1000G (www.ncbi.nlm.nih.gov/variation/tools/1000genomes/) and NHLBI (http://www.nhlbi.nih.gov/) datasets were collected in VCF format. The datasets consisted of a total of 18,014 alleles from high-coverage exome sequences derived from 28 ethnic groups in 19 countries. The MEFV mutations for FMF were retrieved and selected from literature sources in PubMed (www.ncbi.nlm.nih.gov/pubmed), OMIM (http://www.ncbi.nlm.nih.gov/omim) and the autoinflammatory mutation database INFEVERS (http://fmf.igh.cnrs.fr/ISSAID/infevers/index.php) (Milhavet et al. 2008). The detected variants were then classified by pathogenicity, mutation type, allele frequency, countries, and racial groups. Information on mutation types, positions, reference sequences, and pathogenicity was retrieved from OMIM and NCBI dbSNP (http://www.nlm.nih.gov/SNP/) to generate exome-based epidemiology. ExAC Browser (http://exac.broadinstitute.org/) was additionally searched for the mutation alleles of MEFV.
Pairwise proportion tests of data consistency between two different exome resources
To project the performance of risk prediction based on analyses of exome sequence studies, exome-based estimates were statistically compared with the clinical prevalence survey. Evidence of data consistency was based on significant differences in pairwise comparisons between populations if two estimates differed significantly (two-sample test for equality of proportions with continuity correction). The standard hypothesis test was H0: π1 = π2 against the alternative (two-sided) 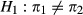 . The pairwise prop test can be used to test the null hypothesis that the proportions (probabilities of success) in two groups are the same. This test is referred to as a z-test because the statistics were as follows:
. The pairwise prop test can be used to test the null hypothesis that the proportions (probabilities of success) in two groups are the same. This test is referred to as a z-test because the statistics were as follows:
where 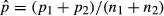 and indices (1, 2) refer to the first and second line of the table. In a two-way contingency table where H0: π1 = π2, this should yield comparable results to those of the ordinary χ2 test.
and indices (1, 2) refer to the first and second line of the table. In a two-way contingency table where H0: π1 = π2, this should yield comparable results to those of the ordinary χ2 test.
Results
Strategy for epidemiological research on MEFV mutations using population exome sequences
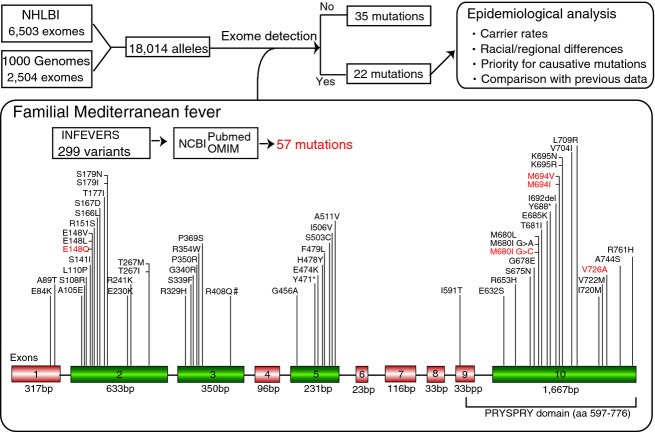
As a first step toward determining the genetic epidemiology of recessive FMF, genotyping datasets were collected from the 1000G and NHLBI projects for the causative variations (Fig.1). The datasets included the exome and its surrounding intronic sequences for 2504 individuals of 26 ethnic origins (ACB, African Caribbeans in Barbados; ASW, American’s of African Ancestry in SW; BEB, Bengali from Bangladesh; CEU, Utah Residents (CEPH) with Northern and Western European ancestry; CHB, Han Chinese in Beijing, China; CHS, Southern Han Chinese; CDX, Chinese Dai in Xishuangbanna, China; CLM, Colombian from Medellin; ESN, Esan in Nigeria ; FIN, Finnish in Finland; GBR, British in England; GIH, Gujarati Indian from Houston, Texas; GWD, Gambian in Western Divisions in The Gambia; IBS, Iberian population in Spain; ITU, Indian Telugu from the UK; JPT, Japanese in Tokyo, Japan; KHV, Kinh in Ho Chi Minh City, Vietnam; LWK, Luhya in Webuye; MSL, Mende in Sierra Leone; MXL, Mexican ancestry from Los Angeles; PEL, Peruvians from Lima, Peru; PJL, Punjabi from Lahore, Pakistan; PUR, Puerto Rico from Puerto Rica; STU, Sri Lankan Tamil from the UK; TSI, Toscani in Italia; YRI, Yoruba in Ibadan) and 6503 individuals of two ethnic origins (AA, African Americans; EA, European Americans) (Table1). Caucasians constituted 20.1% and 66.1% of subjects from the 1000G and NHLBI groups, respectively, whereas Africans constituted 26.4% and 33.9% of subjects, respectively (Table1). East Asian, South Asian, and Hispanic populations, which were represented only in the 1000G project, constituted 20.1%, 19.5%, and 13.9% of the group, respectively (Table1). Six samples were from within the United States, and others were from 16 countries, such as China, Japan, Colombia, Mexico, Puerto Rico, England, Germany, Kenya, and so on (Table1).
Figure 1.
Strategy of epidemiological research on FMF using population exome sequences. A flowchart used to study epidemiology shows the process of mutation detection using the 1000G and NHLBI datasets. A total of 18,014 alleles were screened for 57 MEFV mutations linked to FMF. A schematic representation of FMF-associated mutations in the MEFV gene is shown below. MEFV mutations most frequently found in FMF patients reside in exon 10 (depicted in green), which encodes the C-terminal PRYSPRY domain. Exons 2, 3, and 5 contain a substantial number of rare mutations (depicted in green). The five founder mutations are indicated in red. # R408Q and P369S mutations had been reported in cis as a single allele resulting in a variable clinical symptoms and R408Q is not likely to be a disease-causing variant (Bell et al. 2011).
