Abstract
Multilayer assembly is a commonly used technique to construct multilayer polydimethylsiloxane (PDMS)-based microfluidic devices with complex 3D architecture and connectivity for large-scale microfluidic integration. Accurate alignment of structure features on different PDMS layers before their permanent bonding is critical in determining the yield and quality of assembled multilayer microfluidic devices. Herein, we report a custom-built desktop aligner capable of both local and global alignments of PDMS layers covering a broad size range. Two digital microscopes were incorporated into the aligner design to allow accurate global alignment of PDMS structures up to 4 in. in diameter. Both local and global alignment accuracies of the desktop aligner were determined to be about 20 μm cm−1. To demonstrate its utility for fabrication of integrated multilayer PDMS microfluidic devices, we applied the desktop aligner to achieve accurate alignment of different functional PDMS layers in multilayer microfluidics including an organs-on-chips device as well as a microfluidic device integrated with vertical passages connecting channels located in different PDMS layers. Owing to its convenient operation, high accuracy, low cost, light weight, and portability, the desktop aligner is useful for microfluidic researchers to achieve rapid and accurate alignment for generating multilayer PDMS microfluidic devices.
I. INTRODUCTION
Soft lithography is a widely used technique for rapid prototyping of polydimethylsiloxane (PDMS)-based microfluidic devices owing to its simplicity and low cost.1 Microfluidic device fabricated using soft lithography replicates structure features on a hard master mold defined normally by photolithography, which is typically in 2D. By carefully designing microchannel geometries, 2D PDMS microfluidic devices generated by soft lithography have realized a broad array of functionalities in controlled microfluidic environments, including diffusion, mixing, flow focusing, separation, and droplet generation.2 Assembled multilayer microfluidic devices with 3D channel networks and structures, however, are required to enable additional flow control and complex device design, architecture, and functionality. For example, widely used PDMS microfluidic pneumatic valves and pumps are enabled by additional air channels positioned below flow channels in different PDMS layers of a multilayer microfluidic device.3 The emerging field of organs-on-chips also depends on intricate 3D microstructures and microfluidic environments to recapitulate 3D physiological and pathological microenvironments in vivo.4 These important applications call for convenient, precise, and standardized fabrication techniques that can be extended from standard 2D soft lithography for generating 3D PDMS microfluidic devices.
Currently, there are two major approaches for fabrication of 3D microfluidic devices: direct 3D fabrication and multilayer assembly. In direct 3D fabrication,5,6 monolithic 3D structures are generated in a single step. For example, in direct laser writing, a glass substrate is ablated pointwise by focused femtosecond laser.5 By moving the laser focal point along a prescribed 3D curve, a 3D microfluidic channel can be generated. In direct ink writing,6 3D geometries of microscale channels are determined by direct-write assembly of layered fugitive inks. However, both methods are very selective on substrate materials (not compatible with PDMS) and require specialized equipments for fabrication implementation. More recently, 3D printing technology has been used to fabricate monolithic 3D microfluidic devices directly from thermoplastic7 or photocurable resins.8 These recent advances have demonstrated the great potential of 3D printing for generating complex 3D microscale structures that are hard to achieve otherwise. However, the current resolution of 3D printing (∼100 μm) is still suboptimal for most microfluidic applications. In addition, the low speed of 3D printing prevents large-scale fabrication of microfluidic devices for practical applications.
Alternatively, 3D PDMS microfluidic devices can be fabricated by stacking and bonding multiple PDMS layers with 2D structures, a technique called multilayer assembly. In multilayer assembly, multiple PDMS layers containing 2D structures are aligned and bonded together to form a function microfluidic device embedded with intricate 3D channel networks and structures. Compared with direct 3D fabrication techniques, multilayer assembly requires dissecting 3D fluidic geometry into multiple 2D layers, which can complicate device design and fabrication processes. Despite this drawback, multiplayer assembly does not require any special equipment other than those needed by soft lithography and thus greatly simplifies fabrication processes for 3D PDMS microfluidic devices.9–11 Moreover, a distinct advantage of multiplayer assembly over direct 3D fabrication techniques is the easiness of handling 2D PDMS layers, making multilayer assembly the most widely used technique to fabricate 3D PDMS microfluidic devices for large-scale microfluidic integration,12 organs-on-chips,13,14 and high-throughput cell separation.15
The resolution of PDMS microfluidic devices fabricated by multiplayer assembly is largely dependent on the alignment accuracy when stacking and bonding multiple structured 2D PDMS layers. Alignment by hand is widely used for quick assembly of 3D PDMS microfluidic devices.16 However, intrinsic large alignment error and performance variation make it suboptimal for integrated multilayer microfluidic devices requiring high alignment accuracy.17 Conventional mask aligners used in microfabrication cleanroom designed specifically for Si-based microfabrication can achieve alignment accuracy down to submicron. However, these mask aligners cannot be directly used for alignment of PDMS-based microfluidic devices, due to notable differences between thin, rigid, and standard-sized Si wafers and thick, deformable PDMS layers of irregular sizes. Moreover, mask aligners are large and expensive machines dedicated for cleanroom usages only. Combined together, these issues render conventional mask aligners hardly useful for microfluidics researchers working extensively with PDMS structures to generate 3D microfluidic devices.
