Abstract
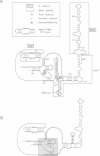
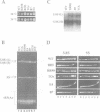
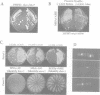
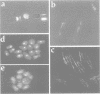
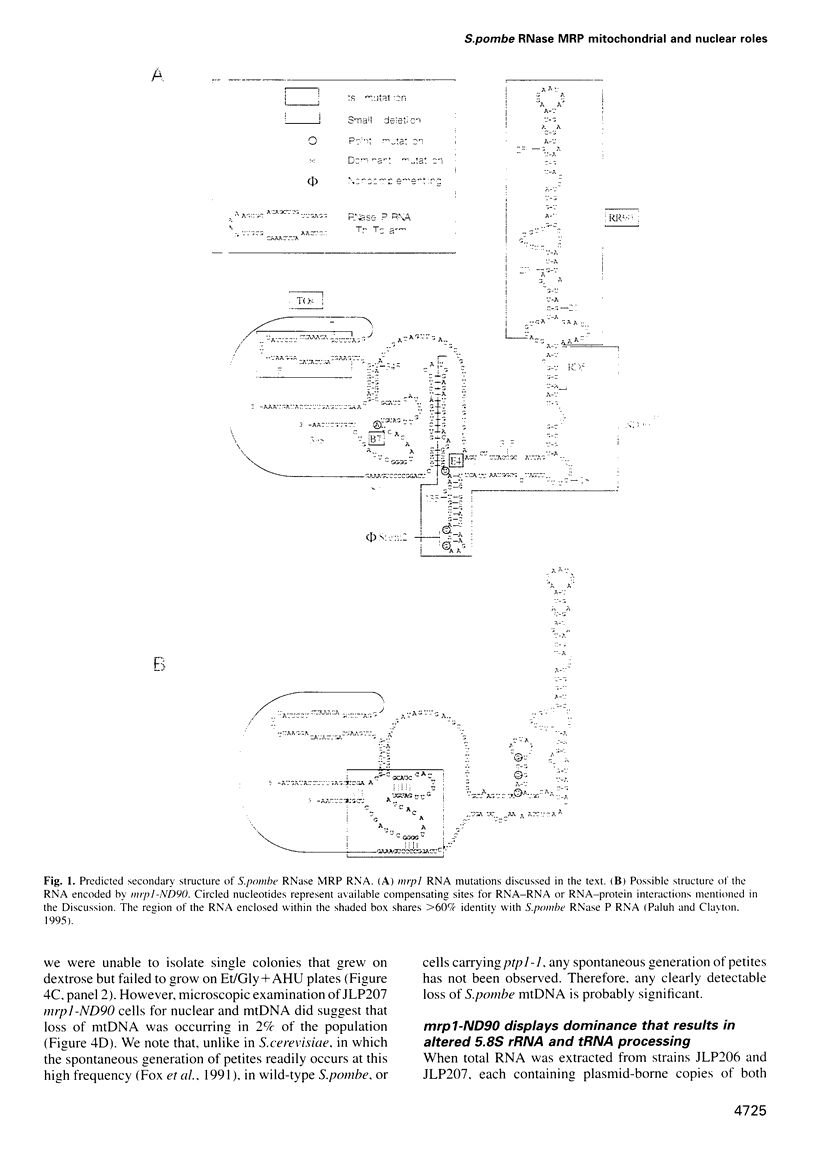
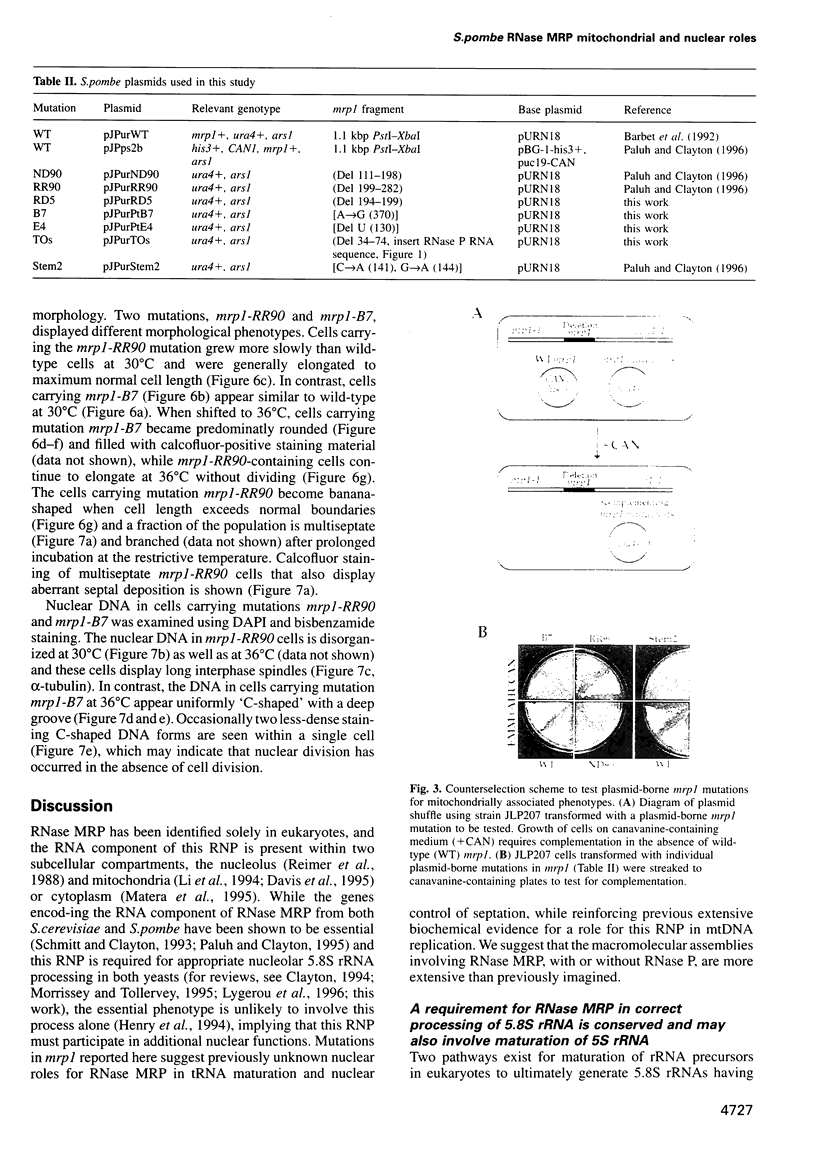
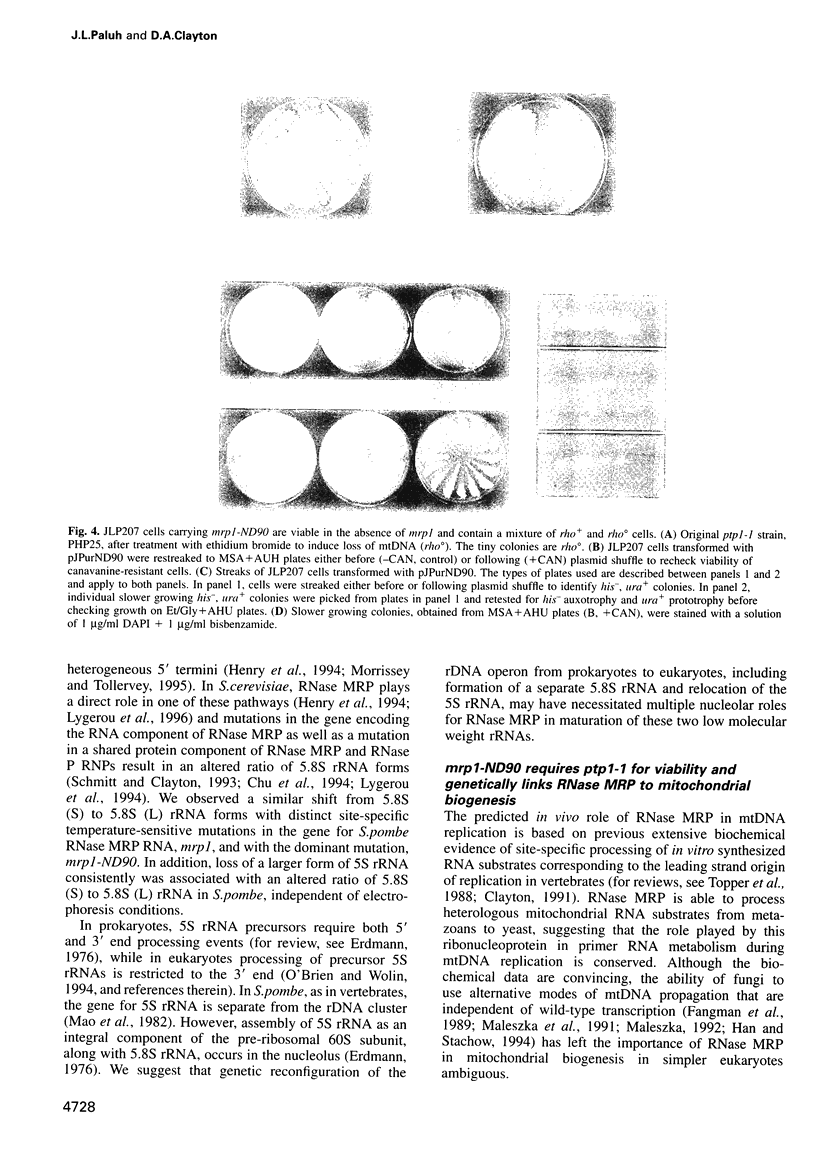
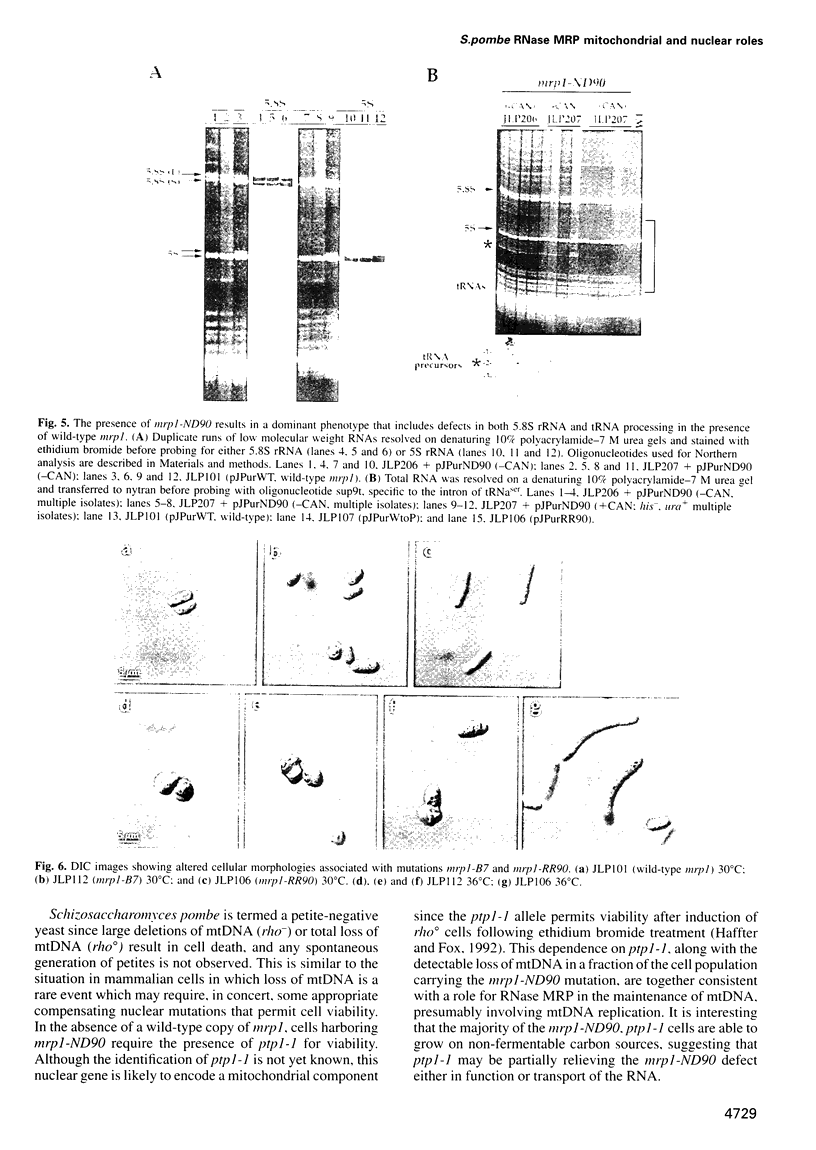
The essential gene for RNase MRP RNA, mrp1, was identified previously in Schizosaccharomyces pombe by homology to mammalian RNase MRP RNAs. Here we describe distinct site-specific mutations in RNase MRP RNA that support a conserved role for this ribonucleoprotein in nucleolar 5.8S rRNA processing. One characterized mutation, mrp1-ND90, displays dominance and results in accumulation of unspliced precursor RNAs of dimeric tRNA(Ser)-tRNA(Met)i, suggesting a novel nuclear role for RNase MRP in tRNA processing. Cells carrying the mrp1-ND90 mutation, in the absence of a wild-type copy of mrp1, additionally require the mitochondrially associated nuclear mutation ptp1-1 for viability. Analysis of this mrp1 mutation reinforces previous biochemical evidence suggesting a role for RNase MRP in mitochondrial DNA replication. Several mutations in mrp1 result in unusual cellular morphology, including alterated nuclear organization, and are consistent with a broader nuclear role for RNase MRP in regulating a nuclear signal for septation; these results are a further indication of the multifunctional nature of this ribonucleoprotein.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W. J., Carr A. M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992 May 1;114(1):59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Bennett J. L., Jeong-Yu S., Clayton D. A. Characterization of a Xenopus laevis ribonucleoprotein endoribonuclease. Isolation of the RNA component and its expression during development. J Biol Chem. 1992 Oct 25;267(30):21765–21772. [PubMed] [Google Scholar]