Table 1.
Population disposition (ethnicity and male/female ratio)
| Population | Total no. | Percentage (in each project) | Percentage (in total) |
|---|---|---|---|
| 1000 genomes | |||
| AFR (African) | 661 | 26.4 | 7.34 |
| AMR (Ad Mixed American) | 347 | 13.9 | 3.85 |
| EAS (East Asian) | 504 | 20.1 | 5.60 |
| EUR (European) | 503 | 20.1 | 5.58 |
| SAS (South Asian) | 489 | 19.5 | 5.43 |
| ACB (African Caribbeans in Barbados) | 96 | 3.83 | 1.07 |
| ASW (Americans of African Ancestry in SW USA) | 61 | 2.44 | 0.68 |
| BEB (Bengali from Bangladesh) | 86 | 3.43 | 0.95 |
| CDX (Chinese Dai in Xishuangbanna, China) | 93 | 3.71 | 1.03 |
| CEU (Utah Residents (CEPH) with Northern and Western European ancestry) | 99 | 3.95 | 1.10 |
| CHB (Han Chinese in Bejing, China) | 103 | 4.11 | 1.14 |
| CHS (Southern Han Chinese) | 105 | 4.19 | 1.17 |
| CLM (Colombians from Medellin, Colombia) | 94 | 3.75 | 1.04 |
| ESN (Esan in Nigeria) | 99 | 3.95 | 1.10 |
| FIN (Finnish in Finland) | 99 | 3.95 | 1.10 |
| GBR (British in England and Scotland) | 91 | 3.63 | 1.01 |
| GIH (Gujarati Indian from Houston, Texas) | 103 | 4.11 | 1.14 |
| GWD (Gambian in Western Divisions in The Gambia) | 113 | 4.51 | 1.25 |
| IBS (Iberian population in Spain) | 107 | 4.27 | 1.19 |
| ITU (Indian Telugu from the UK) | 102 | 4.07 | 1.13 |
| JPT (Japanese in Tokyo, Japan) | 104 | 4.15 | 1.15 |
| KHV (Kinh in Ho Chi Minh City, Vietnam) | 99 | 3.95 | 1.10 |
| LWK (Luhya in Webuye, Kenya) | 99 | 3.95 | 1.10 |
| MSL (Mende in Sierra Leone) | 85 | 3.39 | 0.94 |
| MXL (Mexican Ancestry from Los Angeles USA) | 64 | 2.56 | 0.71 |
| PEL (Peruvians from Lima, Peru) | 85 | 3.39 | 0.94 |
| PJL (Punjabi from Lahore, Pakistan) | 96 | 3.83 | 1.07 |
| PUR (Puerto Ricans from Puerto Rico) | 104 | 4.15 | 1.15 |
| STU (Sri Lankan Tamil from the UK) | 102 | 4.07 | 1.13 |
| TSI (Toscani in Italia) | 107 | 4.27 | 1.19 |
| YRI (Yoruba in Ibadan, Nigeria) | 108 | 4.31 | 1.20 |
| NHLBI | |||
| EA (European American) | 4300 | 66.1 | 47.7 |
| AA (African American) | 2203 | 33.9 | 24.5 |
| Total | 9007 | 100 | |
Genetic screening for a possible carrier state was indicated for all individuals. A total of 299 (September 2014) nucleotide variants in MEFV have been reported in INFEVERS (Milhavet et al. 2008), but only a subset is found in patients with typical FMF disease and others are benign polymorphisms including synonymous and nonsynonymous variants. Of 57 MEFV mutations for FMF, 22 were detected in one or both of the two population exome datasets (Fig.1) (the list of MEFV mutations was retrieved from NCBI OMIM, INFEVERS, and PubMed; see also supplementary references). These mutations were present above the minimum variant allele frequency threshold (5.55E-05). MEFV mutations were classified by mutation type, derived allele frequency (DAF), countries, racial group, and clinical impact (Fig.1 and Table2). The other 35 mutations were not detected owing to their low DAF in a total of 9007 individuals (Fig.1).
Table 2.