A variety of custom-built alignment systems and mechanisms have been developed for alignment of PDMS layers when using multilayer assembly to construct 3D microfluidic devices. One approach is to use mechanical jigs to align different PDMS pieces.11,18 Alignment error of multilayer assembly using jigs depends on the jig tolerance and PDMS device size, with alignment accuracy reported ranging from 50 to 100 μm cm−1 (an alignment accuracy of 50–100 μm when aligning 1-cm-wide devices). Since using mechanical jigs for alignment of different PDMS layers requires additional structural features (e.g., defined device size and protrusions and holes as guidance for alignment) on PDMS layers, it can complicate design and fabrication of 3D microfluidic devices. To develop an aligner system compatible with soft lithography, an automated PDMS alignment and bonding system has been developed.17 The design of this system was similar to conventional mask aligners but with the sample mounting parts modified for easy handling of PDMS devices. With a high-accuracy stereoscope and a precise XYZ translation stage, alignment accuracy achieved by this automated aligner was about 50 μm cm−1. However, equipped with only one stereoscope, this system was limited to alignment of PDMS pieces smaller than the field of view of the stereoscope (<1 cm).
Here, we report a high-resolution, custom-built desktop aligner capable of both local and global alignments of PDMS devices covering a broad size range. The desktop aligner was equipped with two digital microscopes to allow accurate global alignments of PDMS structures up to 4 in. in diameter. Both local and global alignment accuracies of the desktop aligner were characterized to be about 20 μm cm−1 using defined alignment marks of different sizes. We further applied the desktop aligner to achieve accurate alignments of different functional PDMS layers in 3D multilayer microfluidics including a lung-on-a-chip device as well as a microfluidic device integrated with vertical passages (vias) connecting channels located in different PDMS layers. The desktop aligner inherited the advantages of conventional cleanroom mask aligners in terms of versatility, easy operation, and high accuracy and at the same time was meticulously engineered to be low cost, light weight, and portable. These highly desirable features make the desktop aligner readily accessible to microfluidics researchers for rapid, convenient, and standardized alignment of different PDMS layers when using multilayer assembly to fabricate 3D microfluidic devices.
II. DESKTOP ALIGNER DESIGN
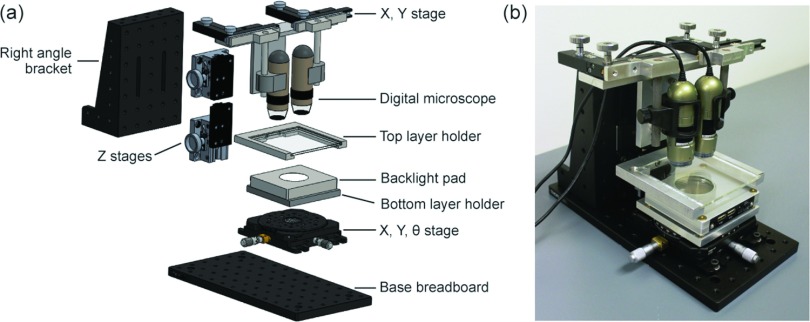
Design of the desktop aligner followed that of conventional mask aligners. Each component of the desktop aligner was carefully designed and engineered to accommodate handling of PDMS layers. Figure 1(a) is the 3D CAD model showing design of the desktop aligner consisting of mechanical positioning stages to place PDMS layers and an imaging system to monitor alignment of the PDMS layers in real time.
FIG. 1.
(a) 3D CAD model showing desktop aligner design. (b) Photograph of assembled desktop aligner placed on a lab bench.
A. Mechanical positioning system
PDMS layers to be aligned were considered rigid when designing the mechanical positioning system for placing the PDMS layers. The alignment process could be abstracted as completely defining relative positions of the two PDMS layers to be aligned, i.e., reducing the degree of freedom (DOF) from six to zero. This could be achieved by mounting one PDMS layer on a static base, while mounting the other on a six-axis platform. To further simplify the mechanical positioning system, we assumed that the two PDMS layers to be aligned were always parallel to each other to reduce the DOF from six to four. This assumption could be easily satisfied by ensuring that all PDMS layers had uniform thicknesses during soft lithography, e.g., by carefully placing PDMS-covered Si masters on a horizontal surface during the PDMS curing step. The other four DOFs were adjustable using translational and rotation stages. Specifically, the bottom PDMS layer holder was mounted onto an XYR platform (#XYR1, Thorlabs), while the top PDMS layer holder was mounted onto a Z translation stage (#123-4710, OptoSigma) (Figure 1(a)). Since accuracies in translational and rotation stages dictated overall alignment accuracy, the XYR platform was selected to have a high resolution of 10 μm. The Z translation stage that was used to control the Z-position of the top PDMS holder and bring the two PDMS layers into conformal contact. Therefore, a dove-and-pinion stage with a relatively low resolution of 100 μm was selected for the Z translation stage. Since PDMS can reversibly bond to smooth glass surfaces under the van der Waals force,1 a piece of 4 in. glass plate was used in conjunction with the top PDMS layer holder to position the top PDMS layer (Figure 1(a)). The bottom PDMS layer was directly placed onto the bottom PDMS layer holder and remained affixed during the entire alignment process. Both the top and bottom PDMS layer holders were fabricated using a manual milling machine.
