Abstract
Purpose:
Pulmonary inflammation is associated with a variety of diseases. Assessing pulmonary inflammation on in vivo imaging may facilitate the early detection and treatment of lung diseases. Although routinely used in thoracic imaging, computed tomography has thus far not been compellingly shown to characterize inflammation in vivo. Alternatively, magnetic resonance imaging (MRI) is a nonionizing radiation technique to better visualize and characterize pulmonary tissue. Prior to routine adoption of MRI for early characterization of inflammation in humans, a rigorous and quantitative characterization of the utility of MRI to identify inflammation is required. Such characterization may be achieved by considering ex vivo histology as the ground truth, since it enables the definitive spatial assessment of inflammation. In this study, the authors introduce a novel framework to integrate 2D histology, ex vivo and in vivo imaging to enable the mapping of the extent of disease from ex vivo histology onto in vivo imaging, with the goal of facilitating computerized feature analysis and interrogation of disease appearance on in vivo imaging. The authors’ framework was evaluated in a preclinical preliminary study aimed to identify computer extracted features on in vivo MRI associated with chronic pulmonary inflammation.
Methods:
The authors’ image analytics framework first involves reconstructing the histologic volume in 3D from individual histology slices. Second, the authors map the disease ground truth onto in vivo MRI via coregistration with 3D histology using the ex vivo lung MRI as a conduit. Finally, computerized feature analysis of the disease extent is performed to identify candidate in vivo imaging signatures of disease presence and extent.
Results:
The authors evaluated the framework by assessing the quality of the 3D histology reconstruction and the histology—MRI fusion, in the context of an initial use case involving characterization of chronic inflammation in a mouse model. The authors’ evaluation considered three mice, two with an inflammation phenotype and one control. The authors’ iterative 3D histology reconstruction yielded a 70.1% ± 2.7% overlap with the ex vivo MRI volume. Across a total of 17 anatomic landmarks manually delineated at the division of airways, the target registration error between the ex vivo MRI and 3D histology reconstruction was 0.85 ± 0.44 mm, suggesting that a good alignment of the ex vivo 3D histology and ex vivo MRI had been achieved. The 3D histology-in vivo MRI coregistered volumes resulted in an overlap of 73.7% ± 0.9%. Preliminary computerized feature analysis was performed on an additional four control mice, for a total of seven mice considered in this study. Gabor texture filters appeared to best capture differences between the inflamed and noninflamed regions on MRI.
Conclusions:
The authors’ 3D histology reconstruction and multimodal registration framework were successfully employed to reconstruct the histology volume of the lung and fuse it with in vivo MRI to create a ground truth map for inflammation on in vivo MRI. The analytic platform presented here lays the framework for a rigorous validation of the identified imaging features for chronic lung inflammation on MRI in a large prospective cohort.
Keywords: framework, reconstruction, multimodal fusion, in vivo imaging signature, in vivo MRI, ex vivo MRI, histopathology, inflammation
1. INTRODUCTION
Pulmonary inflammation is a common condition associated with a variety of lung diseases such as asthma or chronic obstructive pulmonary disorder.1 Manifestations of inflammation include hypertrophy of airway epithelial cells, infiltration and activation of leukocytes, and structural changes to the architecture of the lung.2,3 The quantitative characterization of pulmonary inflammation on in vivo imaging holds the potential to facilitate improved and early characterization of lung diseases, as well as the investigation of anti-inflammatory drugs.4
Recent studies have investigated the ability of in vivo imaging, both computed tomography (CT)5–7 and magnetic resonance imaging (MRI),8–11 to identify pulmonary inflammation in preclinical models. These studies suggest that in vivo imaging may enable characterization of pulmonary inflammation. MRI is of particular interest in this regard as it is a nonradiation modality with a potentially better ability to image pulmonary soft tissue compared to CT.8,9,11–14 There is however a need for computerized decision support and feature analysis tools to define and evaluate quantitative imaging signatures for pulmonary inflammation on in vivo MRI.
Currently, in most instances, the only way to definitively ascertain the presence and spatial extent of diseases is via pathologic examination of stained histology slices (Fig. 1). While surgically excised lung histopathology could serve not only for defining the precise extent and presence of disease, it could also serve as a conduit to map the extent of disease onto the corresponding in vivo imaging via coregistration. Such accurate mapping of disease extent on in vivo imaging paves the way for a rigorous comparison of imaging appearance of disease and normal regions. Furthermore, when image intensities alone are unable to discriminate disease from other confounding tissue regions, computerized feature analysis methods such as textural analysis could help prize out subtle cues to distinguish the similar appearing tissue regions.15–18
FIG. 1.
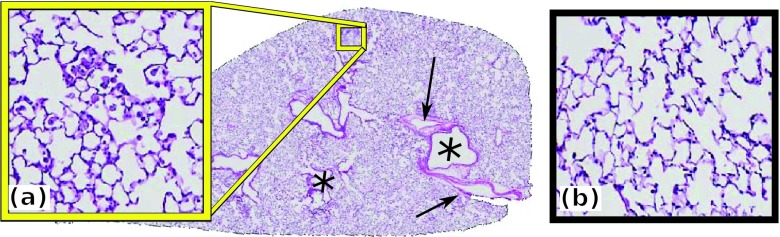
Lung hematoxylin and eosin (H&E) stained slice showing regions of (a) inflammation and (b) normal tissue. Airways (*) and blood vessels (→) are visible.
In this work, we introduce a novel analytic framework to facilitate imaging signature discovery for disease. Our framework was evaluated in the context of initial MRI based characterization of chronic inflammation in a mouse model. Specifically, the framework is comprised of three modules. First, the histology specimen is digitally reconstructed in 3D in order to facilitate its fusion with in vivo MRI. Such reconstruction is required as correspondences between lung histology and in vivo MRI slices may not be ascertainable due to different image viewing and histology cutting planes. Second, the 3D inflammation is mapped from the 3D reconstructed histology volume onto the in vivo MRI by coregistration of the 3D histology volume and the in vivo MRI using the ex vivo MRI as a conduit. Finally, image-derived features are extracted from the in vivo MRI of two mice with a phenotype of chronic pulmonary inflammation and the normal lung of five control mice. A comparison is performed between inflamed regions mapped from histology in two mice with an inflammation phenotype and normal lung from five control mice. By choosing these specific phenotypes, we can evaluate our framework to distinguish inflammation on in vivo MRI from normal lung tissue. The 3D histological reconstruction and its fusion with MRI were qualitatively and quantitatively evaluated in three mice, the two mice with an inflammation phenotype and one control mouse, while the preliminary textural analysis was performed on the latter three mice and an additional four control mice. While we do not claim in this paper to have identified the definitive computer extracted MRI features for diagnosis of pulmonary inflammation, the presented algorithmic pipeline paves the way for future discovery of validation of imaging signatures for diseases, including inflammation.