Estimated carrier rates of FMF by race, ethnicity, and country using 1000G and NHLBI
| Disease name | Familial mediterranean fever (FMF) | ||||||
|---|---|---|---|---|---|---|---|
| OMIM entry | #249100 | ||||||
| Gene name | MEFV | ||||||
| mRNA ID | NM_024596.3 | ||||||
| Variant name | p.Glu84Lys | p.Leu110Pro | p.Glu148Gln | p.Arg151Ser | p.Glu230Lys | p.Thr267Ile | p.Arg329His |
| dbSNP | rs150819742 | rs11466018 | rs3743930 | rs104895185 | rs104895080 | rs104895081 | rs104895112 |
| ALL | 0.000167 (3/17996) | 0.00330 (59/17878) | 0.0440 (770/17494) | 0.000225 (4/17778) | 0.000392 (7/17870) | 0.000111 (2/18002) | 0.00178 (32/18002) |
| 1 in _ | 5998.7 | 303.0 | 22.7 | 4444.5 | 2552.9 | 9001.0 | 562.6 |
| NHLBI ALL | 0 (0/12988) | 0 (0/12870) | 0.0110 (137/12486) | 0 (0/12770) | 0 (0/12862) | 0.0000770 (1/12994) | 0.00177 (23/12994) |
| EA | 0 (0/8698) | 0 (0/8530) | 0.0111 (93/8354) | 0 (0/8460) | 0 (0/8506) | 0.000116 (1/8600) | 0.000116 (21/8600) |
| AA | 0 (0/4390) | 0 (0/4340) | 0.0104 (44/4232) | 0 (0/4310) | 0 (0/4356) | 0 (0/4394) | 0.000455 (2/4394) |
| 1000 Genome ALL | 0.000599 (3/5008) | 0.0118 (59/5008) | 0.126 (633/5008) | 0.000799 (4/5008) | 0.00140 (7/5008) | 0.000200 (1/5008) | 0.00160 (9/5008) |
| AFR | 0 (0/1322) | 0 (0/1322) | 0.0204 (27/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) |
| AMR | 0 (0/694) | 0 (0/694) | 0.0115 (8/694) | 0 (0/694) | 0 (0/694) | 0 (0/694) | 0.00144 (1/694) |
| EAS | 0.00298 (3/1008) | 0.0585 (59/1008) | 0.289 (291/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) |
| EUR | 0 (0/1006) | 0 (0/1006) | 0.00895 (9/1006) | 0 (0/1006) | 0.000994 (1/1006) | 0 (0/1006) | 0.00199 (2/1006) |
| SAS | 0 (0/978) | 0 (0/978) | 0.305 (298/978) | 0.00409 (4/978) | 0.00613 (6/978) | 0.00102 (1/978) | 0.00511 (5/978) |
| ACB | 0 (0/192) | 0 (0/192) | 0.0105 (2/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) |
| ASW | 0 (0/122) | 0 (0/122) | 0.0328 (4/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) |
| BEB | 0 (0/172) | 0 (0/172) | 0.343 (59/172) | 0.00581 (1/172) | 0.0174 (3/172) | 0.00581 (1/172) | 0 (0/172) |
| CDX | 0 (0/186) | 0.0269 (5/186) | 0.328 (61/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) |
| CEU | 0 (0/198) | 0 (0/198) | 0.0101 (2/198) | 0 (0/198) | 0.00505 (1/198) | 0 (0/198) | 0 (0/198) |
| CHB | 0 (0/206) | 0.0777 (16/206) | 0.286 (59/296) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| CHS | 0 (0/210) | 0.0714 (15/210) | 0.290 (61/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) |
| CLM | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0.00532 (1/188) |
| ESN | 0 (0/198) | 0 (0/198) | 0.0160 (3/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| FIN | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| GBR | 0 (0/182) | 0 (0/182) | 0.0275 (5/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) |
| GIH | 0 (0/206) | 0 (0/206) | 0.427 (88/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| GWD | 0 (0/226) | 0 (0/226) | 0.00443 (1/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) |
| IBS | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) |
| ITU | 0 (0/204) | 0 (0/204) | 0.260 (53/204) | 0.00490 (1/204) | 0.0147 (3/204) | 0 (0/204) | 0 (0/204) |
| JPT | 0.014 (3/208) | 0.0577 (12/208) | 0.216 (45/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) |
| KHV | 0 (0/198) | 0.0556 (11/198) | 0.328 (65/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| LWK | 0 (0/198) | 0 (0/198) | 0.0707 (14/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| MSL | 0 (0/170) | 0 (0/170) | 0.00588 (1/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| MXL | 0 (0/128) | 0 (0/128) | 0.0156 (2/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) |
| PEL | 0 (0/170) | 0 (0/170) | 0.0118 (2/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| PJL | 0 (0/192) | 0 (0/192) | 0.260 (50/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0.0260 (5/192) |
| PUR | 0 (0/208) | 0 (0/208) | 0.0192 (4/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) |
| STU | 0 (0/204) | 0 (0/204) | 0.235 (48/204) | 0.00980 (2/102) | 0 (0/204) | 0 (0/204) | 0 (0/204) |
| TSI | 0 (0/214) | 0 (0/214) | 0.00935 (2/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0.00935 (2/214) |
| YRI | 0 (0/216) | 0 (0/216) | 0.00926 (2/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) |
| Variant name | p.Ser339Phe | p.Pro369Ser | p.Arg408Gln | p.Gln474Lys | p.Ser503Cys | p.Ala511Val | p.Ile591Thr |
|---|---|---|---|---|---|---|---|
| dbSNP | rs104895157 | rs11466023 | rs11466024 | rs104895104 | rs190705322 | rs144270019 | rs11466045 |
| ALL | 0.000198 (3/18001) | 0.00967 (174/18002) | 0.00889 (160/18002) | 0.000111 (2/18002) | 0.000198 (3/18002) | 0.000198 (3/18002) | 0.00900 (162/18002) |
| 1 in _ | 6000.3 | 103.5 | 112.5 | 9001.0 | 6000.7 | 6000.3 | 111.1 |