B. Optical system
The optical system is a critical component in the desktop aligner for monitoring relative positions and alignment of alignment marks on the top and bottom PDMS layers. To reduce cost while increasing portability of the desktop aligner, hand-held compact digital microscopes (AD4113 T, Dino-lite Digital Microscope) with a resolution of 3 μm were used for real-time monitoring of alignment (Figure 1(a)). Each digital microscope was mounted onto a small XY translation stage (#910, National Optical) with a resolution of 100 μm to position the two digital microscopes right above alignment marks on the top and bottom PDMS layers. The two microscope-mounting XY stages were both mounted on a single Z translation stage (#123-4710, OptoSigma) so that the Z translation stage could be adjusted to simultaneously focus the two microscopes on the alignment marks (Figure 1(a)). Positions of the two microscopes on the small XY translation stages were adjusted and fixed such that they were always perpendicular to the top and bottom PDMS layer holders.
To obtain clear images for transparent PDMS layers, a backlight pad (BL-ZW1, Dino-lite Digital Microscope) was integrated onto the bottom PDMS layer holder of the desktop aligner (Figure 1(a)). During alignment, the bottom PDMS layer was placed directly on top of the backlight pad to provide transmitted light. The digital microscopes were also integrated with reflected light for imaging non-transparent layers (e.g., PDMS coated on a silicon wafer).
C. System integration
To provide a steady support for the desktop aligner, a monolithic optical breadboard with a size of 12 in. × 6 in. (MB612F, Thorlabs) was used as the base of the desktop aligner. The XYR stage and the bottom PDMS layer holder were mounted directly onto the base breadboard. A large right angle bracket (AP90RL, Thorlabs) was also mounted onto the base breadboard to serve as a vertical plane for mounting the top PDMS layer holder and the optical system. This strategic positioning resulted in a highly compact design of the desktop aligner, with an overall size of 12 in. × 6 in. × 10 in. (L × W × H). Owing to high tolerance and reliable mounting enabled by standard optomechanics components, the whole desktop aligner was steady and robust. Because of its small volume, the desktop aligner was highly portable and could be easily placed onto conventional laboratory benches or in biosafety cabinets where sterilization might be required. Figure 1(b) illustrates the assembled desktop aligner on a laboratory bench. During operation of the aligner, the two digital microscopes were connected to a laptop through USB for monitoring the alignment process.
D. Alignment procedure
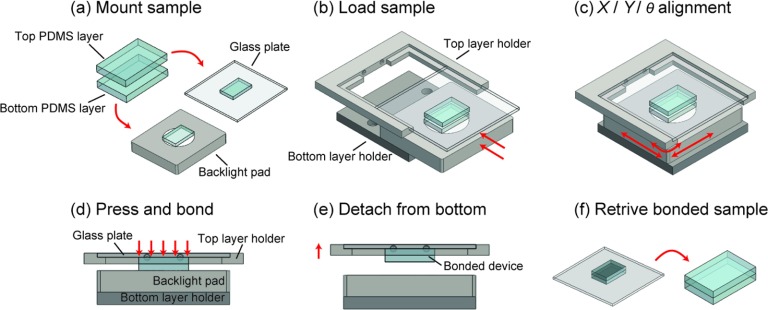
Prior to alignment, the two PDMS layers to be aligned were treated briefly with oxygen plasma for permanent bonding. This step should be skipped if reversible bonding of the two PDMS layers were desired. After this optional plasma treatment, the top PDMS layer was reversibly bonded to the 4 in. glass plate with structured features and alignment marks facing downward. The bottom PDMS layer was placed directly onto the backlight pad (Figure 2(a)). In practice, the PDMS layers were always placed at the center of the glass plate and the backlight pad to facilitate rapid alignment. The glass plate attached with the top PDMS layer was then inserted into the top PDMS layer holder. The backlight pad with the bottom PDMS layers on top was inserted into the bottom PDMS layer holder (Figure 2(b)).
FIG. 2.
Alignment procedure using the desktop aligner. (a) Top and bottom PDMS layers to be aligned were first mounted onto a 4 in. glass plate and a backlight pad, respectively. (b) The glass plate and the backlight pad were then inserted into the and bottom layer holders of the aligner, respectively. (c) Digital microscopes were focused onto the alignment marks on the PDMS layers before the X/Y/θ stage beneath the backlight pad was carefully adjusted such that the alignment marks on the top and bottom PDMS layers completely overlapped with each other. (d) The top and bottom PDMS layers were bonded with each other by slightly lowering the top layer holder. (e) The top layer holder was raised to allow bonded PDMS layers to detach from the backlight pad. (f) The glass plate was retrieved from the top layer holder, and the bonded PDMS device was gently peeled off from the glass plate.
