Abstract
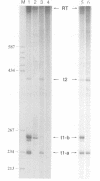
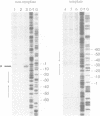
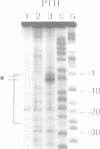
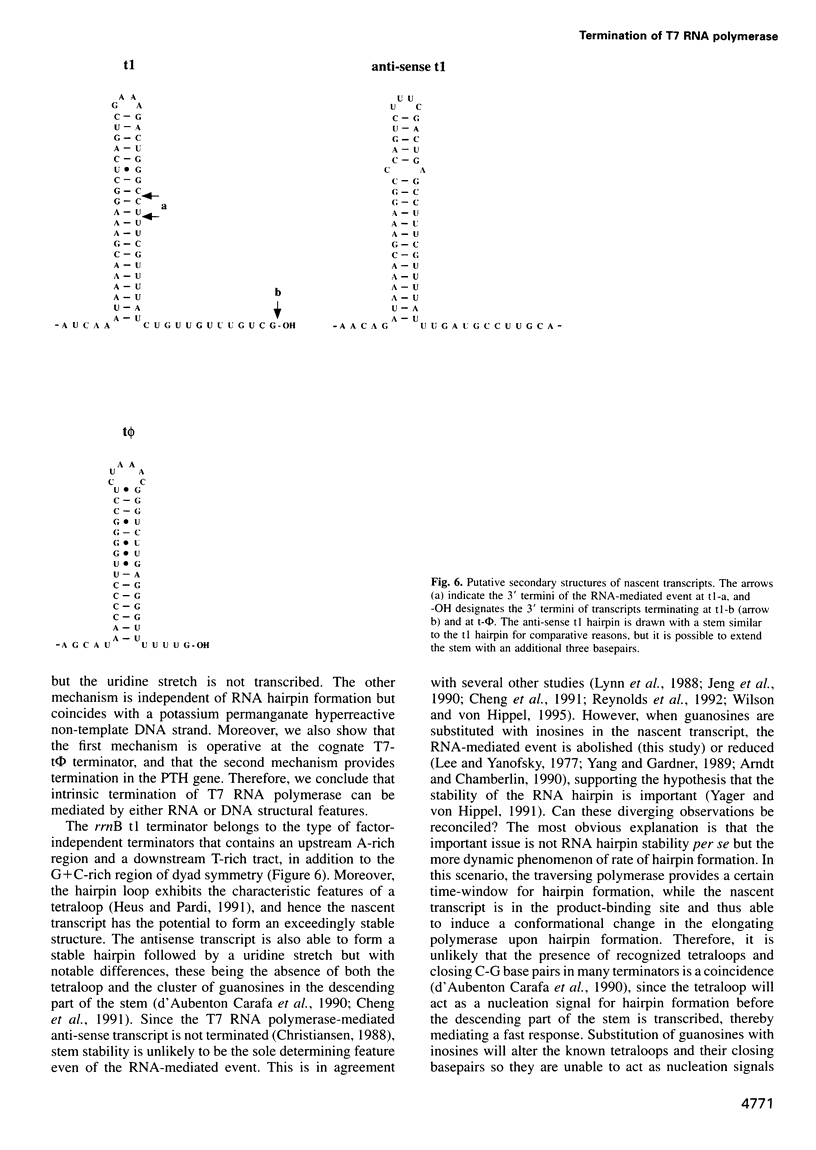
Intrinsic termination of T7 RNA polymerase transcription occurs at different signals in vitro. One type of signal is similar to that mediating factor-independent termination of Escherichia coli RNA polymerase, whereas the other type does not involve RNA hairpin formation. By examining the termination behaviour of T7 RNA polymerase at the E.coli rrnB operon t1 terminator, at the T7-t(phi) terminator, at the human preproparathyroid hormone gene terminator on both single- and double-stranded templates, and in the presence of GTP or ITP during transcription, we show that the termination event can be mediated by either RNA or DNA structural features. Moreover, by using co-transcriptional probing with potassium permanganate, we present evidence for the presence of transcription-induced hyperreactive thymidines on the non-template strand in the DNA-mediated event, and a putative sequence motif is identified. We conclude that intrinsic termination of T7 RNA polymerase transcription in vitro can be mediated either by a hairpin in the nascent RNA or by a sequence motif including hyperreactive thymidines in the non-template DNA strand.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmann C. R., Solow-Cordero D. E., Chamberlin M. J. RNA cleavage and chain elongation by Escherichia coli DNA-dependent RNA polymerase in a binary enzyme.RNA complex. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3784–3788. doi: 10.1073/pnas.91.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt K. M., Chamberlin M. J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990 May 5;213(1):79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992 1993;88:1–21. [PubMed] [Google Scholar]
- Cheng S. W., Lynch E. C., Leason K. R., Court D. L., Shapiro B. A., Friedman D. I. Functional importance of sequence in the stem-loop of a transcription terminator. Science. 1991 Nov 22;254(5035):1205–1207. doi: 10.1126/science.1835546. [DOI] [PubMed] [Google Scholar]
- Christiansen J. The 9S RNA precursor of Escherichia coli 5S RNA has three structural domains: implications for processing. Nucleic Acids Res. 1988 Aug 11;16(15):7457–7476. doi: 10.1093/nar/16.15.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Platt T. A model for transcription termination suggested by studies on the trp attenuator in vitro using base analogs. Cell. 1980 Jul;20(3):739–748. doi: 10.1016/0092-8674(80)90320-7. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Jeng S. T., Gardner J. F., Gumport R. I. Transcription termination by bacteriophage T7 RNA polymerase at rho-independent terminators. J Biol Chem. 1990 Mar 5;265(7):3823–3830. [PubMed] [Google Scholar]