| NHLBI ALL | 0.000153 (2/12993) | 0.00562 (73/12994) | 0.00569 (74/12994) | 0.0000770 (1/12994) | 0 (0/12994) | 0.000231 (3/12994) | 0.0108 (140/12994) |
| EA | 0.000233 (2/8600) | 0.00674 (58/8600) | 0.00674 (58/8600) | 0.000116 (1/8600) | 0 (0/8600) | 0.000349 (3/8600) | 0.0152 (131/8600) |
| AA | 0 (0/4393) | 0.00341 (15/4394) | 0.00364 (16/4394) | 0 (0/4394) | 0 (0/4394) | 0 (0/4394) | 0.00205 (9/4394) |
| 1000 Genome ALL | 0.000200 (1/5008) | 0.0202 (101/5008) | 0.0172 (86/5008) | 0.000200 (1/5008) | 0.000599 (3/5008) | 0 (0/5008) | 0.00439 (22/5008) |
| AFR | 0 (0/1322) | 0.00227 (3/1322) | 0.00150 (2/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) |
| AMR | 0 (0/694) | 0.00721 (5/694) | 0.00721 (5/694) | 0 (0/694) | 0 (0/694) | 0 (0/694) | 0.00432 (3/694) |
| EAS | 0 (0/1008) | 0.0675 (68/1008) | 0.0546 (55/1008) | 0 (0/1008) | 0.00298 (3/1008) | 0 (0/1008) | 0 (0/1008) |
| EUR | 0.000994 (1/1006) | 0.00398 (4/1006) | 0.00398 (4/1006) | 0 (0/1006) | 0 (0/1006) | 0 (0/1006) | 0.0179 (18/1006) |
| SAS | 0 (0/978) | 0.0215 (21/978) | 0.0205 (20/978) | 0.00102 (1/978) | 0 (0/978) | 0 (0/978) | 0.00102 (1/978) |
| ACB | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) |
| ASW | 0 (0/122) | 0.00820 (1/122) | 0.00820 (1/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) |
| BEB | 0 (0/172) | 0.0291 (5/172) | 0.0291 (5/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0.00581 (1/172) |
| CDX | 0 (0/186) | 0.0860 (16/186) | 0.0806 (15/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) |
| CEU | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0.0152 (3/198) |
| CHB | 0 (0/206) | 0.0583 (12/206) | 0.0388 (8/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| CHS | 0 (0/210) | 0.0571 (12/210) | 0.0476 (10/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) |
| CLM | 0 (0/188) | 0.00532 (1/188) | 0.00532 (1/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0.0106 (2/188) |
| ESN | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| FIN | 0 (0/198) | 0.0151 (3/198) | 0.0160 (3/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0.0202 (4/198) |
| GBR | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0.0220 (4/182) |
| GIH | 0 (0/206) | 0.00485 (1/206) | 0.00485 (1/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| GWD | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) |
| IBS | 0 (0/214) | 0.00467 (1/214) | 0.00467 (1/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0.00935 (2/214) |
| ITU | 0 (0/204) | 0.0441 (9/204) | 0.0392 (8/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) |
| JPT | 0 (0/208) | 0.0577 (12/208) | 0.0529 (11/208) | 0 (0/208) | 0.0144 (3/208) | 0 (0/208) | 0 (0/208) |
| KHV | 0 (0/198) | 0.0808 (16/198) | 0.0556 (11/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| LWK | 0 (0/198) | 0.00505 (1/197) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| MSL | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| MXL | 0 (0/128) | 0.00781 (1/128) | 0.00781 (1/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0.00781 (1/128) |
| PEL | 0 (0/170) | 0.0118 (2/170) | 0.0118 (2/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| PJL | 0 (0/192) | 0.0104 (2/192) | 0.0104 (2/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) |
| PUR | 0 (0/208) | 0.00481 (1/208) | 0.00481 (1/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) |
| STU | 0 (0/204) | 0.0196 (4/204) | 0.0196 (4/204) | 0.00490 (1/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) |
| TSI | 0.00467 (1/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0.0234 (5/214) |
| YRI | 0 (0/216) | 0.00463 (1/216) | 0.00463 (1/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) |
| Variant name | p.Glu632Ser | p.Arg653His | p.Met680Ile | p.Met694Val | p.Lys695Arg | p.Val722Met | p.Val726Ala | p.Ala744Ser |
|---|---|---|---|---|---|---|---|---|
| dbSNP | rs104895128 | rs104895085 | rs28940580 | rs61752717 | rs104895094 | rs104895201 | rs28940579 | rs61732874 |
| ALL | 0.0000555 (1/18002) | 0.0000555 (1/18002) | 0.0000555 (1/18002) | 0.000222 (4/18001) | 0.00233 (42/18002) | 0.0000555 (1/18002) | 0.00139 (25/18002) | 0.00144 (26/18002) |
| 1 in _ | 18002.0 | 18002.0 | 18002.0 | 4500.3 | 428.6 | 18002.0 | 720.1 | 692.3 |
| NHLBI ALL | 0.0000770 (1/12994) | 0 (0/12994) | 0.0000770 (1/12994) | 0.000231 (3/12994) | 0.00254 (33/12994) | 0.0000770 (1/12994) | 0.00185 (24/12994) | 0.00131 (17/12994) |
| EA | 0.000116 (1/8600) | 0 (0/8600) | 0.000116 (1/8600) | 0.000349 (3/8600) | 0.00384 (33/8600) | 0.000116 (1/8600) | 0.00267 (23/8600) | 0.00186 (16/8600) |
| AA | 0 (0/4394) | 0 (0/4394) | 0 (0/4394) | 0 (0/4394) | 0 (0/4394) | 0 (0/4394) | 0.000228 (1/4394) | 0.000228 (1/4394) |
| 1000 Genome ALL | 0 (0/5008) | 0.000200 (1/5008) | 0 (0/5008) | 0.000200 (1/5008) | 0.00180 (9/5008) | 0 (0/5008) | 0.000200 (1/5008) | 0.00180 (9/5008) |