The remainder of the paper is organized as follows. First, we discuss previous work (Sec. 2) and provide an overview of our methodology (Sec. 3). A detailed methodological description of our framework is provided in Sec. 4, while the results are presented and discussed in Sec. 5. Finally, in Sec. 6, we present concluding remarks and future directions.
2. PREVIOUS WORK
In this paper, we present a pipeline of algorithmic steps in order to facilitate discovery of in vivo imaging signatures for disease. Specifically, in this paper, we evaluate this framework for the problem of identifying computer extracted MRI features associated with pulmonary inflammation in mice. Since our framework involves both 3D histologic reconstruction and radiology-pathology coregistration, we discuss previous related work in the context of these two areas.
Recently, a few approaches have been presented for coregistration of ex vivo histology and in vivo imaging data. Some approaches attempted to directly map the 2D histology slices onto the in vivo imaging by first determining and establishing slice19–22 or landmark correspondences.23 Alternative approaches inspired by the actual process of histology sample preparation have also been proposed.24 Yet, when slice correspondences between the ex vivo histology and in vivo imaging datasets do not exist or are difficult to identify, 3D reconstruction techniques may allow for creation of a 3D histology volume and enable volumetric coregistration with corresponding in vivo imaging.25 Such techniques are particularly useful in preclinical studies, where finely cut histologic sections corresponding to the in vivo imaging may be available. Some approaches have employed one-to-one registration26–29 of histology slices by utilizing rigid26,27 or deformable28,29 transforms. However, one-to-one registration of adjacent slices is prone to propagation of registration errors between slices, resulting in a progressive shift along the Z-axis. Alternative approaches have included one-to-many30,31 or many-to-many registration schemes32 either using a coarse-to-fine deformable transforms30 or natural gradients.31 Yet these methods have not been applied in the context of a multibody registration of multiple objects relative to each other.
Once the coregistration of the histology volume and the in vivo MRI has been accomplished, mapping of disease extent on the in vivo imaging can be established. This then paves the way for application of computerized feature analysis to identify imaging features to distinguish disease presence from confounders. Such methods are needed as image intensity alone may be insufficient to capture subtle differences between normal and diseased regions.15–18,33,34
3. BRIEF OVERVIEW AND NOVEL CONTRIBUTIONS
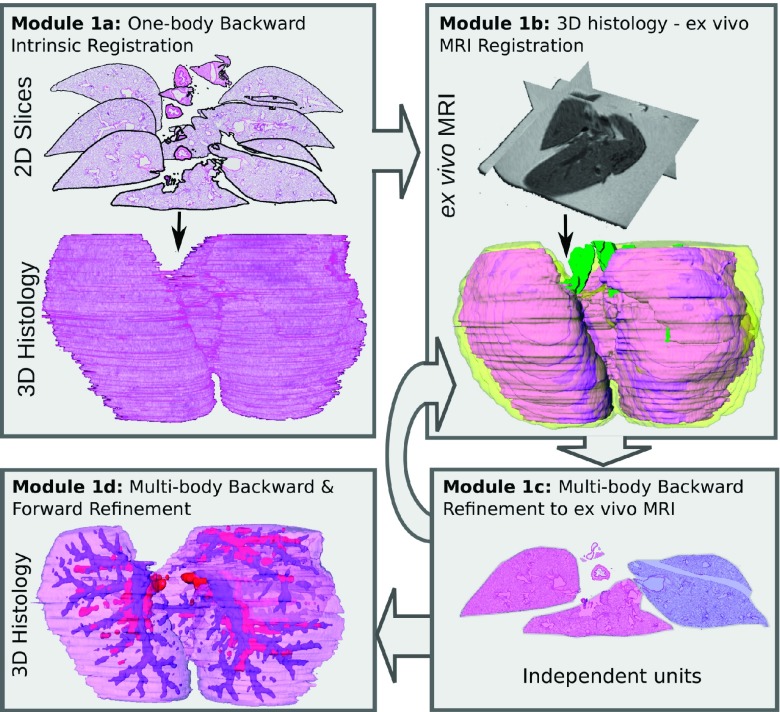
The current study seeks to develop the algorithmic pipeline to pave the way for quantitative characterization of in vivo imaging signatures of disease. In this work, we specifically look at the use case of characterization of pulmonary inflammation on MRI in a mouse model for evaluating the framework. Figure 2 illustrates the three main modules of our framework and the novel aspects of our approach.
FIG. 2.
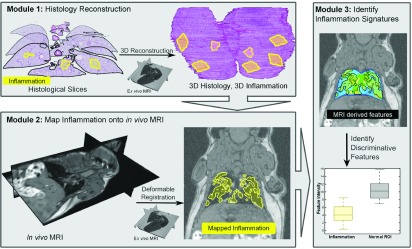
Overview of our algorithmic pipeline for characterization of pulmonary inflammation. Module 1: 3D histology volume is reconstructed from the 2D histology sections using ex vivo MRI as a conduit; the reconstruction also results in the generation of a 3D map of inflammation (yellow). Module 2: Volumetric coregistration of 3D histology and in vivo MRI volumes allows for the mapping of inflammation onto in vivo imaging. Module 3: Computer extracted image features of inflammation can then be identified from the in vivo MRI.
Module 1 (Sec. 4.B): The 3D histology volume is reconstructed from the 2D histology sections using an iterative alignment scheme that progressively increases the optimization complexity from a single body registration to a multibody registration of individual lobes. This reconstruction scheme was designed to facilitate the alignment of slices between each other within the 3D volume, while simultaneously correcting for lobe movement relative to each other during histology sample preparation. Module 1 introduces a novel multiresolution reconstruction of the 3D histology volume based on a one-to-many multibody refinement of lung lobes. This approach involves making the individual lobes “spatially aware” of each other during the registration,35 the spatial prior information coming from the ex vivo MRI.
Module 2 (Sec. 4.C): The 3D histology volume is registered to the in vivo MRI using both an affine and deformable transform with the ex vivo MRI serving as a conduit for the registration (similar to Module 1). The registration of the histologically reconstructed volume with the in vivo MRI enables the mapping of the extent of lung inflammation from the 3D histology volume onto the in vivo MRI. This novel registration approach limits the influence of imaging artifacts, such as elastic deformations on account of the absence of neighboring organs, as well as tissue sample preparation that causes tissue shrinkage.
Module 3 (Sec. 4.D): Textural features36–38 are extracted from in vivo MRI and compared between the inflamed and not inflamed regions. The preliminary feature analysis considered in this module serves as a means of showcasing how the presented framework can enable feature discovery.
4. METHODOLOGY
4.A. Data
Seven mice were included in this study: two surfactant protein D knockout (Sftpd−/−) mice which have a demonstrated phenotype of chronic pulmonary inflammation and five C57BL/6J wild type (WT) mice (normal controls) (Table I). The mice allow us to compare image-derived features between inflamed and noninflamed regions. The lung MRI was acquired with a 1 Tesla (T) M2 High Energy Performance MRI System (Aspect Magnet Technologies Ltd.). A T1-weighted MR gradient recalled echo sequence was used. Imaging parameters include TE/TR = 3.5/15 ms, flip angle 30∘, 0.55 mm slice thickness. MR images were taken from in vivo lungs of anesthetized mice, under isoflurane, gated for peak inspiration.
