Abstract
Targeted approaches have revealed frequent epigenetic alterations in ovarian cancer, but the scope and relation of these changes to histologic subtype of disease is unclear. Genome-wide methylation and expression data for 14 clear cell carcinoma (CCC), 32 non-CCC, and 4 corresponding normal cell lines were generated to determine how methylation profiles differ between cells of different histological derivations of ovarian cancer. Consensus clustering showed that CCC is epigenetically distinct. Inverse relationships between expression and methylation in CCC were identified, suggesting functional regulation by methylation, and included 22 hypomethylated (UM) genes and 276 hypermethylated (HM) genes. Categorical and pathway analyses indicated that the CCC-specific UM genes were involved in response to stress and many contain hepatocyte nuclear factor (HNF) 1 binding sites, while the CCC-specific HM genes included members of the estrogen receptor alpha (ERalpha) network and genes involved in tumor development. We independently validated the methylation status of 17 of these pathway-specific genes, and confirmed increased expression of HNF1 network genes and repression of ERalpha pathway genes in CCC cell lines and primary cancer tissues relative to non-CCC specimens. Treatment of three CCC cell lines with the demethylating agent Decitabine significantly induced expression for all five genes analyzed. Coordinate changes in pathway expression were confirmed using two primary ovarian cancer datasets (p<0.0001 for both). Our results suggest that methylation regulates specific pathways and biological functions in CCC, with hypomethylation influencing the characteristic biology of the disease while hypermethylation contributes to the carcinogenic process.
Keywords: ovarian clear cell carcinoma, HNF1, ERalpha, methylation, epigenetic characterization
Introduction
Ovarian cancer is the fifth most common cause of cancer death in women 1 and has the worst mortality rate of all gynecologic cancers 2. Epithelial ovarian cancers are a heterogeneous disease with distinct clinicopathological and molecular features 3 and are classified pathologically into four major histologic subtypes based entirely on tumor cell morphologic criteria: serous adenocarcinoma (SAC), mucinous adenocarcinoma (MAC), endometrioid adenocarcinoma (EAC) and clear cell carcinoma (CCC). Since these histologic subtypes of ovarian cancer have unique clinical characteristics and behavior, histology-specific biomarkers and individualized therapeutic strategies are needed that would improve therapeutic options and outcomes. Ovarian CCC is distinct from the other major histologic types of epithelial ovarian cancer 4, 5 and often arises from endometriosis. The clinical outcome of advanced stage CCC is generally poor and this may be related to the relative resistance to conventional platinum- or taxane-based chemotherapies 6-8. Although the different histologic subtypes of ovarian cancer have been associated with genetic defects, such as TP53 mutations in high grade SAC, PTEN and CTNNBI mutations in EAC, and KRAS mutations in MAC, the molecular features of CCC have remained elusive 9. Recent genome-wide technologies have revealed frequent ARID1A mutations 10, 11 and overexpression of HNF1B 12 in ovarian CCC. We previously identified an ovarian CCC-specific gene signature characterized by activation of the HNF1B pathway. Furthermore, some of these signature genes are regulated by DNA methylation 13.
DNA methylation is an epigenetic mechanism of gene regulation that plays a crucial role in many biological processes including X-chromosome inactivation, genomic imprinting, aging, and cancer 14. In cancer cells, global DNA hypomethylation and aberrant hypermethylation of tumor suppressor genes are well-established mechanisms that contribute to cancer cell transformation 15. Analysis of individual genes has led to a large number now reported as targets of DNA methylation in ovarian cancer 1, 16. However, genome-wide methylation profiles may be more instructive since the methylation status of any particular single gene has not been shown to be predictive, diagnostic or prognostic for this disease 15, 17. Higher resolution elucidation of DNA methylation profiles may lead to discovery of biomarkers for diagnosis, improved methods of classification and disease monitoring as well as to a better understanding of cancer biology. Though several comprehensive DNA methylation analyses have been reported in ovarian cancer, limitations have included a relatively small number of samples, lack of focus on histological subtypes, and in limited number and selection of genes analyzed 14, 17-20. Only four genes (14-3-3 sigma as SFN, PYCARD, WT1 and HNF1B) have been reported as aberrantly methylated in CCC 1, 13, 16, 21. Therefore epigenetic features that more fully characterize histological subtypes including CCC have yet to be identified.