| AFR | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) | 0 (0/1322) |
| AMR | 0 (0/694) | 0 (0/694) | 0 (0/694) | 0.00144 (1/694) | 0.00288 (2/694) | 0 (0/694) | 0 (0/694) | 0.00576 (4/694) |
| EAS | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) | 0 (0/1008) |
| EUR | 0 (0/1006) | 0.000994 (1/1006) | 0 (0/1006) | 0 (0/1006) | 0.00696 (7/1006) | 0 (0/1006) | 0.000994 (1/1006) | 0.00497 (5/1006) |
| SAS | 0 (0/978) | 0 (0/978) | 0 (0/978) | 0 (0/978) | 0 (0/978) | 0 (0/978) | 0 (0/978) | 0 (0/978) |
| ACB | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) |
| ASW | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) | 0 (0/122) |
| BEB | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) | 0 (0/172) |
| CDX | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) | 0 (0/186) |
| CEU | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| CHB | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| CHS | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) | 0 (0/210) |
| CLM | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0 (0/188) | 0.0106 (2/188) |
| ESN | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| FIN | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0.0253 (5/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| GBR | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) | 0 (0/182) |
| GIH | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) | 0 (0/206) |
| GWD | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) | 0 (0/226) |
| IBS | 0 (0/214) | 0.00467 (1/214) | 0 (0/214) | 0 (0/214) | 0.00467 (1/214) | 0 (0/214) | 0 (0/214) | 0.0187 (4/214) |
| ITU | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) |
| JPT | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) |
| KHV | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| LWK | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) | 0 (0/198) |
| MSL | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| MXL | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) | 0 (0/128) |
| PEL | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0.00588 (1/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) | 0 (0/170) |
| PJL | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) | 0 (0/192) |
| PUR | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0 (0/208) | 0.00962 (2/208) | 0 (0/208) | 0 (0/208) | 0.00962 (2/208) |
| STU | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) | 0 (0/204) |
| TSI | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0 (0/214) | 0.00467 (1/214) | 0 (0/214) | 0.00467 (1/214) | 0.00467 (1/214) |
| YRI | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) | 0 (0/216) |
Carrier rate variability by mutation type and ethnicity
Among the 22 MEFV mutations detected, the most common was E148Q, with a frequency of 1 in 22.7 (4.40%) (Table2). The allele frequencies of the other four founder mutations were as follows: M680I, 0.00555%; M694I, 0%; M694V, 0.0222%; and V726A, 0.139% (Table2). The prevalence rates are theoretically estimated to be equivalent to the squares of these carrier rates. Previous studies demonstrate that E148Q is not likely to be a disease-causing mutation (Tchernitchko et al. 2003, 2006; Giancane et al. 2015) and the high carrier rate of E148Q in this research (Table2) was consistent with the previous data. Thus, E148Q is excluded from following analysis.
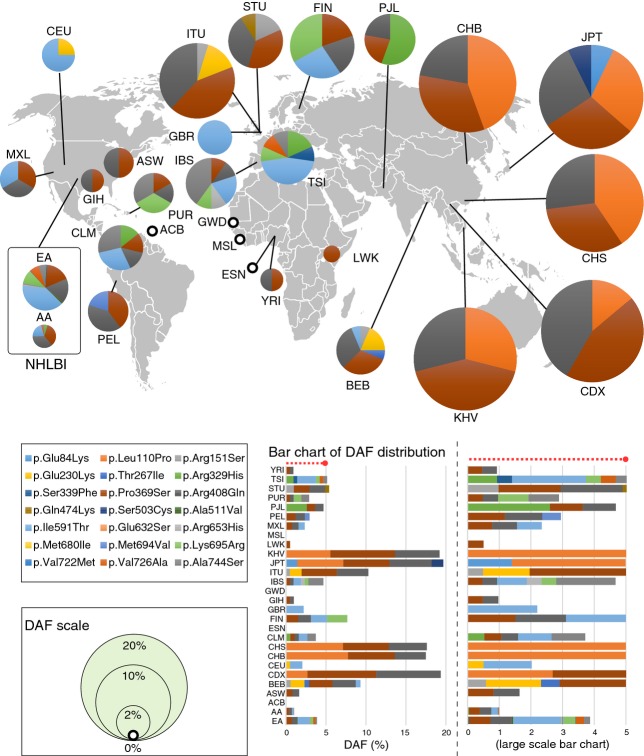
Carrier frequencies for disease-causing variants vary significantly by racial and ethnic groups (Lazarin et al. 2013). Figure2 shows a global map of the DAF distribution of the 21 MEFV mutations for FMF except E148Q. Among the other four founder mutations, M680I was Caucasian-specific, and V726A was much more prevalent in Caucasian (Table2). In East Asian populations, the average DAF was 18.7% (CDX, 19.4%; CHB, 17.5%; CHS, 17.6%; JPT, 19.7%) (Fig.2 and Table2). Also, the major variant in Europeans (NHLBI: 1.52%; 1000G: 1.79%) and Hispanics (1000G: 0.432%) was I591T. A recently available database, ExAC, with more than 60,000 exomes was additionally searched for 57 MEFV mutations (Table3) although this database does not provide the detailed information about country and ethnic group of individuals. A total of 31 MEFV mutations were detected (Table3) and the similar DAF distribution was obtained between ExAC and 1000 Genome + NHLBI data (Table2 and 3) as a whole.
Figure 2.
Geographical distribution of derived allele frequencies for MEFV mutations of FMF. The pie areas are proportional to derived allele frequency (DAF) of the 21 (except E148Q) MEFV mutations of FMF. The 1000G and NHLBI (26 + 2) populations are displayed separately. The bar chart of DAF distribution is described in the lower right panel.
Table 3.