TABLE I.
Description of the acquisition and preparation protocols for the multimodal radiology and pathology datasets used for evaluating our analytic framework. The 3D reconstruction of the histology was performed at 0.5x, while the in vivo MRI was resampled to have consistent, close to isotropic voxels sizes of 250 μm3 for feature analysis. The lungs of two Sftpd−/− mice and one WT mouse were fixed, carefully sliced, and had H&E staining. These histology images were utilized to assess the accuracy of the 3D histology reconstruction approach and the quality of the histology-MRI fusion. The remaining four WT mice were only used for the validation of the textural analysis module. Since the latter four mice are control animals without an inflammation phenotype, no mapping of histology onto the imaging is required and hence was not performed.
| Mouse | Count | Modality | Sequence | Resolution (μm3) | Voxels | Annotations |
|---|---|---|---|---|---|---|
| 2 | In vivo MRI | T1 GRE | M × M × 500 | 256 × 256 × N | Lung, blood | |
| M ∈ {156, 234} | N ∈ {32, 34} | vessels | ||||
| M × M × 500 | M × M × N | Lung, airways | ||||
| Sftpd−/− | 2 | Ex vivo MRI | T1 GRE | M ∈ {125, 156} | M ∈ {256, 512} | |
| N ∈ {20, 34} | ||||||
| M × M × N | Lung, airways | |||||
| 2 | Histology | 0.75 × 0.75 × 110 | M ∈ {5000, 15 000} | Blood vessels | ||
| N ∈ {62, 74} | Inflammation | |||||
| 1 | In vivo MRI | T1 GRE | 234 × 234 × 500 | 256 × 256 × 32 | Lung | |
| 1 | Ex vivo MRI | T1 GRE | 156 × 156 × 500 | 256 × 256 × 34 | Lung, airways | |
| WT | 1 | Histology | 0.75 × 0.75 × 110 | M × M × 79 | Lung, airways | |
| M ∈ {5000, 15 000} | Blood vessels | |||||
| M × M × N | M × M × N | Lung | ||||
| 4 | In vivo MRI | T1 GRE | M ∈ {156, 500} | M ∈ {128, 256} | ||
| N ∈ {200, 500} | N ∈ {30, 128} |
After the in vivo MRI acquisition, the mouse lung was extracted and the inflation was fixed to the same volume as the in vivo MRI lung with 4% paraformaldehyde and 2% sucrose. Ex vivo lungs were imaged using the same sequence protocol as the in vivo MRI (Table I). In order to use the ex vivo MRI as a conduit for the histology reconstruction, the ex vivo images were acquired after the lung was fixed (via inflation with fixative). Since the ex vivo scan shows an inflated lung and to facilitate the coregistration of in vivo and ex vivo MRI, we also used an inflated lung for the in vivo MRI acquisition. The fixed lung was then embedded in paraffin and 5 μm sections were cut with a spacing of 110 μm (Table I). These sections were stained with hematoxylin and eosin (H&E) (Fig. 1) and digitized at 10x magnification using the Olympus VS120-SL scanning microscopy system.
The specific physiologic characteristics captured by each of the acquired modalities are summarized below.
-
1.
Whole body 1 T in vivo MRI shows the in vivo lung at a lower resolution, allowing for the visualization of arteries within the lung as hyperintense densities. Other organs, e.g., heart or liver, are spatially located in the proximity of the lung, resulting in its elastic compression.
-
2.
Ex vivo MRI shows the lung at better resolution compared to the in vivo MRI and allows for the visualization of large bronchi (hypointense regions). The same protocol used for in vivo imaging was also used for ex vivo imaging. The ex vivo acquisition provides a conduit to coregister in vivo MRI with ex vivo histology as they share similar attributes.
-
3.
Ex vivo H&E stained histology slices are obtained from the entire lung of three mice, two Sftpd−/− and one WT. The lungs of the remaining four WT mice did not undergo histology preparation, as they lack inflammation, and thus, they do not require histology mapping on in vivo imaging. The histology images have the highest resolution, allowing for the annotation of inflammation, blood vessels, and airways. Inflammation was identified as hypertrophy of airway epithelial cells and leukocyte infiltration39 and was manually delineated by an expert with substantial expertise in lung pathology. Large airways, blood vessels, and lung lobes were identified using an automatic active contour approach,40 parameterized to segment connected regions of interest. The results of the automated segmentation were then further manually partitioned into different anatomic classes: airways, blood vessels, or lung lobes. In spite of careful sample preparation, the histology slices may suffer from preparation artifacts, e.g., folding and shrinking. The histology slices were downsampled to a 0.5x magnification for further processing and reconstruction.
4.B. Module 1: Reconstruction of the 3D histology volume
The 3D reconstruction follows an iterative approach (Fig. 3), in which the complexity of the registration is progressively increased. See Table II for a summary of notation used in this manuscript and Secs. 4.C and 4.D.
FIG. 3.
Submodules of the histology reconstruction procedure (see Sec. 4.B for details).
TABLE II.
Notations used in this paper.
| Symbol | Definition |
|---|---|
| N | Number of histology slices |
| 3D histology volume | |
| Histology slice, i ∈ {1, …, N} | |
| Unit m in | |
| Airways | |
| Ex vivo MRI | |
| Ex vivo MRI slice, i ∈ {1, …, N} | |
| In vivo MRI | |
| ψ | Optimized scoring function |
| Histology scoring function | |
| Ex vivo MRI scoring function | |
| MI | Mutual information |
| Rigid transformation | |
| wj | Weight of the adjacent slice |
| Weight of ex vivo term |
The reconstruction involves optimization of the following scoring function for each histological slice , where N represents the total number of slices:
| (1) |
where is the rigid transformation of the slice . The term quantifies the intrinsic alignment of to neighboring k and k′ slices located either lower and, respectively, higher in the Z stack. The Z stack refers to the third dimension in the reconstructed volume , where the first and second dimensions are defined relative to the 2D histology slice coordinate frame. The second term, , encodes the alignment of with the corresponding slice in the ex vivo MRI, , while represents the weight of the ex vivo term. Module 1 has four submodules.
-
1.Module 1a. One-body backward intrinsic registration ensures that each slice is optimized within the reconstruction, ∀i ∈ {2, …, N}, relative to the adjacent k = 7, slices located lower in the Z stack (backward registration). The ex vivo MRI is not considered in the scoring function as it has not yet been registered relative to the 3D histology reconstruction (i.e., ). Specifically, in Module 1a, Eq. (1) becomes
where wj = exp(−(j2/4)) controls the influence of adjacent slices based on their proximity within the Z-stack.(2) -
2.