In this study, our initial objective was to determine if epigenomic patterns could distinguish the various histologic subtypes of ovarian cancer, including CCC. We identified an ovarian CCC methylation profile that was distinct from the other histologic subtypes of ovarian cancer. Through analysis of genes showing strong associations between DNA methylation and levels of transcription, we identified important biological mechanisms contributing to characteristic features of ovarian CCC 4 that appear to be deregulated through coordinate epigenetic modifications in this disease.
Materials and Methods
Detailed methods are provided in the Supplementary Materials.
Cell lines and clinical samples
The immortalized non-cancerous cell lines were maintained as described 22,23, 24. Ovarian cancer cell lines, including 14 CCC and 32 non-CCC cell lines (Supplementary Table 1), were cultured in RPMI1640/FBS/pen-strep medium, (Sigma-Aldrich Co., St. Louis, MO) with 10% heat inactivated fetal bovine serum (v/v; Invitrogen) in an atmosphere of 5% CO2 at 37°C. The short tandem repeat (STR) genotypes of all ovarian cancer cell lines were analyzed to authenticate the cell lines using either the PowerPlex® 1.2 System (Promega, Madison, WI) at The Fragment Analysis Facility of Johns Hopkins University or the AmpFℓSTR® Identifiler® Plus PCR Amplification Kit (Applied Biosystems, Carlsbad, CA) at the University of Colorado Cancer Center, DNA Sequencing and Analysis Core. The STR genotypes of ovarian cancer cell lines that are available from ATCC, RIKEN BioResource Center Cell Bank, or the Japanese Collection of Research Bioresources (JCRB) Cell Bank were identical to the source genotypes as reported within their respective STR databases and all other non-commercially available cell lines were shown to be derived from females with unique genotypes.
Tissue specimens were derived from 85 patients with ovarian cancer treated at Duke University Medical Center, all of whom provided written informed consent (13 CCC, 53 SAC, 11EAC, and 8 MAC) and from eight patients without malignant disease (4 ovarian surface epithelium samples and 4 fallopian tube epithelium samples). For all tumor specimens, representative sections were mounted on slides and stained with hemotoxylin and eosin to confirm that at least 60% of the cellular content comprised cancer cells with less than 20% necrosis. Tumors were histologically classified according to World Health Organization (WHO) criteria.
Extraction of Genomic DNA and Bisulfite Treatment
Genomic DNA was extracted from cells cultured in 10 cm dishes to 70-90% confluence using the QIAamp® DNA Mini Kit (Qiagen Inc., Valencia, CA) or from 5-20 mg of clinical tissues using the AllPrep® DNA/RNA/Protein Mini Kit (Qiagen Inc.) for methylation beadchip analysis. Puregene Reagents (Qiagen; Valencia, CA) were used for DNA extraction after Decitabine treatment (see below). Five hundred ng of genomic DNA were bisulfite modified using the EZ DNA Methylation™ Kit (Zymo Research Co., Irvine, CA) according to the manufacturer's protocol for the Infinium methylation assay, and 800 ng of genomic DNA was bisulfite modified using the same kit but according to standard protocol for methylation analysis of individual genes.
Illumina Infinium HumanMethylation BeadChip Assay
Bisulfite-converted genomic DNA was analyzed using Illumina’s Infinium HumanMethylation27 BeadChip for cell line samples (GEO Accession Number GSE51688) and HumanMethylation450 Beadchip for clinical tissue specimens (GEO Accession Number GSE51820) (Illumina Inc., San Diego, CA). The HumanMethylation450 BeadChip includes 90% of the content contained on the HumanMethylation27 BeadChip. Chip processing was performed according to the manufacturer’s protocol by Expression Analysis Inc., (Durham, NC), an Illumina Certified Service Provider. Data were extracted using Illumina® GenomeStudio™ v2010.3 software (Illumina Inc.). Methylation values for each CpG locus are represented as β-values, a quantitative measure of DNA methylation, with levels ranging from 0 (completely unmethylated) to 1 (completely methylated). Bisulfite conversion efficiency (i.e., the conversion of non-CpG cytosines to uracils) was assessed for all samples by Pyrosequencing (see below) prior to data generation using the Illumina BeadChips.