Estimated carrier rates of FMF using ExAC
| Mutation | dbSNP | East Asian | South Asian | Latino | European (Non-Finnish) | European (Finnish) | African | Other |
|---|---|---|---|---|---|---|---|---|
| p.Glu84Lys | rs150819742 | 0.00151 (13/8588) | 0.0000609 (1/16418) | 0 (0/11480) | 0 (0/64378) | 0 (0/6316) | 0 (0/9498) | 0 (0/864) |
| p.Leu110Pro | rs11466018 | 0.08465 (717/8470) | 0.000793 (13/16386) | 0.000349 (4/11458) | 0.000159 (10/62936) | 0 (0/6602) | 0 (0/9144) | 0.00115 (1/870) |
| p.Ser141Ile | rs104895130 | 0 (0/7806) | 0 (0/15912) | 0 (0/10018) | 0.0000183 (1/54662) | 0 (0/5548) | 0 (0/7550) | 0.00132 (1/756) |
| p.Glu148Gln | rs3743930 | 0.315 (2275/7222) | 0.302 (4688/15536) | 0.0218 (184/8460) | 0.0197 (961/48764) | 0.00129 (6/4668) | 0.0184 (124/6734) | 0.0716 (49/684) |
| p.Glu148Val | rs104895076 | 0 (0/6392) | 0.000474 (7/14762) | 0 (0/8234) | 0.0000416 (2/48076) | 0 (0/4594) | 0 (0/6636) | 0 (0/668) |
| p.Arg151Ser | rs104895185 | 0 (0/5646) | 0.000712 (10/14052) | 0 (0/7110) | 0 (0/43120) | 0 (0/3968) | 0 (0/5938) | 0 (0/612) |
| p.Ser166Leu | 0 (0/1864) | 0.000897 (9/10030) | 0 (0/1514) | 0 (0/14194) | 0 (0/930) | 0 (0/2048) | 0 (0/264) | |
| p.Glu167Asp | rs104895079 | 0 (0/1704) | 0 (0/9872) | 0 (0/1378) | 0.0000756 (1/13236) | 0 (0/842) | 0 (0/1896) | 0 (0/246) |
| p.Ser179Asn | 0 (0/1368) | 0.00179 (17/9516) | 0 (0/1070) | 0.0000881 (1/11352) | 0 (0/644) | 0 (0/1552) | 0 (0/228) | |
| p.Glu230Lys | rs104895080 | 0 (0/8570) | 0.00431 (71/16486) | 0.000260 (3/11540) | 0.0000907 (6/66162) | 0 (0/6606) | 0 (0/10240) | 0 (0/898) |
| p.Thr267Ile | rs104895081 | 0 (0/8644) | 0.000363 (6/16512) | 0.000173 (2/11574) | 0.000135 (9/66732) | 0 (0/6612) | 0 (0/10392) | 0 (0/906) |
| p.Arg329His | rs104895112 | 0 (0/8630) | 0.00228 (37/16258) | 0.000522 (6/11504) | 0.00223 (147/65936) | 0 (0/6396) | 0.000293 (3/10248) | 0.00112 (1/892) |
| p.Ser339Phe | rs104895157 | 0 (0/8610) | 0.000185 (3/16246) | 0 (0/11474) | 0.000274 (18/65636) | 0 (0/6394) | 0.000197 (2/10148) | 0 (0/890) |
| p.Arg354Trp | rs104895116 | 0 (0/8562) | 0.0000613 (1/16300) | 0 (0/11482) | 0.0000611 (4/65466) | 0 (0/6434) | 0 (0/10158) | 0 (0/880) |
| p.Pro369Ser | rs11466023 | 0.0716 (616/8608) | 0.0147 (241/16442) | 0.00468 (54/11536) | 0.00975 (645/66174) | 0.0155 (101/6522) | 0.00397 (41/10318) | 0.0189 (17/902) |
| p.Arg408Gln | rs11466024 | 0.0541 (465/8600) | 0.0144 (237/16428) | 0.00434 (50/11518) | 0.00973 (639/65696) | 0.0153 (101/6596) | 0.00419 (42/10016) | 0.0190 (17/896) |
| p.Gln474Lys | rs104895104 | 0 (0/8652) | 0.0000606 (1/16512) | 0 (0/11578) | 0 (0/66724) | 0 (0/6614) | 0 (0/10404) | 0 (0/908) |
| p.Phe479Leu | rs104895083 | 0 (0/8654) | 0 (0/16512) | 0 (0/11578) | 0.0000599 (4/66740) | 0 (0/6614) | 0 (0/10406) | 0 (0/908) |
| p.Ser503Cys | rs190705322 | 0.00162 (14/8654) | 0 (0/16512) | 0 (0/11578) | 0 (0/66738) | 0 (0/6614) | 0 (0/10402) | 0 (0/908) |
| p.Ala511Val | rs144270019 | 0 (0/8654) | 0 (0/16512) | 0 (0/11578) | 0.0000749 (5/66718) | 0 (0/6614) | 0 (0/10400) | 0 (0/908) |
| p.Ile591Thr | rs11466045 | 0.000116 (1/8594) | 0.00335 (55/16402) | 0.00290 (33/11374) | 0.0147 (976/66284) | 0.0215 (141/6564) | 0.00176 (18/10254) | 0.0100 (9/898) |
| p.Glu632Ser | rs104895128 | 0 (0/8636) | 0 (0/14474) | 0 (0/11556) | 0.0000456 (3/65754) | 0 (0/6614) | 0 (0/10308) | 0 (0/862) |
| p.Arg653His | rs104895085 | 0 (0/8654) | 0.0000608 (1/16444) | 0 (0/11576) | 0.0000300 (2/66624) | 0 (0/6614) | 0.000289 (3/10386) | 0 (0/906) |
| p.Gly678Glu | rs104895088 | 0 (0/8654) | 0 (0/16512) | 0 (0/11576) | 0.0000450 (3/66740) | 0 (0/6614) | 0 (0/10406) | 0 (0/908) |
| p.Met680Ile (G→C) | rs28940580 | 0 (0/8654) | 0 (0/16512) | 0 (0/11576) | 0.000150 (10/66740) | 0 (0/6614) | 0 (0/10406) | 0.00110 (1/908) |
| p.Met680Ile (G→A) | rs28940580 | 0 (0/8654) | 0 (0/16512) | 0 (0/11576) | 0.0000300 (2/66740) | 0 (0/6614) | 0 (0/10406) | 0 (0/908) |
| p.Met694Val | rs61752717 | 0 (0/8654) | 0 (0/16512) | 0.000432 (5/11576) | 0.000285 (19/66738) | 0 (0/6614) | 0.0000961 (1/10406) | 0.00330 (3/908) |
| p.Lys695Arg | rs104895094 | 0 (0/8654) | 0.0000606 (1/16512) | 0.00225 (26/11576) | 0.00791 (528/66740) | 0.0161 (107/6614) | 0.0000961 (1/10406) | 0.00551 (5/908) |
| p.Val722Met | rs104895201 | 0 (0/8654) | 0 (0/16512) | 0 (0/11578) | 0.0000450 (3/66736) | 0 (0/6614) | 0 (0/10406) | 0 (0/908) |
| p.Val726Ala | rs28940579 | 0 (0/8654) | 0.000121 (2/16512) | 0.000173 (2/11578) | 0.0000450 (217/66736) | 0 (0/6614) | 0 (0/10406) | 0.00330 (3/908) |
| p.Ala744Ser | rs61732874 | 0 (0/8654) | 0.000363 (6/16512) | 0.00181 (21/11578) | 0.00214 (143/66728) | 0.000756 (5/6614) | 0.000385 (4/10404) | 0.00110 (1/906) |
| Total (All/except E148Q) | 0.529/0.214 | 0.347/0.0450 | 0.0397/0.0179 | 0.0679/0.0482 | 0.0704/0.0692 | 0.0297/0.0123 | 0.137/0.0658 |
Consistency of data between two different exomes