To fasten the alignment process, rough alignment was first conducted by adjusting the X, Y, and θ of the XYR platform while monitoring positions of the top and bottom PDMS layers by naked eyes. The top PDMS layer holder was then lowered to place the top PDMS layer in close proximity (<1 mm) to the bottom PDMS layer. Care should be taken during this step to avoid direct contact between the top and bottom PDMS layers, which might lead to permanent bonding of the PDMS layers before successful alignment. Height of the digital microscopes was slowly adjusted to focus on the alignment marks on the bottom PDMS layer, which remained affixed during the entire alignment and bonding process. By monitoring live video captured by the digital microscopes, we could determine easily how far the alignment marks on the top and bottom PDMS layers were away from each other and adjust appropriately the X, Y, and θ of the XYR platform (Figure 2(c)). For global alignment, this alignment process could be an iterative process till both the alignment marks on the left and right overlapped concurrently. After the alignment was accomplished, the two PDMS layers were bonded together by slightly lowering the top PDMS layer holder (Figure 2(d)). The top PDMS layer holder was raised again with the bonded PDMS device attached to the glass plate on the top PDMS layer holder (Figure 2(e)). The glass plate was retrieved from the top PDMS layer holder, and the bonded PDMS device was gently peeled off from the glass plate (Figure 2(f)). A brief baking step at 110 °C could be employed to strengthen covalent bonding between the PDMS layers.
For multilayer assembly of PDMS devices smaller than 1 cm, the field of view of a single digital microscope would be large enough for imaging the whole device. In this case, the alignment procedure was the same as the procedure described above, except that only one digital microscope was used during alignment.
III. CALIBRATION OF ALIGNMENT ACCURACY
Detailed calibration of the desktop aligner was carried out to characterize its performance including both local and global alignment accuracies. It should be noted that alignment accuracy under one digital microscope would dictate the overall alignment accuracy of the aligner and was thus determined first. Such calibration of local alignment accuracy was also needed for alignment of multilayer PDMS microfluidic devices smaller than 1 cm that could fit the field of view of a single digital microscope used in this work. For global alignment of larger multilayer PDMS microfluidic devices or even whole wafers, both digital microscopes needed to be used simultaneously, in which case global alignment accuracy would determine the overall alignment across the entire device or wafer.
A. Local alignment accuracy under one microscope
To quantify alignment accuracy, PDMS layers containing square-shaped alignment marks were fabricated by soft lithography. Briefly, Si molds were first fabricated using contact lithography and deep reactive ion etching (DRIE; SPTS Pegasus). Si molds were primed with (tridecafluoro-1, 1, 2, 2,-tetrahydrooctyl)-1-trichlorosilane (United Chemical Technologies) for 1 hr under vacuum to facilitate subsequent release of cured PDMS from the molds. PDMS precursor with 10:1 (wt:wt) base to curing agent ratio was prepared and poured onto Si molds, followed by degassing in a vacuum desiccator. PDMS precursor was baked in a 60 °C oven overnight before peeled off from molds and cut into pieces.
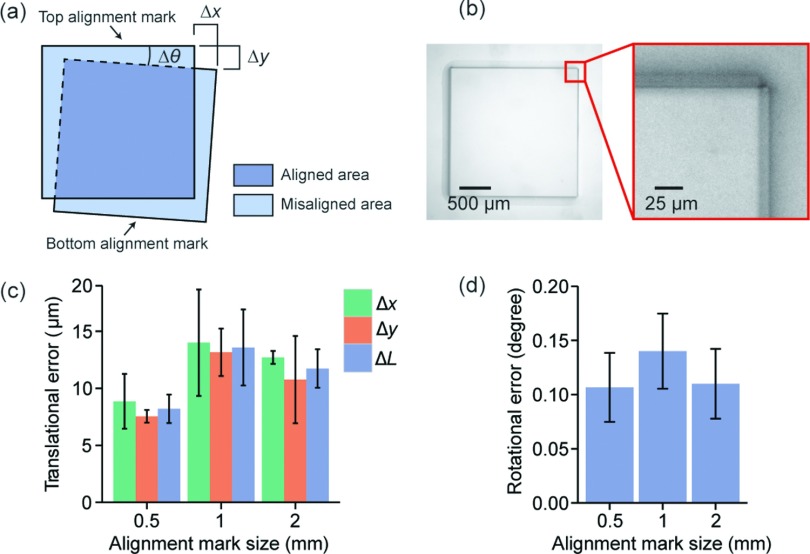
For calibration of alignment accuracy, the top and bottom PDMS layers were identical and both contained square-shaped alignment marks of different sizes. A perfect alignment would result in a complete overlap of the square-shaped alignment marks from the top and bottom PDMS layers. In practice, there would be translational and angular shifts between the two PDMS layers. Alignment accuracy was thus characterized by the maximum shift (misalignment) of alignment marks in the X (Δx) and Y directions (Δy) and the angular shift (Δθ) measured using AxioVision software (Carl Zeiss Microscopy) (Figure 3(a)).
FIG. 3.