Module 1b. 3D histology-ex vivo MRI registration ensures that the lung segmented from the ex vivo MRI is coregistered to the 3D histology volume, , using an affine transformation. A three-level pyramid registration scheme within the ITK-based package elastix41 was used to optimize the normalized mutual information, employed as the scoring function.
-
3.Module 1c. MultiBody backward refinement to ex vivo MRI ensures that independent lobular units m are considered and their rigid transforms are individually optimized. During histology sample preparation (fixing and staining), the five lobes may move relative to each other. The five lobes were split into two independent units, m ∈ {1, 2}, the left and right lung, forming , with their own optimized rigid transformation . More than two lobular units can be considered. Eq. (1) thus becomes
where m, n ∈ {1, 2}, m≠n represent independent units. is defined using Eq. (2) while encodes the mutual information of the MRI slice, and the entire histology slice, composed of the transformed unit and not transformed units , where n≠m, .(3) While only the optimized unit m is considered in , all independent units, , are considered in the second term, . This helps to ensure that each unit is aware of the location of the other units and thus helps to limit their overlap.
-
4.Module 1d. Multibody backward and forward refinement ensures that the independent units are optimized relative to the lobular units in the adjacent slices i + j, j ∈ { − k, k′}, j≠0. As opposed to Module 1c, in Module 1d, we consider the lobular units of adjacent slices located not only lower but also higher in the Z stack. Specifically, Eq. (3) becomes
(4)
Module 1 iterates between the different submodules 1a–d using the following scheme. In iteration 1, the procedures defined in submodule 1a are performed. In iteration 2, the procedures defined in submodules 1b, 1c are performed. In iteration 3, the procedures defined in submodules 1b, 1c are reiterated to refine the reconstruction relative to the ex vivo MRI. Finally, in iteration 4, the steps in submodule 1d are employed to refine the final reconstruction. At each iteration, the transformation of each histology slice , or respective lobular unit, is refined.
Module 1 is evaluated by assessing the accuracy of the 3D histology volume reconstruction and registration with ex vivo MRI via the following measures.
-
1.
The intrinsic alignment of the histology slices: , where the dice similarity coefficient (DSC) is defined as D(A, B) = 2|A∩B|/(|A| + |B|) and |A| represents the cardinality of the set A.
-
2.
Alignment of the reconstruction, , with the ex vivo MRI, : .
-
3.The alignment of L landmarks corresponding to the airway tree divisions within histology, , and ex vivo MRI, , l ∈ {1, …, L},
(5)
A good reconstruction and fusion are suggested by a large DSC (max value 1) and reduced RMSD (min value: 0).
4.C. Module 2: Map disease ground truth from histology onto in vivo MRI volume
The linear alignment of and was achieved using as a conduit in the affine registration. To maintain the higher resolution offered by the histology, the registrations were performed using an isotropically upsampled in vivo MRI. is warped to using a B-spline based free-form deformation in a three-level pyramid registration scheme that optimizes the normalized mutual information, where the finer pyramid has a grid spacing of 4 mm. The optimized deformable transformation that warps onto is also applied to project the 3D inflammation ground truth on to , thus creating the in vivo ground truth for inflammation. We evaluated the registration of with by quantifying the alignment via .
4.D. Module 3: Identify computer extracted features associated with inflammation
Following standardization of lung intensities via landmark-based histogram alignment,42 78 features were derived from the in vivo MRI in both Sftpd−/− and control mice after the in vivo MRI was resampled to a consistent 250 μm voxel size. The computer derived features extracted from are summarized in Table III.
TABLE III.
Computer extracted MRI derived features capture different types of information, e.g., quantifying the smoothness, heterogeneity, or directional patterns. These features are not intended as a comprehensive compendium of textural features, but rather as an illustration of the types of feature interrogation of the diseased regions that can be facilitated via the newly presented histologic reconstruction and radiology–pathology coregistration pipeline.
These features attempt to capture subtle subvisual differences in image intensity that may not be visibly discernible on the original MRI. For instance, Haralick features37 capture co-occurring intensity statistics, while the Gabor filters38 are steerable wavelets that emphasize and capture oriented gradient patterns in the image. First and second order statistics36 are able to characterize image smoothness and identify edges.
As previously mentioned, the goal of this work was not so much to validate imaging signatures for lung inflammation, but so much as to pave the framework to facilitate feature discovery. With this in mind, we evaluated our framework with some well established image texture features to identify their association with inflammation in the lung. These features were largely drawn from classical textural operators including first and second order statistics,36 steerable filters,38 or Haralick features,37 with the goal of being able to characterize the heterogeneous appearance of inflammation. Manifestations of inflammation include accumulation of foamy appearing alveolar macrophages and of peribronchial and perivascular infiltrates in the lung.2,3 We expect that these accumulations modify the visible smoothness of the lung, which could be captured via first and second order statistical texture features.36 Moreover, such infiltrates may appear with similar intensity patterns at different locations within the lung, suggesting the need of Haralick features to identify such correlated patterns. The discontinuous accumulation of leukocytes may result in the creation of heterogeneous patches with borders that may be emphasized by Gabor filter features.
In order to compare the appearance profiles of inflamed and noninflamed regions and identify those textural features that are most discriminating between inflammation and noninflammation, we evaluate the difference between feature value distributions via the Bhattacharyya distance43
| (6) |
where p and q are the discrete probability density function of the image derived features for the inflammation and normal regions, respectively. B represents the number of bins, while pb and qb are the normalized frequency of textural feature responses within each bin B.
As inflammation has a discontinuous spatial distribution, we choose to compare image derived features within inflamed regions in the Sftpd−/− mice and with an anatomically corresponding region of interest in the control mice. The regions of inflammation were identified in the Sftpd−/− mice as described in Secs. 4.B and 4.C.