To determine the validity of this methodology, I examined the extent of differences in the two exome-based prevalence rates by comparing DAFs in African and European ancestries between the 1000G and NHLBI datasets (Table S1). A pairwise proportions test was used to test the null hypothesis that the proportions in the two estimates were identical. This formula is referred to as a Z test because the statistic is as follows:
where 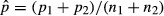 and the indices (1, 2) refer to the first and second lines of the table. A pairwise proportion test between the two exome resources showed no significant differences between the two different exomes (43 cases; P ≫ 0.05), except in one case (P < 0.05) (Table S1). This finding suggests that exome-based predictions are free of most confounding factors (such as diagnostic criteria, instrument, skill, and screening rate) and may be a more objective indicator.
and the indices (1, 2) refer to the first and second lines of the table. A pairwise proportion test between the two exome resources showed no significant differences between the two different exomes (43 cases; P ≫ 0.05), except in one case (P < 0.05) (Table S1). This finding suggests that exome-based predictions are free of most confounding factors (such as diagnostic criteria, instrument, skill, and screening rate) and may be a more objective indicator.
Discussion
Although genetic research into FMF began in 1990s (The French International FMF consortium 1997; The International FMF Consortium 1997), we still lack a complete picture of its genetic variation, carrier frequency, and penetrance. Exome-based epidemiology is a promising alternative method for genetic epidemiology because it provides information on both common and rare mutations in large numbers of individuals. Using exome data from a total of 9007 individuals from the two largest population exomes, I established a reliable epidemiology of FMF mutations with a small margin of error.
Estimated prevalence rates are considerably higher than those seen in clinical practice
There are only little data of carrier frequencies for comparison with this study because previous studies analyzed individuals mainly from some Mediterranean countries. Hofer et al. (2006) reported FMF prevalence rates in individuals from Western countries on a mass scale. The relative frequencies of the mutations found in this study [NHLBI EA, 4.98% (3.87%; except E148Q); 1000G EUR, 5.27% (4.38%; except E148Q) (Table2)] are considerably higher than the results of Hofer et al. (2006) who reported a prevalence of as 2.5 per 100,000 (0.004%) people even if E148Q is excluded in this research. In another study in Japanese populations, carrier frequencies [1000G JPT, 41.3% (19.7%; except E148Q) (Table2)] were much higher than the clinical incidence rate (0.000417%; http://www.nanbyou.or.jp/entry/3238). The variant T267I was frequently detected in Bangladesh (0.581% Table2), where FMF patients were not reported. These results suggest that some MEFV mutations, including E84K, L110P, E148Q, T267I, P369S, R408Q, S503C, and I591T, are polymorphisms, not disease-causing mutations. It has been proposed that E148Q is likely to be a polymorphism, not a disease-causing mutation, and has low penetrance (Tchernitchko et al. 2003, 2006) and my findings agree with these research results. Furthermore, the result here suggested the nonpenetrance of some other mutations. There is often the case where the causative mutations are determined too easily without analyzing potential effect of mutations (Cooper et al. 2013; van Rheenen et al. 2014; Siemiatkowska et al. 2014). Discordance between DAF and incidence rate may be caused by another infrequent mutation closely linked to these mutations. Another promising hypothesis is that FMF is a multifactorial and polygenic genetic disorder associated with the effects of multiple genes in combination with lifestyle and environmental factors.
In conclusion, exome-based epidemiology revealed the country-by-country carrier rates of FMF with respect to each mutation and provided a clue to understand the penetrance and screening priority of each mutation. An unexpectedly high carrier rate of FMF in Europeans and Asians raises the strong possibility that some MEFV mutations may be benign variants with few or no pathological significance. This study highlights the need for caution in interpreting genetic tests in FMF patients. Similar method could be used to uncover the incomplete or no penetrance of mutations in genetic disorders.
Acknowledgments
The author thanks the 1000 Genomes Project, the NHLBI GO Exome Sequencing Project and ExAC project for the available datasets.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Comparison of epidemiological data between two different exomes (1000G and NHLBI).