Characterization of local alignment accuracy using one digital microscope. (a) Schematic showing definition of alignment errors. Translational alignment errors were characterized by the maximum alignment shifts of the top and bottom square-shaped alignment marks in the X and Y directions (Δx and Δy) and their average ΔL(ΔL = (Δx + Δy)/2). The rotational alignment error was defined by the angular shift (Δθ). (b) Representative micrograph showing alignment of 2-mm square-shaped alignment marks on the top and bottom PDMS layers. The top right corner of the alignment marks was magnified to show alignment details. (c) Translational alignment errors with relationship to square-shaped alignment mark sizes. (d) Rotational alignment error as a function of square-shaped alignment mark sizes. Error bars in (c) and (d) represent standard error of the mean calculated from 3 different devices (n = 3).
To investigate the effect of feature size on alignment accuracy, square-shaped alignment marks with widths of 0.5 mm, 1 mm, and 2 mm were fabricated and aligned under the same microscope magnification (60×). For each mark size, there was no significant difference (p > 0.05, paired student t-test) between alignment errors in either X- or Y-direction, supporting that alignment accuracy was not biased toward a specific direction (Figure 3(c)). In addition, translational alignment accuracy was not dependent on alignment mark size (p > 0.05, one-way ANOVA test). To compare with alignment platforms reported previously, we further defined translational accuracy of the aligner (ΔL) as ΔL = (Δx + Δy)/2. Translational accuracy ΔL of the desktop aligner was 8.2 ± 1.2 μm for 0.5-mm marks, 13.6 ± 3.3 μm for 1-mm marks, and 11.7 ± 1.7 μm for 2-mm marks, very comparable with results reported previously using more complex aligner designs.17 The translational errors were below 20 μm in all measurements. Similar to translational errors, rotational error was not dependent on mark size either (p > 0.05, one-way ANOVA test). The rotational error was 0.11 ± 0.03° for 0.5-mm mark, 0.14 ± 0.03° for 1-mm mark, and 0.11 ± 0.03° for 2-mm mark (Figure 3(d)). Overall, rotational error of the desktop aligner was consistently below 0.2° or 3.5 × 10−3 rad.
B. Global alignment accuracy under two microscopes
During alignment, mismatch in rotational directions of the top and bottom PDMS layers would cause translational alignment errors along the X and Y directions. For a device with a size of w, translational error resulted from rotational mismatch Δθ can be estimated to be about w × Δθ. Notably, translational error induced by rotation mismatch increased linearly with device size. Considering alignment of a 3-cm long microfluidic device with a rotational error of 0.2°, translational alignment error resulted from rotational mismatch alone would be as large as 100 μm. To address such issue of misalignment propagation due to rotational mismatch, multiple alignment marks are needed for proper global alignments of large microfluidic devices, also a common practice for wafer-scale alignment using conventional mask aligners.
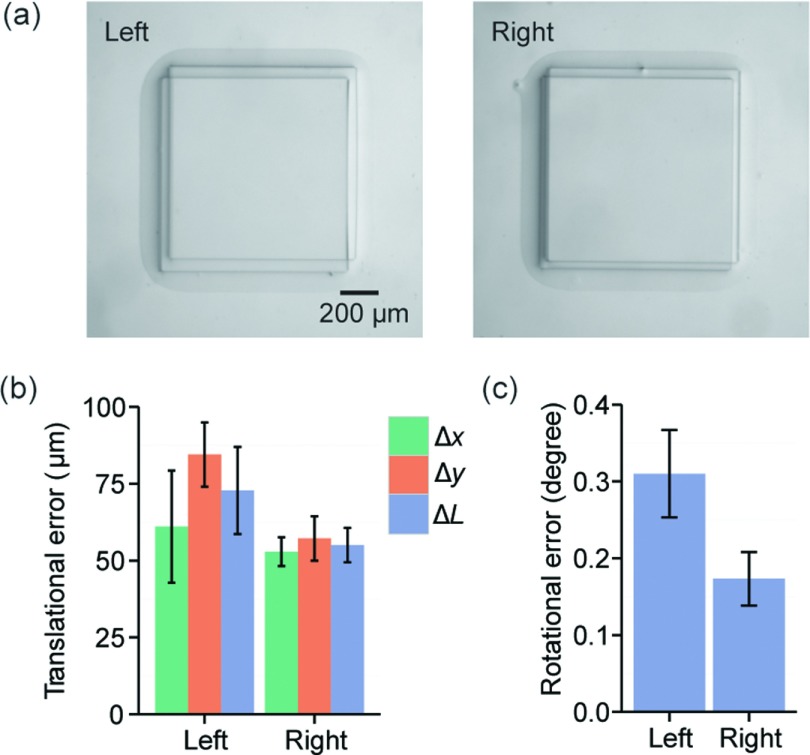
To examine global alignment using the desktop aligner, two identical large PDMS layers containing 1-mm wide square-shaped alignment marks positioned 3 cm apart were aligned using the two digital microscopes simultaneously. Microscopic images showing mark alignments suggested that alignment errors resulted from global alignments of large PDMS devices using two microscopes were noticeably greater than those from local alignments of small PDMS devices under a single microscope (Figure 4(a)). We further quantified translational and angular alignment errors, and indeed, the translational accuracy of the aligner ΔL under the global alignment mode was 72.8 ± 24.5 μm and 55.0 ± 5.6 μm for the left and right alignment marks, respectively (Figure 4(b)). The translational errors were below 100 μm in all measurements. There was no significant difference between translational errors on the left and right alignment marks (p > 0.05, one-way ANOVA test), attributable to the fact that during global alignment, we normally would make effort to balance alignment on both left and right sides of PDMS devices. The global alignment accuracy was thus calculated to be about 21.3 ± 2.6 μm cm−1. Previous studies have suggested that the global alignment accuracy during PDMS layer alignment is primarily affected by PDMS shrinkage during curing.19 Under the curing condition used in this work, the shrinkage ratio of PDMS is about 1%, with a variance of 0.2%.20 This translates to an alignment error of 20 μm cm−1 when aligning two PDMS layers, very comparable to the value obtained from our experiments. To achieve higher global alignment accuracy, it has been suggested that one may need more sophisticated techniques such as sandwich mold fabrication for soft lithography.20,21
FIG. 4.