- Bothwell A. L., Garber R. L., Altman S. Nucleotide sequence and in vitro processing of a precursor molecule to Escherichia coli 4.5 S RNA. J Biol Chem. 1976 Dec 10;251(23):7709–7716. [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987 Feb;6(2):409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Nurse P. How fission yeast fission in the middle. Cell. 1996 Jan 26;84(2):191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chant J. Septin scaffolds and cleavage planes in Saccharomyces. Cell. 1996 Jan 26;84(2):187–190. doi: 10.1016/s0092-8674(00)80972-1. [DOI] [PubMed] [Google Scholar]
- Chu S., Archer R. H., Zengel J. M., Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. A nuclear function for RNase MRP. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Structure and function of the mitochondrial genome. J Inherit Metab Dis. 1992;15(4):439–447. doi: 10.1007/BF01799602. [DOI] [PubMed] [Google Scholar]
- Dairaghi D. J., Clayton D. A. Bovine RNase MRP cleaves the divergent bovine mitochondrial RNA sequence at the displacement-loop region. J Mol Evol. 1993 Oct;37(4):338–346. doi: 10.1007/BF00178864. [DOI] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Davis A. F., Jeong-Yu S., Clayton D. A. Distribution of RNase MRP RNA during Xenopus laevis oogenesis. Mol Reprod Dev. 1995 Nov;42(3):359–368. doi: 10.1002/mrd.1080420313. [DOI] [PubMed] [Google Scholar]
- Egel R., Willer M., Kjaerulff S., Davey J., Nielsen O. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 1994 Oct;10(10):1347–1354. doi: 10.1002/yea.320101012. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Ruusala T. Budding yeast CAN1 gene as a selection marker in fission yeast. Nucleic Acids Res. 1991 Mar 11;19(5):1150–1150. doi: 10.1093/nar/19.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Fangman W. L., Henly J. W., Churchill G., Brewer B. J. Stable maintenance of a 35-base-pair yeast mitochondrial genome. Mol Cell Biol. 1989 May;9(5):1917–1921. doi: 10.1128/mcb.9.