Methylation Specific Polymerase Chain Reaction (MS-PCR)
DNA methylation of estrogen receptor alpha (ERalpha) network genes (ESR1, BMP4, DKK1, SOX11, SNCG and MOSC1) was validated using MS-PCR. Primer sequences and PCR conditions are provided in Supplemental Table 2a.
Pyrosequencing
All primers for pyrosequencing assays were designed using PSQ assay design software version 1.0.6 (Biotage, Uppsala, Sweden). The pyrosequencing assays were designed such that we analyzed the same CpG sites as those present on the Illumina BeadChip for eleven genes (ESR1, HNF1A, HNF1B, C14orf105, KIF12, MIA2, PAX8, SERPINA6, SGK2, SRC and TM4SF4) and for adjacent CpG sites for the F2 gene. Primer sequences and PCR conditions are shown in Supplemental Table 2b. Methylation values for each CpG site were calculated using Pyro Q-CpG software 1.0.9 (Biotage).
Real Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Two micrograms of total RNA prepared from 36 ovarian cancer cell lines (13 CCC and 23 non-CCC; using RNA Stat60, Teltest; Friendswood TX) or 49 frozen tissues (13 CCC and 36 non-CCC; using the AllPrep® DNA/RNA/Protein Mini Kit; Qiagen Inc.) were used to generate cDNA in a 40 μl volume with the Superscript II kit (Invitrogen). Two μl of the cDNA was used as template for PCR using TaqMan assays in a 20 μl volume (Applied Biosystems) for HNF1B (Hs01001602), SGK2 (Hs00367639), C14orf105 (Hs00216847), F2 (Hs01011988), ESR1 (Hs00174860), CRIP1 (Hs00832816), SOX11 (Hs00846583), IGFBP4 (Hs01057900) and BMP4 (Hs00370078). B2M (Hs 00187842) was used as an internal control. Relative expression was calculated using the delta delta Ct method.
5-Aza-2'-Deoxycytidine (Decitabine) Treatment
RMG-2, RMG-5, and KOC-7C cell lines were seeded into 6-well plates and the following day they were treated in triplicate by adding RPMI1640/FBS/pen-strep media (mock) or the same media containing 5μM 5-aza-2'-deoxycytidine (Decitabine; Sigma-Aldrich Co). The media was replaced daily (with/without Decitabine as relevant) for three days after which the cells had reached 70% to 90% confluence and were harvested for DNA extraction as described above or RNA extraction using RNA Stat60 (Teltest; Friendswood TX). Real time RT-PCR and pyrosequencing analysis were performed as described above.
Bioinformatics
Identification of genes showing differential methylation
Identification of genes differentially methylated between groups was done by comparing β-values of these groups using unpaired t-tests with thresholds of p <0.01 and difference of average β-values >0.2 (20% methylation) for the comparison between ovarian cancer cell lines and normal cells or between CCC and non-CCC. Genes annotated by gene name or gene ID (Illumina annotation file) were used for the analyses.
Unsupervised consensus clustering
ConsensusCluster software 25 was used to evaluate the similarity between genes or specimens.
Identification of genes regulated by DNA methylation in ovarian cancer
Two cell line microarray datasets [Gene Expression Omnibus (GEO) Accession Number: GSE25428 and GSE29175 13, 26] were used for the identification of candidate genes whose mRNA expression levels are regulated by DNA methylation in ovarian cancer. The ComBat normalizing algorithm was used before performing any analysis to reduce the likelihood of batch effects 27. Candidate genes functionally regulated by methylation were identified based on the following criteria: 1) the p-value of unpaired Student t-tests is less than 0.01; 2) the average β-value difference is more than 0.2 (20% methylation) between 14 CCC cell lines and 32 non-CCC cell lines; and 3) a significant inverse correlation (p <0.05) between expression microarray values and β-values from the Infinium assay for the 42 ovarian cancer cell lines with gene expression data.