Characterization of global alignment accuracy using two digital microscopes. (a) Representative micrographs showing alignments of a pair of 1-mm square-shaped alignment marks positioned 3 cm apart on the top and bottom PDMS layers. (b) Translational alignment errors determined from the left and right alignment marks, as indicated. (c) Rotational alignment errors determined from the left and right alignment marks, as indicated. Error bars in (b) and (c) represent standard error of the mean calculated from 3 different devices (n = 3).
Rotational error resulted from global alignments of large PDMS devices was 0.31 ± 0.06° and 0.17 ± 0.03° for the left and right alignment marks, respectively, slightly greater than those observed in local alignment (Figure 4(c)). Although there was no significant difference between average rotational errors on the left and right alignment marks (p > 0.05, one-way ANOVA test), in each individual alignment, the left and right rotation errors were not identical, likely due to the flexibility of PDMS layers, leading to their deformation during the alignment procedure. Such non-linear effect could be more prominent for large PDMS device. Taking the average rotation error on both left and right alignment marks () and the distance between the alignment marks (w = 3 cm), the translational error should be at least 126 μm, almost twice as large as the translation accuracy ΔL obtained experimentally, supporting the importance and necessity to apply two microscopes simultaneously for global alignments of large multilayer PDMS devices.
IV. APPLICATIONS OF DESKTOP ALIGNER
A. Lung-on-a-chip device
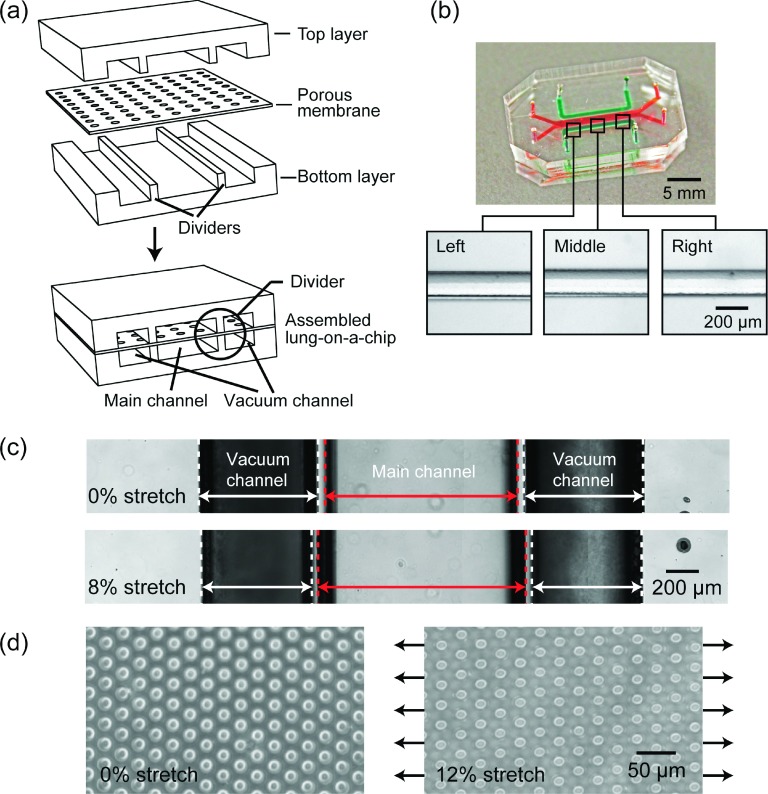
To demonstrate its utility for fabrication of multilayer PDMS microfluidic devices, we applied the desktop aligner to achieve accurate alignment of different functional PDMS layers in a lung-on-a-chip device.13 Such lung-on-a-chip device developed recently has received a significant attention as it has been shown to reconstitute the critical functional alveolar-capillary interface of the human lung, useful for expanding the capabilities of cell culture models and providing low-cost alternatives to animal and clinical studies for drug screening and toxicology applications. This lung-on-a-chip device comprised top and bottom PDMS layers containing microchannels and a thin, porous PDMS membrane that was sandwiched between the two PDMS layers (Figure 5(a)).13 Importantly, both the top and bottom PDMS layers contained a main cell culture channel in the middle and two vacuum actuation channels on the sides. There were two dividers in each PDMS layer to separate the three channels apart. The opposite sides of the porous PDMS membrane sandwiched between the top and bottom PDMS layers would be used for adhesion of alveolar epithelial cells and microvascular endothelial cells. During device operation, the side channels were periodically activated by vacuum, leading to stretches of the porous membrane and thus cells attached to it. It would be critically important that the dividers on the top and bottom PDMS layers were precisely aligned and bonded together. Otherwise, the lung-on-a-chip device would subject to vacuum leakage and operational failure.