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Folley L. S., Mulero J. J., McMullin T. W., Thorsness P. E., Hedin L. O., Costanzo M. C. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Gold H. A., Craft J., Hardin J. A., Bartkiewicz M., Altman S. Antibodies in human serum that precipitate ribonuclease P. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5483–5487. doi: 10.1073/pnas.85.15.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold H. A., Topper J. N., Clayton D. A., Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989 Sep 22;245(4924):1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Haffter P., Fox T. D. Nuclear mutations in the petite-negative yeast Schizosaccharomyces pombe allow growth of cells lacking mitochondrial DNA. Genetics. 1992 Jun;131(2):255–260. doi: 10.1093/genetics/131.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I. M., Hyams J. S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988 Mar;89(Pt 3):343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Han Z., Stachow C. Analysis of Schizosaccharomyces pombe mitochondrial DNA replication by two dimensional gel electrophoresis. Chromosoma. 1994 Jun;103(3):162–170. doi: 10.1007/BF00368008. [DOI] [PubMed] [Google Scholar]
- Hashimoto C., Steitz J. A. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J Biol Chem. 1983 Feb 10;258(3):1379–1382. [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J. P., Petfalski E., Kearsey S., Tollervey D. The 5' end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994 May 15;13(10):2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R., Cao L. G., Wang Y. L., Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol. 1995 Dec;131(6 Pt 2):1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992 Oct;11(10):3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Smagula C. S., Parsons W. J., Richardson J. A., Gonzalez M., Hagler H. K., Williams R. S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994 