Categorical Analyses
The biological characteristics of the CCC-specific methylation signature were evaluated using the enrichment of Molecular Signatures Database (MSigDB) gene sets (v2.5 updated April 7 2008) 28 with the R package allez 1.0 29. Briefly, for each gene set, the proportion of the annotated genes in the CCC hypermethylated or hypomethylated gene sets was compared to that for all probeset genes. A gene set was considered significantly enriched if the z-score was more than 4.0 30.
Pathway Analyses
The relationship of hypermethylated or hypomethylated genes to particular pathways was evaluated using MetaCore™ software (GeneGo; http://www.genego.com/), an integrated knowledge database and software suite for evaluation of the association between a particular gene list and known pathways.
Heatmap of beta-values and expression
Absolute β-values are represented by color gradient intensity, from white; β-value =0, to red; β-value =1, using Java TreeView (http://jtreeview.sourceforge.net/). Average-linkage hierarchical clustering was performed in published expression microarray datasets representing clinical ovarian cancer samples (GSE6008 and GSE2109 31, 32) using genes transcriptionally regulated by DNA methylation with Cluster version 3.0. (http://rana.lbl.gov/eisen/).
Statistical Analyses
The student t test was used to compare continuous variables between groups. Fisher’s exact test was used for analysis of categorical variables. Correlations between expression microarray values and methylation β-values from the Infinium assay or between β-values and % methylation values from pyrosequencing were evaluated using Pearson’s correlation. P-values less than 0.05 were considered statistically significant.
Results
Divergent Methylation Profiles Between Ovarian Cancer Cells and Non-cancerous Cells
By comparing the β-values in ovarian cancer with those in non-cancerous cells, 2,003 genes (2,561 CpG sites) of a total possible 14,495 genes (27,578 CpG sites) (13.8%) in ovarian cancer cell lines showed methylation levels that differed relative to non-cancerous specimens (Supplementary Table 3). Of these differentially methylated genes, 1,870 genes (2,404 CpG sites; 93.4%) were hypermethylated (HM) whereas only 139 genes (157 CpG sites; 7.0%) were hypomethylated (UM) in ovarian cancer cell lines.
In the clinical samples analyzed with the larger probeset, 3,432 genes (6,919 CpG sites) of 21,231 genes (365,860 CpG sites) (16.2%) showed methylation levels that differed compared to normal specimens. Of the differentially methylated genes, 2,383 genes (4 830 CpG sites; 65.6%) were HM whereas 1,253 genes (2,089 CpG sites; 34.5%) were UM in ovarian cancer tissues (Supplementary Table 3).
CCCs Possess a Unique Methylation Profile
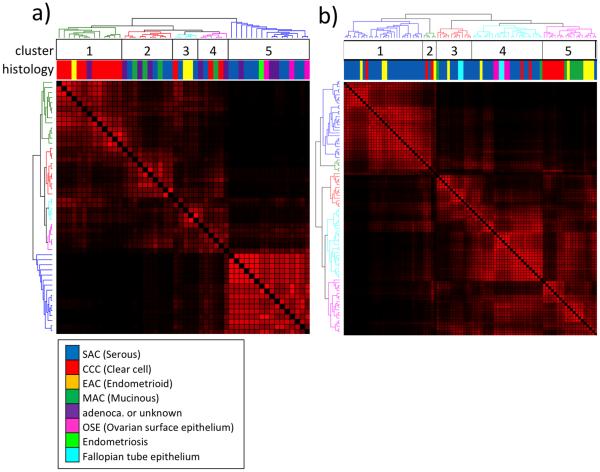
Unsupervised consensus clustering generated a distinct CCC-specific cluster in the cell line dataset (Figure 1a). The non-cancerous cell lines clustered together with nine of sixteen SAC (56.3%) cell lines. This data indicates CCCs possess a distinct epigenomic pattern as compared to the other histologic types of ovarian cancer and non-cancerous specimens. Similar to the clustering results in the cell lines, a distinctive CCC-specific cluster was again generated in the analyses of clinical specimens. Also like the cell lines, 25 of 51 (49.0%) SAC clinical specimens clustered together with normal specimens (Figure1b). Among the tissue specimens, six of eight MAC and five of eleven EAC samples were grouped within the cluster containing the majority of the CCC samples. However, both the MAC and EAC sub-clusters are distinct from the CCC sub-cluster, indicating that the methylation profiles of MAC and EAC share higher similarity to CCC as compared to the other tissue specimens.