FIG. 5.
(a) Schematic showing design and assembly of a PDMS-based tri-layer lung-on-a-chip device consisting of identical top and bottom layers separated by a porous membrane. The circled area highlights dividers with a width of 150 μm that separate main and vacuum channels in the top and bottom layers. (b) Photograph showing a functional lung-on-a-chip device assembled using the desktop aligner. Highlighted rectangular regions are magnified to show precise alignments of dividers in the top and bottom PDMS layers across the whole lung-on-a-chip device. (c) Bright field images showing displacements of the dividers before (top) and after (bottom) activation of vacuum in the vacuum channels, resulting in 8% uniaxial stretch of the thin porous membrane sandwiched between the top and bottom PDMS layers. (d) Circular through holes on the porous membrane before (left) and after (right) 12% uniaxial stretch. Note that after stretch, circular through holes deformed into oval shaped holes.
As the width of the dividers was 150 μm, it was difficult to align them completely by hand under a stereoscope. Given that the overall size of the lung-on-a-chip device was 1 cm × 2.5 cm, we applied the desktop aligner and used the global alignment mode for its alignment and assembly. Figure 5(b) shows aligned dividers from the top and bottom PDMS layers using the desktop aligner, with alignment mismatch hardly observable by naked eye. The maximum misalignment between dividers from the top and bottom PDMS layers was 23.2 ± 1.7 μm, which was about 15% of the divider width. Moreover, after connecting the side vacuum actuation channels to vacuum, the dividers and the porous membrane were successfully stretched (Figures 5(c) and 5(d)), supporting the proper functionality of the lung-on-a-chip device.
B. Microfluidic device integrated with vias
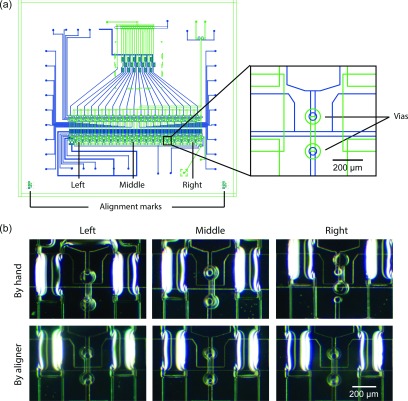
We further applied the desktop aligner to achieve accurate global alignment for a 2-layer microfluidic device integrated with vias connecting channels located in different PDMS layers. The 3 cm × 3 cm microfluidic device contained a flow layer and a control layer (Figure 6(a)). The most critical structures on this multilayer microfluidic device were 50-μm-diameter vias on the control layer and 100-μm-diameter vias on the flow layers. To form functional interconnections between the two PDMS layers, the smaller vias in the control layer needed to be properly positioned inside the larger vias in the flow layer. As shown in Figure 6(c), the 2-layer microfluidic device aligned using the desktop aligner had consistent small alignment errors (<30 μm) across the entire device (Figure 6(c)), and all vias on the control layer were properly located inside the vias on the flow layer. In distinct comparison, the microfluidic device aligned by hand under a stereoscope showed very significant spatial variations of alignment errors across the entire device. While the vias in the device center were properly aligned, those on the device edges were misaligned, resulting in device fabrication failure. This spatial variation of alignment accuracy was attributable to the unsteady sample positioning in alignment by hand and the limited field of view of the stereoscope, under which only a small portion of the 2-layer microfluidic device could be seen.
FIG. 6.
(a) Schematic of a microfluidic device integrated with vertical passages (vias) connecting channels located in different PDMS layers (i.e., flow and control layers). The flow and control layers are plotted in green and blue, respectively. The whole microfluidic device was 3 cm × 3 cm. Vias on the control layer (D = 50 μm, blue) needed to be aligned with vias on the flow layer (D = 100 μm, green) to form functional interconnections. (b) (Top) Micrographs showing alignments by hand of vias on the control and flow layers. While vias in the middle of the device were properly aligned, those on the left and right edges of the device were completely misaligned. (Bottom) Micrographs showing alignments of vias on the control and flow layers using the desktop aligner. Global alignments using the desktop aligner ensured that alignment errors were consistent across the entire device, with all vias on the control layer located within vias on the flow layer.