Mar;124(6):871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Séraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996 Apr 12;272(5259):268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z., Mitchell P., Petfalski E., Séraphin B., Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994 Jun 15;8(12):1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Maleszka R. Electrophoretic profiles of mitochondrial plasmids in Neurospora suggest they replicate by a rolling circle mechanism. Biochem Biophys Res Commun. 1992 Aug 14;186(3):1669–1673. doi: 10.1016/s0006-291x(05)81600-6. [DOI] [PubMed] [Google Scholar]
- Maleszka R., Skelly P. J., Clark-Walker G. D. Rolling circle replication of DNA in yeast mitochondria. EMBO J. 1991 Dec;10(12):3923–3929. doi: 10.1002/j.1460-2075.1991.tb04962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Appel B., Schaack J., Sharp S., Yamada H., Söll D. The 5S RNA genes of Schizosaccharomyces pombe. Nucleic Acids Res. 1982 Jan 22;10(2):487–500. doi: 10.1093/nar/10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massardo D. R., Manna F., Schäfer B., Wolf K., Del Giudice L. Complete absence of mitochondrial DNA in the petite-negative yeast Schizosaccharomyces pombe leads to resistance towards the alkaloid lycorine. Curr Genet. 1994 Jan;25(1):80–83. doi: 10.1007/BF00712972. [DOI] [PubMed] [Google Scholar]
- Matera A. G., Frey M. R., Margelot K., Wolin S. L. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995 Jun;129(5):1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison J. M., Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985 Apr;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrissey J. P., Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995 Feb;20(2):78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- O'Brien C. A., Wolin S. L. A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev. 1994 Dec 1;8(23):2891–2903. doi: 10.1101/gad.8.23.2891. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Clayton D. A. Schizosaccharomyces pombe RNase MRP RNA is homologous to metazoan RNase MRP RNAs and may provide clues to interrelationships between RNase MRP and RNase P. Yeast. 1995 Oct;11(13):1249–1264. doi: 10.1002/yea.320111305. [DOI] [PubMed] [Google Scholar]