5. EXPERIMENTAL RESULTS AND DISCUSSION
5.A. 3D histology reconstruction
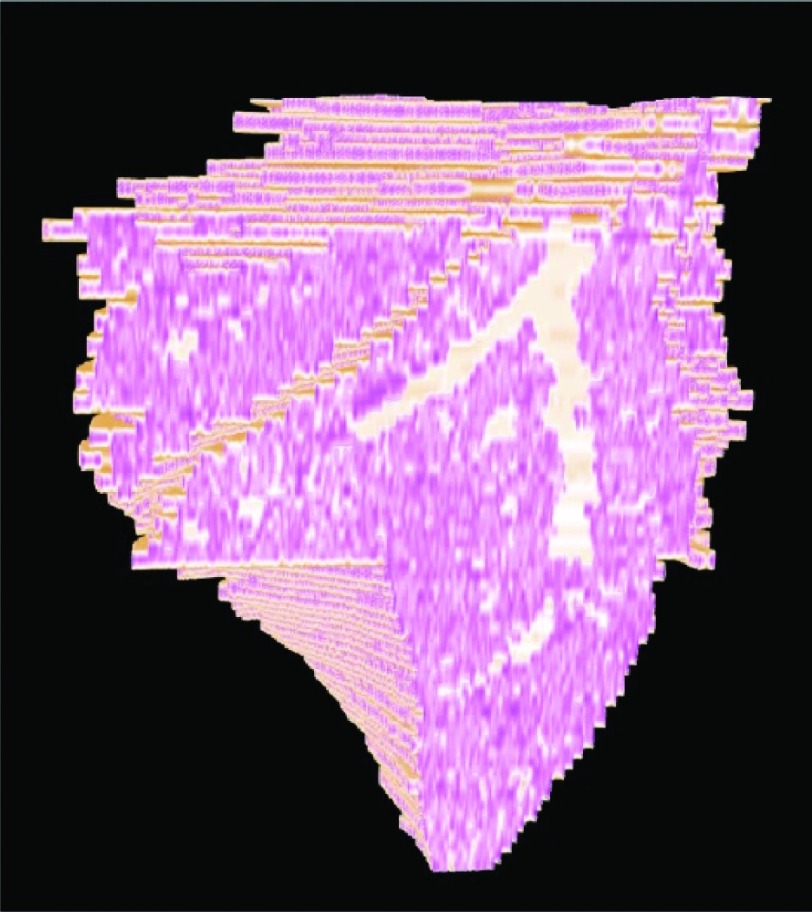
Figure 4 depicts the final results of successfully aligning the 2D H&E slices into a 3D histology volume without introducing noticeable “drift” between slices. The exterior surface of the 3D reconstruction (brown in Fig. 4) appears to be smooth, without significant zig-zag patterns visible at the edges of the reconstruction [see arrow in Fig. 4(b)].
FIG. 4.
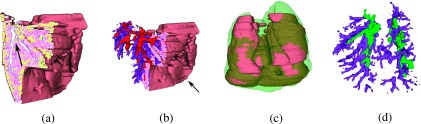
Histology volume reconstruction, ; (a) cut through shows the 3D continuity of the lung outline (brown), airways (blue outline, see arrow), and inflammation (yellow); (b) same as (a) with completely reconstructed airways (purple) and blood vessels (red); arrows point to reduced zig-zag pattern which is a qualitative indication of a good alignment; (c) overlay of and ex vivo MRI, (green); (d) alignment of airways, histology (purple), ex vivo MRI (green).
The airways appear as continuous 3D structures [arrow in Fig. 4(a) and Fig. 5 (Multimedia view)] within the histology reconstruction [purple in Figs. 4(b) and 4(d)], suggesting minimal alignment errors between slices as illustrated in Figs. 4(a)–4(c) and assessed quantitatively by [Fig. 6(a)]. The extracted airways appear to closely overlap in 3D between the histology and the ex vivo MRI lung, as illustrated in Figs. 4(d). Moreover, the high degree of reconstruction accuracy is also reflected quantitatively in the low RMSD = 0.85 ± 0.44 mm between the 17 landmark points on the 3D histology reconstruction and ex vivo MRI of the three mice for which the reconstruction was performed.
FIG. 5.
FIG. 6.
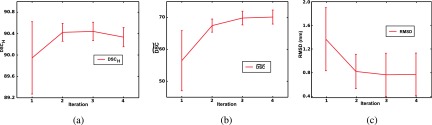
Progression of 3D reconstruction quality per iteration shown for three mice: (a) , (b) , and (c) landmark RMSD; iteration 1 - Module 1a, iteration 2 - Modules 1b,1c; iteration 3 - Repeat Modules 1b, 1c; iteration 4 - Module 1d.
The reconstruction approach was evaluated for the three mice (two Sftpd−/− mice and one control WT) at each iteration (Fig. 6). The reconstruction shows a high following Module 1a (first iteration), suggesting that the one-to-many registration without spatial constraints is able to closely coregister the histology slices. Yet, both the and RMSD show their worse performance when the registration is not constrained by .
Following the execution of Modules 1b, 1c (second iteration) in which the lobe units are simultaneously registered with constraints provided by ex vivo MRI, the intrinsic and and landmark RMSD significantly improve, possibly reflecting the benefit of the second iteration.
In the third iteration, Modules 1b and 1c are rerun to refine the transformation of the ex vivo MRI and subsequently of the lobular units. Fig. 6 shows minimal improvement in , but substantial improvement of . Moreover, the decrease in RMSD suggests that the refinement of the ex vivo MRI transformation relative to the 3D histology reconstruction is required to further improve the reconstruction accuracy.
After Module 1d (fourth iteration), reach their maxima, while the RMSD deviation is minimized to 0.85 ± 0.44 mm. decreases slightly.
5.B. Fusion of 3D histology to in vivo MRI
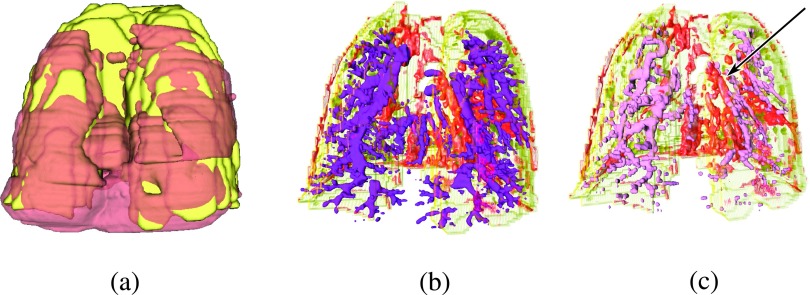
Figure 7(a) shows the 3D histology volume, (brown), overlaid onto in vivo MRI, (yellow). The airway tree (purple) in Fig. 7(b) is shown relative to the blood vessels (red) extracted from to depict the intertwining of the two systems. As expected, the two systems run in parallel to each other, without overlap as qualitatively seen in Fig. 7(b). Alignment accuracy is assessed qualitatively by visually investigating the blood vessel alignment, which in these figures appear to suggest close correspondences between blood vessels in and (pink) [see arrow in Fig. 7(c)].
FIG. 7.
Visualization of the 3D histology volume, (brown), and in vivo MRI volume, (yellow), alignment; (a) overlay of and ; (b) overlay of airways from (dark purple) onto blood vessels from (red). (c) Overlay of blood vessels from (red) and blood vessels from (pink). Arrow points to area of visually assessed close overlap.
Table IV summarizes the quantitative evaluation via dice similarity coefficient, DSC, after each of the affine and deformable registration steps. DSC in the twoSftpd−/− mice is computed between and and not surprisingly shows an improvement in alignment following deformable registration. For the five control mice, DSC reflects the quality of registration that allows for the identification of the anatomically corresponding region of interest.
TABLE IV.
Mean and standard deviation of DSC between reconstructed histology volumes and in vivo MRI lung volumes, .