Fig. 1.
Consensus clustering of DNA methylation profiles for 46 ovarian cancer and four non-cancerous cell lines (A), and 83 clinical ovarian cancer specimens and eight normal counterparts (B). Red-black coloration represents the similarity of methylation profiles between samples, from similar to divergent methylation patterns, respectively. The same specimens are ordered identically in the individual rows and columns. The colored dendogram indicates the five different clusters and the color bar indicates the histological subtype derivation of each specimen.
To evaluate the similarity between cell lines and clinical samples, the number of differentially methylated probes that overlap between these datasets was determined among the 25,978 CpG probes in common between the HumanMethylation27 and HumanMethylation450 BeadChips. Of these evaluable CpG probes, 1,042 and 77 CpGs showed hypermethylation in CCC compared to non-CCC cell lines and patient specimens, respectively, with a statistically significant overlap of 25 probes (p <0.0001). We also found that the cell lines and clinical samples had 54 and 53 CpG probes showing hypomethylation, respectively, in CCC relative to non-CCC, with six CpG probes in common, a finding that is also statistically significant (p <0.0001). These 25 hypermethylated and six hypomethylated probes are shown in Supplementary Tables 4a and 4b. These results support that there are specific methylation profiles in cell lines that are also detectable in patient samples.
Identification of Genes Transcriptionally Regulated by DNA Methylation in CCC
Next, we examined the biological significance of the CCC-specific DNA methylation. For this purpose, the genes most likely to be transcriptionally regulated by DNA methylation in CCC were identified using cell line data because of the availability of both methylation and expression microarray data and the similarity of methylation status between the cell lines and clinical specimens. First, 856 genes (1,042 CpG sites) showed increased methylation, and 44 genes (54 CpG sites) showed decreased methylation in CCC as compared to non-CCC (Supplementary Figure 1a). Next, potential functional relationships between expression and methylation of the 856 HM and 44 UM CCC genes was assessed by determining if there were correlations between these values. Of 625 evaluable HM genes, 276 genes showed a significant inverse correlation (r <-0.3051, p <0.05) between methylation and expression. Of the 33 evaluable UM genes, 22 showed a significant inverse correlation (r <-0.3217, p <0.05) between methylation and expression (Supplementary Figure 1b). We considered these 276 HM and 22 UM genes as candidate CCC-specific epigenetically regulated genes (Supplementary Table 5a and 5b, respectively).
Hypomethylation of the HNF1 Pathway and Hypermethylation of Estrogen Receptor Pathway Genes in CCC
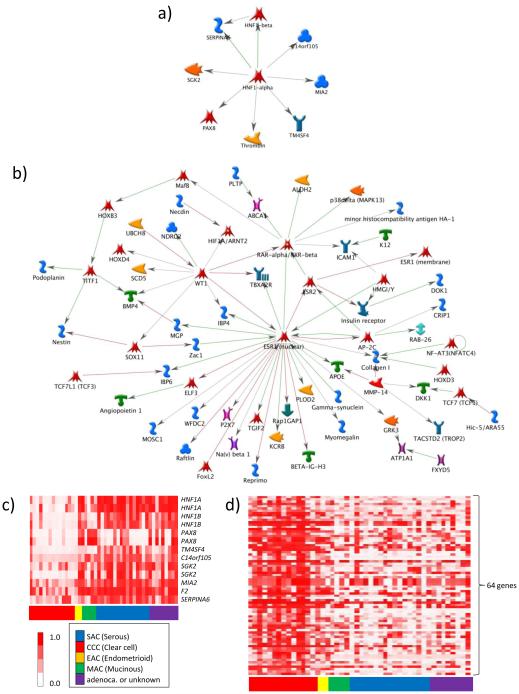
Biological functions of the 22 CCC-specific UM genes were characterized by analyzing the targeted pathways and ‘biological process’ Gene Ontology (GO) terms. Pathway analysis using MetaCore™ indicated nine of these 22 UM genes (41%) are members of the HNF1 pathway (Figure 2a). Furthermore, the nine CCC-specific UM HNF1 pathway genes are not randomly deregulated but rather synchronously altered by methylation in CCC (Figure 2c), suggesting that hypomethylation in CCC targets HNF1 pathway genes. Analyses using allez showed significant enrichment of all HNF1 binding motifs among the CCC UM genes including categorical terms V$HNF1_01, RGTTAMWNATT_V$HNF1_01, V$HNF1_Q6, and V$HNF1_C (p <0.00001; Supplementary Table 6). Allez showed that the 22 CCC UM genes were enriched for stress response-related GO terms such as “response to oxidative stress” (z-score >4.0; Table 1, top).