- Piperno G., Fuller M. T. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985 Dec;101(6):2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin J., Frendewey D. A mutation in a single gene of Schizosaccharomyces pombe affects the expression of several snRNAs and causes defects in RNA processing. EMBO J. 1990 Feb;9(2):525–534. doi: 10.1002/j.1460-2075.1990.tb08139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Rossmanith W., Karwan R. RNase MRP and RNase P share a common substrate. Nucleic Acids Res. 1993 Jul 11;21(14):3239–3243. doi: 10.1093/nar/21.14.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G., Raska I., Scheer U., Tan E. M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988 May;176(1):117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Bennett J. L., Dairaghi D. J., Clayton D. A. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. FASEB J. 1993 Jan;7(1):208–213. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994 Nov 1;8(21):2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Clayton D. A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes Dev. 1992 Oct;6(10):1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Topper J. N., Clayton D. A. Characterization of human MRP/Th RNA and its nuclear gene: full length MRP/Th RNA is an active endoribonuclease when assembled as an RNP. Nucleic Acids Res. 1990 Feb 25;18(4):793–799. doi: 10.1093/nar/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Yuan Y., Tan E., Reddy R. The 40-kilodalton to autoantigen associates with nucleotides 21 to 64 of human mitochondrial RNA processing/7-2 RNA in vitro. Mol Cell Biol. 1991 Oct;11(10):5266–5274. doi: 10.1128/mcb.11.10.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]