|
Affine | Deformable |
|---|---|---|
| Sftpd−/− | 56.0 (0.7) | 73.7 (0.9) |
| Control | 62.2 (8.3) | 75.9 (5.4) |
5.C. Feature characterization
Figure 8 illustrates a Gabor wavelet representation. This feature was ranked first according to the Bhatacharyya distance (Table V) reflecting the most substantial difference in distribution between the Sftpd−/− inflammation [Fig. 8(a)] and the corresponding volume of interest in the control mouse [Fig. 8(b)]. These differences are also reflected in the box and whiskers plot in Fig. 8(c). The statistical significance of these differences was not evaluated in this preliminary study due to the small sample size. The difference in Gabor features between inflamed and noninflamed regions appears to suggest that inflammation may influence the appearance of linear patterns within the in vivo MRI. Clearly, independent validation in a large cohort is needed to establish the statistical significance of these findings.
FIG. 8.
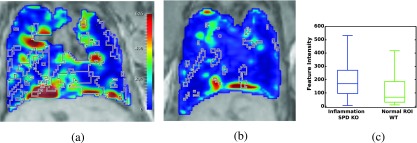
Preliminary evaluation of the framework in characterizing imaging signatures of inflammation. The most discriminative feature (Gabor) is shown using the same colormap in (a) Sftpd−/− lung with mapped inflammation (outlined with the black and white line) from 3D histology reconstruction; (b) WT lung with volume of interest (outlined with the black and white line); (c) feature distributions show modest separation between the computer extracted features derived from the inflamed and noninflamed regions.
TABLE V.
Top five scoring features ranked according to the Bhattacharyya distance.
| Feature | Parameters | Rank |
|---|---|---|
| Gabor | Angle: 0 | 1 |
| Gabor | Angle: 2.74 | 2 |
| Gabor | Angle: 0.39 | 3 |
| Gabor | Angle: 0.78 | 4 |
| Gabor | Angle: 2.35 | 5 |
Table V shows the five-top ranked features according to the Bhattacharyya distance,43 while Table VI shows the three-top ranked features, according to the same criterion, but within each feature class. Based on the Bhatacharyya distance criteria, the MRI intensity ranked 16 out of 79 features (Table VI), suggesting that several computer extracted features were more specific for identification of inflammation compared to the original signal intensity.
TABLE VI.
Top three features within each feature group ranked according to the Bhattacharyya distance.
| Feature | Parameters | Rank |
|---|---|---|
| Angle: 0.00 | 1 | |
| Gabor | Angle: 2.75 | 2 |
| Angle: 0.39 | 3 | |
| Energy | 10 | |
| Haralick | Correlation | 11 |
| IDM | 12 | |
| Median | 8 | |
| First order | Range | 13 |
| Mean | 17 | |
| Gradient magnitude | 38 | |
| Second order | Gradient X | 44 |
| Sobel X | 56 | |
| MRI intensity | 16 |
6. CONCLUDING REMARKS
We introduced a general analytic framework for 3D histologic reconstruction, multimodal fusion of radiology and pathology in order to facilitate computer based feature interrogation of disease appearance on in vivo imaging. The framework enables the mapping of disease extent from the histology onto the in vivo imaging, creating the disease ground truth required for further feature analysis. We evaluated our framework in a preliminary study aimed at characterizing the in vivo MRI signature of inflammation in a preclinical mouse model. Our evaluation showed that potential candidate in vivo imaging computer extracted MRI features of lung inflammation may be identified using our framework.
Our methodology comprised of multiple individual modules including (1) reconstruction of 3D histology volume using ex vivo MRI as a conduit, (2) coregistration of the 3D histology volume with the in vivo MRI, and (3) textural feature comparison between diseased and normal regions. Qualitative and quantitative results suggest that the individual modules have a high degree of accuracy. Our framework yielded (1) accurate intrinsic alignment of the 2D histologic slices within the reconstruction, (2) proper alignment of the 3D histology and ex vivo MRI, and (3) accurate alignment of the 3D histology and in vivo MRI on the three mice, two Sftpd−/− and one control, considered in our study. Despite the high variability in in vivo MRI imaging parameters, resolution, and field of view, our framework was able to identify preliminary textural features that appear to be associated with pulmonary inflammation in the seven mice, two Sftpd−/− and five controls in our evaluation cohort.
Some challenges may influence the accuracy of the fusion of 2D histology and in vivo MRI lung. First, the lung is a soft tissue which is prone to major elastic deformation caused by the neighboring organs within the in vivo MRI. Moreover, the histology preparation causes deformation and shrinkage of the tissue. In order to account for these challenges, we used the ex vivo MRI as a conduit in the registration and employed a pyramid anisotropic affine registration in the 3D histology reconstruction, and a deformable registration during the histology - in vivo MRI fusion. The reconstruction procedure is further complicated by the lung being composed of five lobes distributed between the left and right lungs. The lobes are not attached and thus are capable of moving relative to each other during tissue excise and histological preparation. Our multibody refinement approach used during the reconstruction was implemented to overcome the possible movements of the lobes relative to each other.
The framework described in this paper was evaluated in an established mouse model of chronic pulmonary inflammation. Inflammation was identified on histology and subsequently mapped onto in vivo MRI for two Sftpd−/− mice. A similar registration approach was used to define the volume of interest in the control mice, to generate a noninflamed volume of interest that corresponds anatomically and spatially to the regions of high inflammation likelihood, as shown by the Sftpd−/− mice. The noninflamed volume of interest could not have been mapped from histology, since the entire volume of the lung is normal in the control animals. We anticipate that the choice of the volume of interest is not essential as the entire volume is noninflamed in the control animals, yet we made every attempt to control for anatomical and interindividual variations by considering elastic registrations within five animals. Although, as proof-of-concept and for evaluation purposes, we performed a 3D histology reconstruction and fusion with MRI in one control animal, we considered it unnecessary for the remaining four control mice.
A total of three animals were used to evaluate the 3D histology reconstruction and its fusion with MRI. The evaluation showed good landmark and/or volumetric alignment between the 3D reconstructed histology lung and the MRI-outlined lung, indicating a proper ground truth mapping of inflammation from histology onto MRI. MRI features were extracted from a total of seven mice and were utilized to evaluate differences between inflamed and noninflamed regions. While statistical significance was not evaluated due to the small sample size, the expression patterns of some of the features warrant a subsequent in depth feature analysis. Our initial results represent preliminary data and could potentially pave the way for the use of the fusion framework in interrogation of in vivo imaging signatures of lung inflammation. In future work, we intend to validate the image features identified in this preliminary study on a large, independent validation cohort.
We believe our unique reconstruction and image analysis framework may be extended to include additional histological stains, molecular biomarkers, or other imaging modalities, e.g., micro-CT, to enable a comprehensive study of inflammation and other lung conditions.