- Johnson T. L., Chamberlin M. J. Complexes of yeast RNA polymerase II and RNA are substrates for TFIIS-induced RNA cleavage. Cell. 1994 Apr 22;77(2):217–224. doi: 10.1016/0092-8674(94)90314-x. [DOI] [PubMed] [Google Scholar]
- Kainz M., Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992 Feb 14;255(5046):838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry. 1990 Jan 9;29(1):269–278. doi: 10.1021/bi00453a037. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S. P., Kasper L. M., Gardner J. F. Contributions of RNA secondary structure and length of the thymidine tract to transcription termination at the thr operon attenuator. J Biol Chem. 1988 Jan 5;263(1):472–479. [PubMed] [Google Scholar]
- Macdonald L. E., Durbin R. K., Dunn J. J., McAllister W. T. Characterization of two types of termination signal for bacteriophage T7 RNA polymerase. J Mol Biol. 1994 Apr 29;238(2):145–158. doi: 10.1006/jmbi.1994.1277. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Muller D. K., Coleman J. E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988 May 31;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- McDowell J. C., Roberts J. W., Jin D. J., Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994 Nov 4;266(5186):822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Nudler E., Kashlev M., Nikiforov V., Goldfarb A. Coupling between transcription termination and RNA polymerase inchworming. Cell. 1995 May 5;81(3):351–357. doi: 10.1016/0092-8674(95)90388-7. [DOI] [PubMed] [Google Scholar]
- Reynolds R., Bermúdez-Cruz R. M., Chamberlin M. J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992 Mar 5;224(1):31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- Ring B. Z., Roberts J. W. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994 Jul 29;78(2):317–324. doi: 10.1016/0092-8674(94)90300-x. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Sousa R., Patra D., Lafer E. M. Model for the mechanism of bacteriophage T7 RNAP transcription initiation and termination. J Mol Biol. 1992 Mar 20;224(2):319–334. doi: 10.1016/0022-2836(92)90997-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Wilson K. S., von Hippel P. H. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A. 1995 Sep 12;92(19):8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager T. D., von Hippel P. H. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991 Jan 29;30(4):1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- Zhou W., Reines D., Doetsch P. W. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995 Aug 25;82(4):577–585. doi: 10.1016/0092-8674(95)90030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aubenton Carafa Y., Brody E., Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990 Dec 20;216(4):835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]