Fig. 2.
MetaCore™ pathway analyses of the 22 CCC-specific UM genes (A) and the 276 CCC-specific HM genes (B), and heatmaps showing methylation profiles of nine HNF1 pathway genes (C) and 64 ERalpha network genes (D) in 46 ovarian cancer cell lines. Of the 22 CCC-specific UM genes for which expression and methylation are inversely correlated, nine were identified as members of the HNF1 transcriptional network (A). Of 276 CCC-specific HM genes for which expression and methylation are inversely correlated, 65 were members of the ERalpha (encoded by ESR1) network (B). In the heatmaps, red-white coloration represents the Infinium beta-value, from completely methylated to completely unmethylated, respectively. Genes within the HNF1 pathway (C) and ERalpha network (D) show synchronous hypomethylation and hypermethylation, respectively, in CCC versus non-CCC cell lines.
Table 1.
Categorical analysis of ‘biological process’ gene ontology (GO) terms for the 22 UM genes (top) and 276 HM genes (bottom) in CCCa
| 22 CCC-specific UM genes | |||
|---|---|---|---|
| MSigID.c5.bp | n.probes | z.score | p value |
|
POSITIVE_REGULATION_OF
_TRANSCRIPTION__DNA_DEPENDENT† |
202 | 8.78 | <0.000001 |
|
POSITIVE_REGULATION_OF_RNA
_METABOLIC_PROCESS* |
205 | 8.70 | <0.000001 |
|
POSITIVE_REGULATION_OF
_TRANSCRIPTION† |
242 | 7.90 | <0.000001 |
|
POSITIVE_REGULATION_OF_NUCLEOBASE_
NUCLEOSIDE_NUCLEOTIDE_AND_NUCLEIC _ACID_METABOLIC_PROCESS* |
260 | 7.58 | <0.000001 |
|
POSITIVE_REGULATION_OF
_CELLULAR_METABOLIC_PROCESS* |
396 | 5.83 | <0.000001 |
|
POSITIVE_REGULATION_OF
_METABOLIC_PROCESS* |
407 | 5.73 | <0.000001 |
|
PROTEIN_IMPORT_INTO_NUCLEUS
_TRANSLOCATION† |
21 | 4.66 | 0.000002 |
| RESPONSE_TO_OXIDATIVE_STRESS* | 78 | 4.65 | 0.000002 |
| TYROSINE_PHOSPHORYLATION_OF
_STAT_PROTEIN |
26 | 4.14 | 0.000017 |
| GLUTAMINE_FAMILY_AMINO_ACID
_METABOLIC_PROCESS |
27 | 4.06 | 0.000025 |
| 276 CCC-specific HM genes | |||
| MSigID.c5.bp | n.probes | z.score | p value |
|
NEGATIVE_REGULATION_OF
_CYTOSKELETON_ORGANIZATION _AND_BIOGENESIS‡ |
18 | 7.19 | <0.000001 |
|
MICROTUBULE_POLYMERIZATION
_OR_DEPOLYMERIZATION‡ |
21 | 6.58 | <0.000001 |
| PYRIMIDINE_NUCLEOTIDE
_METABOLIC_PROCESS |
15 | 5.86 | <0.000001 |
| CELL_CYCLE_ARREST_GO_0007050 ¶ | 115 | 4.78 | 0.000001 |
| SENSORY_ORGAN_DEVELOPMENT ‡ | 23 | 4.53 | 0.000003 |
| AEROBIC_RESPIRATION | 24 | 4.40 | 0.000005 |
|
NEGATIVE_REGULATION
_OF_CELL_CYCLE¶ |
157 | 4.33 | 0.000008 |
|
NEGATIVE_REGULATION_OF
_CELLULAR_COMPONENT _ORGANIZATION_AND_BIOGENESI‡ |
45 | 4.05 | 0.000026 |
The 22 CCC-specific UM genes are enriched for GO terms related to stress response (*) as well as terms related to transcriptional activation (†). The 276 CCC-specific HM genes are enriched for terms related to tumor suppression (¶) and terms related to development- and organogenesis (‡).