ACKNOWLEDGMENTS
The authors would like to thank Derek Adler and Dr. Ed Yurkow at the Rutgers Molecular Imaging Center for their imaging expertise. Research reported in this publication was supported by the Department of Defense (W81XWH-13-1-0487), National Institutes of Health under Award Nos. R01CA136535-01, R01CA140772-01, R21CA167811-01, GM108463, HL086621, ES005022, CA136535, CA140722, CA167811; the National Institute of Diabetes and Digestive and Kidney Diseases under Award No. R01DK098503-02, the DOD Prostate Cancer Synergistic Idea Development Award (No. PC120857); the QED award from the University City Science Center and Rutgers University, the Ohio Third Frontier Technology development Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Jeffery P. K., “Remodeling in asthma and chronic obstructive lung disease,” Am. J. Respir. Crit. Care Med. 164, S28–S38 (2001). 10.1164/ajrccm.164.supplement_2.2106061 [DOI] [PubMed] [Google Scholar]
- 2.Atochina E. N., Beers M. F., Hawgood S., Poulain F., Davis C., Fusaro T., and Gow A. J., “Surfactant protein-D, a mediator of innate lung immunity, alters the products of nitric oxide metabolism,” Am. J. Respir. Cell Mol. Biol. 30, 271–279 (2004). 10.1165/rcmb.2003-0091OC [DOI] [PubMed] [Google Scholar]
- 3.Botas C., Poulain F., Akiyama J., Brown C., Allen L., Goerke J., Clements J., Carlson E., Gillespie A. M., Epstein C., and Hawgood S., “Altered surfactant homeostasis and alveolar type II cell morphology in mice lacking surfactant protein D,” Proc. Natl. Acad. Sci. U. S. A. 95, 11869–11874 (1998). 10.1073/pnas.95.20.11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel-Claussen J., Renne J., Hinrichs J., Schönfeld C., Gutberlet M., Schaumann F., Winkler C., Faulenbach C., Krug N., Wacker F. K., and Hohlfeld J. M., “Quantification of pulmonary inflammation after segmental allergen challenge using turbo-inversion recovery-magnitude magnetic resonance imaging,” Am. J. Respir. Crit. Care Med. 189, 650–657 (2014). 10.1164/rccm.201310-1825OC [DOI] [PubMed] [Google Scholar]
- 5.Chen D. L. and Schuster D. P., “Imaging pulmonary inflammation with positron emission tomography: A biomarker for drug development,” Mol. Pharm. 3, 488–495 (2006). 10.1021/mp060050w [DOI] [PubMed] [Google Scholar]
- 6.Jobse B. N., Johnson J. R., Farncombe T. H., Labiris R., Walker T. D., Goncharova S., and Jordana M., “Evaluation of allergic lung inflammation by computed tomography in a rat model in vivo,” Eur. Respir. J. 33, 1437–1447 (2009). 10.1183/09031936.00087508 [DOI] [PubMed] [Google Scholar]
- 7.Ntziachristos V., “Optical imaging of molecular signatures in pulmonary inflammation,” Proc. Am. Thorac. Soc. 6, 416–418 (2009). 10.1513/pats.200901-003aw [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckmann N., Tigani B., Ekatodramis D., Borer R., Mazzoni L., and Fozard J. R., “Pulmonary edema induced by allergen challenge in the rat: Noninvasive assessment by magnetic resonance imaging,” Magn. Reson. Med. 45, 88–95 (2001). 10.1002/1522-2594(200101)45:1%3C88::AID-MRM1013%3E3.0.CO;2-N [DOI] [PubMed] [Google Scholar]
- 9.Tigani B., Cannet C., Karmouty-Quintana H., Blé F.-X., Zurbruegg S., Schaeublin E., Fozard J. R., and Beckmann N., “Lung inflammation and vascular remodeling after repeated allergen challenge detected noninvasively by MRI,” Am. J. Physiol. 292, L644–L653 (2007). 10.1152/ajplung.00122.2006 [DOI] [PubMed] [Google Scholar]
- 10.Blé F.-X., Cannet C., Zurbruegg S., Karmouty-Quintana H., Bergmann R., Frossard N., Trifilieff A., and Beckmann N., “Allergen-induced lung inflammation in actively sensitized mice assessed with MR imaging,” Radiology 248, 834–843 (2008). 10.1148/radiol.2482071452 [DOI] [PubMed] [Google Scholar]
- 11.Holmes J. H. et al. , “Noninvasive mapping of regional response to segmental allergen challenge using magnetic resonance imaging and [F-18] fluorodeoxyglucose positron emission tomography,” Magn. Reson. Med. 53, 1243–1250 (2005). 10.1002/mrm.20504 [DOI] [PubMed] [Google Scholar]
- 12.Wild J. M., Marshall H., Bock M., Schad L. R., Jakob P. M., Puderbach M., Molinari F., Van Beek E. J. R., and Biederer J., “MRI of the lung (1/3): Methods,” Insights Imaging 3, 345–353 (2012). 10.1007/s13244-012-0176-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biederer J., Beer M., Hirsch W., Wild J., Fabel M., Puderbach M., and Van Beek E. J. R., “MRI of the lung (2/3). Why when how? Insights Imaging 3, 355–371 (2012). 10.1007/s13244-011-0146-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biederer J., Mirsadraee S., Beer M., Molinari F., Hintze C., Bauman G., Both M., Van Beek E. J. R., Wild J., and Puderbach M., “MRI of the lung (3/3)-current applications and future perspectives,” Insights Imaging 3, 373–386 (2012). 10.1007/s13244-011-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viswanath S. E., Bloch N. B., Chappelow J. C., Toth R., Rofsky N. M., Genega E. M., Lenkinski R. E., and Madabhushi A., “Central gland and peripheral zone prostate tumors have significantly different quantitative imaging signatures on 3 Tesla endorectal, in vivo T2-weighted MR imagery,” J. Magn. Reson. Imaging 36, 213–224 (2012). 10.1002/jmri.23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal E., Sirlin C. B., Ooi C., Adler A. S., Gollub J., Chen X., Chan B. K., Matcuk G. R., Barry C. T., Chang H. Y., and Kuo M. D., “Decoding global gene expression programs in liver cancer by noninvasive imaging,” Nat. Biotechnol. 25, 675–680 (2007). 10.1038/nbt1306 [DOI] [PubMed] [Google Scholar]
- 17.Gevaert O., Xu J., Hoang C. D., Leung A. N., Xu Y., Quon A., Rubin D. L., Napel S. K., and Plevritis S., “Non–small cell lung cancer: Identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results,” Radiology 264, 387–396 (2012). 10.1148/radiol.12111607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari P., Kurhanewicz J., and Madabhushi A., “Multi-kernel graph embedding for detection, Gleason grading of prostate cancer via MRI/MRS,” Med. Image Anal. 17, 219–235 (2013). 10.1016/j.media.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chappelow J., Madabhushi A., Rosen M., Tomaszeweski J., and Feldman M., “Multimodal image registration of ex vivo 4 Tesla prostate MRI with whole mount histology for cancer detection,” Proc. SPIE 6512, S1–S12 (2007). 10.1117/12.710558 [DOI] [Google Scholar]