References
- Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Chetrit E. Levy M. Familial mediterranean fever. Lancet. 1998;351:659–664. doi: 10.1016/S0140-6736(97)09408-7. [DOI] [PubMed] [Google Scholar]
- Cazeneuve C, Sarkisian T, Pêcheux C, Dervichian M, Nédelec B, Reinert P, et al. MEFV-gene analysis in armenian patients with familial mediterranean fever: diagnostic value and unfavorable renal prognosis of the M694V homozygous genotype-genetic and therapeutic implications. Am. J. Hum. Genet. 1999;65:88–97. doi: 10.1086/302459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae JJ, Cho YH, Lee GS, Cheng J, Liu PP, Feigenbaum L, et al. Gain-of-function pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C. Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundar M, Emirogullari EF, Kiraz A, Taheri S. Baskol M. Common familial Mediterranean fever gene mutations in a Turkish cohort. Mol. Biol. Rep. 2011;38:5065–5069. doi: 10.1007/s11033-010-0652-7. [DOI] [PubMed] [Google Scholar]
- Giancane G, Ter Haar NM, Wulffraat N, Vastert SJ, Barron K, Hentgen V, et al. Evidence-based recommendations for genetic diagnosis of familial Mediterranean fever. Ann. Rheum. Dis. 2015;74:635–641. doi: 10.1136/annrheumdis-2014-206844. [DOI] [PubMed] [Google Scholar]
- Hofer M, Mahlaoui N. Prieur AM. A child with a systemic febrile illness - differential diagnosis and management. Best Pract. Res. Clini. Rheumatol. 2006;20:627–640. doi: 10.1016/j.berh.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Konstantopoulos K, Kanta A, Deltas C, Atamian V, Mavrogianni D, Tzioufas AG, et al. Familial Mediterranean fever associated pyrin mutations in Greece. Ann. Rheum. Dis. 2003;62:479–481. doi: 10.1136/ard.62.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarin GA, Haque IS, Nazareth S, Iori K, Patterson AS, Jacobson JL, et al. An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet. Med. 2013;15:178–186. doi: 10.1038/gim.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh A, Langevitz P, Zemer D, Zaks N, Kees S, Lidar T, et al. Criteria for the diagnosis of familial Mediterranean fever. Arthritis Rheum. 1997;10:1879–1885. doi: 10.1002/art.1780401023. [DOI] [PubMed] [Google Scholar]
- Majeed HA, El-Khateeb M, El-Shanti H, Rabaiha ZA, Tayeh M. Najib D. The spectrum of familial Mediterranean fever gene mutations in Arabs: report of a large series. Semin. Arthritis Rheum. 2005;34:813–818. doi: 10.1016/j.semarthrit.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Milhavet F, Cuisset L, Hoffman HM, Slim R, El-Shanti H, Aksentijevich I, et al. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum. Mutat. 2008;29:803–808. doi: 10.1002/humu.20720. [DOI] [PubMed] [Google Scholar]
- Neocleous V, Costi C, Kyriakou C, Kyriakides TC, Shammas C, Skordis N, et al. Familial Mediterranean fever associated with MEFV mutations in a large cohort of Cypriot patients. Ann. Hum. Genet. 2015;79:20–27. doi: 10.1111/ahg.12087. [DOI] [PubMed] [Google Scholar]
- Omenetti A, Carta S, Delfino L, Martini A, Gattorno M. Rubartelli A. Increased NLRP3-dependent interleukin 1β secretion in patients with familial Mediterranean fever: correlation with MEFV genotype. Ann. Rheum. Dis. 2014;73:462–469. doi: 10.1136/annrheumdis-2012-202774. [DOI] [PubMed] [Google Scholar]
- van Rheenen W, Diekstra FP, van den Berg LH. Veldink JH. Are CHCHD10 mutations indeed associated with familial amyotrophic lateral sclerosis? Brain. 2014;137:e313. doi: 10.1093/brain/awu299. [DOI] [PubMed] [Google Scholar]
- Shinar Y, Obici L, Aksentijevich I, Bennetts B, Austrup F, Ceccherini I, et al. Guidelines for the genetic diagnosis of hereditary recurrent fevers. Ann. Rheum. Dis. 2012;71:1599–1605. doi: 10.1136/annrheumdis-2011-201271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemiatkowska AM, Schuurs-Hoeijmakers JH, Bosch DG, Boonstra FN, Riemslag FC, Ruiter M, et al. Nonpenetrance of the most frequent autosomal recessive leber congenital amaurosis mutation in NMNAT1. JAMA Ophthalmol. 2014;132:1002–1004. doi: 10.1001/jamaophthalmol.2014.983. [DOI] [PubMed] [Google Scholar]
- Sohar E, Gafni J, Pras M. Heller H. Familial Mediterranean fever: a survey of 470 cases and review of the literature. Am. J. Med. 1967;43:227–253. doi: 10.1016/0002-9343(67)90167-2. [DOI] [PubMed] [Google Scholar]
- Tchernitchko D, Legendre M, Cazeneuve C, Delahaye A, Niel F. Amselem S. The E148Q MEFV allele is not implicated in the development of familial Mediterranean fever. Hum. Mutat. 2003;22:339–340. doi: 10.1002/humu.9182. [DOI] [PubMed] [Google Scholar]
- Tchernitchko DO, Gérard-Blanluet M, Legendre M, Cazeneuve C, Grateau G. Amselem S. Intrafamilial segregation analysis of the p. E148Q MEFV allele in familial Mediterranean fever. Ann. Rheum. Dis. 2006;65:1154–1157. doi: 10.1136/ard.2005.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The French International FMF consortium. A candidate gene for familial Mediterranean fever. Nat. Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- The International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- Touitou I. The spectrum of familial Mediterranean fever (FMF) mutations. Eur. J. Hum. Genet. 2001;9:473–483. doi: 10.1038/sj.ejhg.5200658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of epidemiological data between two different exomes (1000G and NHLBI).