V. CONCLUSION
In this paper, we reported a desktop aligner for multilayer assembly of microfluidic devices with 3D channel networks and structures. The desktop aligner was designed with four highly desired features that made it optimal for microfluidics researchers to achieve both local and global alignments for multilayer assembly of 3D PDMS microfluidic devices. First, the desktop aligner was highly compact and portable, with a total volume of only 12 in. × 6 in. × 10 in.. This suggests that the desktop aligner can be easily integrated into a variety of settings where space may be limited (such as regular laboratory benches or biosafety cabinets). The compact design of the desktop aligner was achieved by strategically positioning and arranging different mechanical and optical components such as digital microscopes, translation stages, and device holders into a compact setup. Second, the desktop aligner, equipped with two digital microscopes, is easy to use and can quickly generate accurate local and global alignment results for PDMS devices covering a broad size range. This advantage was made possible by eliminating unnecessary degree of freedom in the mechanical positioning and imaging subsystems. Third, the desktop aligner can achieve both local and global alignments with a resolution of about 20 μm cm−1, which is sufficient for many important microfluidic device applications that are difficult to realize by manual alignments. Finally, the highly accurate desktop aligner is achieved through an efficient and economic design. With such high portability, accuracy, easiness to use, and low cost, the desktop aligner reported in this work will be useful for microfluidics researchers to achieve rapid and accurate alignment for generating multilayer PDMS microfluidic devices.
Acknowledgments
We acknowledge financial support from the National Science Foundation (Nos. ECCS 1231826 and CBET 1263889), the National Institutes of Health (No. R01 HL119542), the UM-SJTU Collaboration on Biomedical Technologies, and the Michigan Center for Integrative Research in Critical Care (M-CIRCC). We thank Marvin Cressey from the Department of Mechanical Engineering at the University of Michigan for help with machining. The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
REFERENCES
- 1.Anderson J. R., Chiu D. T., Wu H., Schueller O. J., and Whitesides G. M., Electrophoresis 21, 27 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Stone H. A., Stroock A. D., and Ajdari A., Annu. Rev. Fluid Mech. 36, 381 (2004). 10.1146/annurev.fluid.36.050802.122124 [DOI] [Google Scholar]
- 3.Unger M. A., Chou H.-P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 4.Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 5.Liao Y., Song J., Li E., Luo Y., Shen Y., Chen D., Cheng Y., Xu Z., Sugioka K., and Midorikawa K., Lab Chip 12, 746 (2012). 10.1039/c2lc21015k [DOI] [PubMed] [Google Scholar]
- 6.Therriault D., White S. R., and Lewis J. A., Nat. Mater. 2, 265 (2003). 10.1038/nmat863 [DOI] [PubMed] [Google Scholar]
- 7.Kitson P. J., Rosnes M. H., Sans V., Dragone V., and Cronin L., Lab Chip 12, 3267 (2012). 10.1039/c2lc40761b [DOI] [PubMed] [Google Scholar]
- 8.Au A. K., Lee W., and Folch A., Lab Chip 14, 1294 (2014). 10.1039/c3lc51360b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson J. R., Chiu D. T., Jackman R. J., Cherniavskaya O., McDonald J. C., Wu H., Whitesides S. H., and Whitesides G. M., Anal. Chem. 72, 3158 (2000). 10.1021/ac9912294 [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Wu J., Wang L., Xiao K., and Wen W., Lab Chip 10, 1199 (2010). 10.1039/b923101c [DOI] [PubMed] [Google Scholar]
- 11.Chen A. and Pan T., Biomicrofluidics 5, 046505 (2011). 10.1063/1.3670368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorsen T., Maerkl S. J., and Quake S. R., Science 298, 580 (2002). 10.1126/science.1076996 [DOI] [PubMed] [Google Scholar]
- 13.Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328, 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan C. Y., Huang P.-H., Guo F., Ding X., Kapur V., Mai J. D., Yuen P. K., and Huang T. J., Lab Chip 13, 4697 (2013). 10.1039/c3lc90115g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Chen W., Liu G., Lu W., and Fu J., Lab Chip 14, 2565 (2014). 10.1039/c4lc00350k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huh D., Kim H. J., Fraser J. P., Shea D. E., Khan M., Bahinski A., Hamilton G. A., and Ingber D. E., Nat. Protoc. 8, 2135 (2013). 10.1038/nprot.2013.137 [DOI] [PubMed] [Google Scholar]
- 17.Kim J., Baek J., Lee K., and Lee S., Sens. Actuators, A 119, 593 (2005). 10.1016/j.sna.2004.09.023 [DOI] [Google Scholar]
- 18.Jeong J., Chun K., Kim J., and Lee B., in Proceedings of AFRICON, AFRICON’09 (IEEE, 2009). [Google Scholar]
- 19.Lee S. W. and Lee S. S., Microsyst. Technol. 14, 205 (2008). 10.1007/s00542-007-0417-y [DOI] [Google Scholar]
- 20.Badshah M. A., Jang H., Kim Y. K., Kim T.-H., and Kim S.-m., J. Micro/Nanolithogr., MEMS, MOEMS 13, 033006 (2014). 10.1117/1.JMM.13.3.033006 [DOI] [Google Scholar]
- 21.Moraes C., Sun Y., and Simmons C. A., J. Micromech. Microeng. 19, 065015 (2009). 10.1088/0960-1317/19/6/065015 [DOI] [Google Scholar]