- 20.Chappelow J., Bloch B. N., Rofsky N., Genega E., Lenkinski R., DeWolf W., and Madabhushi A., “Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information,” Med. Phys. 38, 2005–2018 (2011). 10.1118/1.3560879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson E., Gaed M., Gómez J. A., Moussa M., Pautler S., Chin J. L., Crukley C., Bauman G. S., Fenster A., and Ward A. D., “3D prostate histology image reconstruction: Quantifying the impact of tissue deformation and histology section location,” J. Pathol. Inf. 4, 31 (2013). 10.4103/2153-3539.120874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nir G., Sahebjavaher R. S., Kozlowski P., Chang S. D., Jones E., Goldenberg L. L., and Salcudean S. E., “Registration of whole-mount histology and volumetric imaging of the prostate using particle filtering,” IEEE Trans. Med. Imaging 33, 1601–1613 (2014). 10.1109/tmi.2014.2319231 [DOI] [PubMed] [Google Scholar]
- 23.Orczyk C., Rusinek H., Rosenkrantz A. B., Mikheev A., Deng F-M., Melamed J., and Taneja S. S., “Preliminary experience with a novel method of three-dimensional co-registration of prostate cancer digital histology and in vivo multiparametric MRI,” Clin. Radiol. 68, e652–e658 (2013). 10.1016/j.crad.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turkbey B., Mani H., Shah V., Rastinehad A. R., Bernardo M., Pohida T., Pang Y., Daar D., Benjamin C., McKinney Y. L., Trivedi H., Chua C., Bratslavsky G., Shih J. H., Linehan W. M., Merino M. J., Choyke P. L., and Pinto P. A., “Multiparametric 3 T prostate magnetic resonance imaging to detect cancer: Histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds,” J. Urol. 186, 1818–1824 (2011). 10.1016/j.juro.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H., Piert M. R., Khan A., Shah R., Hussain H., Siddiqui J., and Meyer C. R., “Registration methodology for histological sections and in-vivo imaging of human prostate,” Acad. Radiol. 15, 1027–1039 (2008). 10.1016/j.acra.2008.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ourselin S., Roche A., Subsol X., Pennec G., and Ayache N., “Reconstructing a 3D structure from serial histological sections,” Image Vision Comput. 19, 25–31 (2001). 10.1016/S0262-8856(00)00052-4 [DOI] [Google Scholar]
- 27.Cifor A., Pridmore T., and Pitiot A., “Smooth 3-D reconstruction for 2-D histological images,” in Information Processing in Medical Imaging (Springer, Berlin Heidelberg, 2009), pp. 350–361. 10.1007/978-3-642-02498-6_29 [DOI] [PubMed] [Google Scholar]
- 28.Alic L., Haeck J. C., Bol K., Klein S., van Tiel S. T., Wielepolski P. A., de Jong M., Niessen W. J., Bernsen M., and Veenland J. F., “Facilitating tumor functional assessment by spatially relating 3D tumor histology and in vivo MRI: Image registration approach,” PLoS One 6, e22835 (2011). 10.1371/journal.pone.0022835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lotz J., Berger J., Müller B., Breuhahn K., Grabe N., Heldmann S., Homeyer B., Lahrmann A., Laue H., Olesch J., Schwier M., Sedlaczek O., and Warth A, “Zooming in: High resolution 3D reconstruction of differently stained histological whole slide images,” Proc. SPIE 9041, 904104-1–904104-7 (2014). 10.1117/12.2043381 [DOI] [Google Scholar]
- 30.Yushkevich P., Avants B., Ng L., Hawrylycz M., Burstein P., Zhang H., and Gee J., “3D mouse brain reconstruction from histology using a coarse-to-fine approach,” in Biomedical Image Registration, Lecture Notes in Computer Science (Springer, Berlin Heidelberg, 2006), pp. 230–237. 10.1007/11784012_28 [DOI] [Google Scholar]
- 31.Wang H., Suh J. W., Das S. R., Pluta J. B., Craige C., and Yushkevich P. A., “Multi-atlas segmentation with joint label fusion,” IEEE Trans. Pattern Anal. Mach. Intell. 35, 611–623 (2013). 10.1109/TPAMI.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feuerstein M., Heibel H., Gardiazabal J., Navab N., and Groher M., “Reconstruction of 3-D histology images by simultaneous deformable registration,” in Medical Image Computing and Computer-Assisted Intervention (Springer, Berlin Heidelberg, 2011), pp. 582–589. 10.1007/978-3-642-23629-7_71 [DOI] [PubMed] [Google Scholar]
- 33.Ginsburg S., Tiwari P., Kurhanewicz J., and Madabhushi A., “Variable ranking with PCA: Finding multiparametric MR imaging markers for prostate cancer diagnosis and grading,” in Prostate Cancer Imaging. Image Analysis and Image-Guided Interventions (Springer, Berlin Heidelberg, 2011), pp. 146–157. 10.1007/978-3-642-23944-1_15 [DOI] [Google Scholar]
- 34.Litjens G., Debats O., Barentsz J., Karssemeijer N., and Huisman H., “Computer-aided detection of prostate cancer in MRI,” IEEE Trans. Pattern Anal. Mach. Intell. 33, 1083–1092 (2014). 10.1109/tmi.2014.2303821 [DOI] [PubMed] [Google Scholar]
- 35.Rusu M. and Birmanns S., “Evolutionary tabu search strategies for the simultaneous registration of multiple atomic structures in cryo-em reconstructions,” J. Struct. Biol. 170, 164–171 (2010). 10.1016/j.jsb.2009.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russ J. C., Image Processing Handbook, 4th ed. (CRC, Inc., Boca Raton, FL, 2002). [Google Scholar]
- 37.Haralick R. M., Shanmugan K., and Dinstein I., “Textural features for image classification,” IEEE Trans. Syst. Man Cybern. SMC-3, 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 38.Bovik A. C., Clark M., and Geisler W. S., “Multichannel texture analysis using localized spatial filters,” IEEE Trans. Pattern Anal. Mach. Intell. 12, 55–73 (1990). 10.1109/34.41384 [DOI] [Google Scholar]
- 39.Rudmann D. G., Preston A. M., Moore M. W., and Beck J. M., “Susceptibility to pneumocystis carinii in mice is dependent on simultaneous deletion of ifn-gamma and type 1 and 2 tnf receptor genes,” J. Immunol. 161, 360–366 (1998). [PubMed] [Google Scholar]
- 40.Lankton S. and Tannenbaum A., “Localizing region-based active contours,” IEEE Trans. Pattern Anal. Mach. Intell. 17, 2029–2039 (2008). 10.1109/TIP.2008.2004611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein S., Staring M., Murphy K., Viergever M. A., and Pluim J. P. W., “Elastix: A toolbox for intensity-based medical image registration,” IEEE Trans. Med. Imaging 29, 196–205 (2010). 10.1109/tmi.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 42.Madabhushi A. and Udupa J. K., “New methods of MR image intensity standardization via generalized scale,” Med. Phys. 33, 3426–3434 (2006). 10.1118/1.2335487 [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharyya A., “On a measure of divergence between two multinomial populations,” Indian J. Stat. 7, 401–406 (1946). [Google Scholar]