Among the 276 CCC-specific HM genes, pathway analysis showed that 64 of these were involved in the estrogen receptor pathway (Figure 2b). Similar to the synchronous decreased methylation of HNF1 pathway genes in CCC, these 64 genes exhibit coordinate hypermethylation in CCC relative to non-CCC (Figure 2d). These results indicate that the estrogen receptor pathway is a target of hypermethylation in CCC. A categorical analyses using allez to identify GO terms associated with the HM genes showed that the 276 genes HM in CCC included many with tumor suppressive functions having GO annotations of “cell cycle arrest” and “negative regulation of cell cycle”, and development- and organogenesis- related genes with annotations of “negative regulation of cytoskeleton organization and biogenesis” (Table 1, bottom).
Validation of Hypomethylation and Increased Expression of HNF1 Pathway Genes in CCC
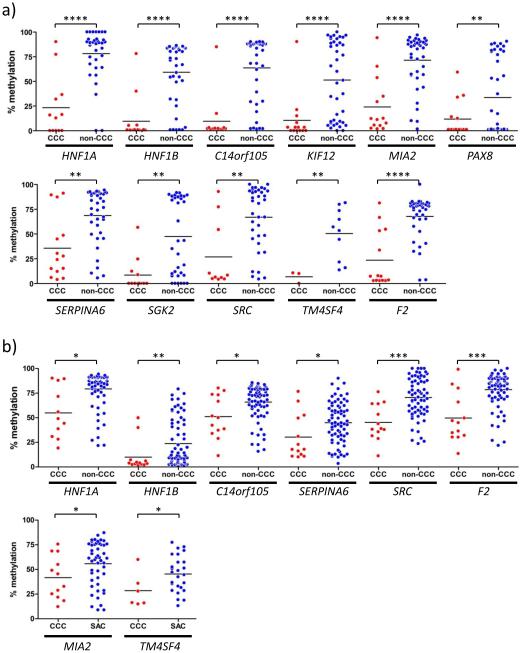
Eleven hypomethylated genes in the HNF1 pathway (HNF1A, HNF1B, C14orf105, KIF12, MIA2, PAX8, SERPINA6, SGK2, SRC, TM4SF4 and F2) identified through analysis of CCC versus non-cancerous cells and from comparisons between CCC versus non-CCC cell lines were analyzed by bisulfite pyrosequencing. The ten genes for which identical CpG sites were analyzed showed very strong positive correlations between the Infinium assayβ-values and the percent methylation as measured by pyrosequencing (r >0.9605, p <0.000001 for all genes; Supplementary Figure 2). Adjacent CpG sites for these same ten genes also showed significant positive correlations between the β-values of the Infinium assay CpG sites and the percent methylation by pyrosequencing, as did the adjacent CpG sites measured for the F2 gene (0.7498< r <0.9813, p <0.000001 for all genes; Supplementary Figure 3). Confirming the results of the methylation beadchip analyses, the pyrosequencing results also showed significantly lower methylation levels in CCC relative to those in non-CCC cell lines for these 11 genes (p <0.01 for all genes; Figure 3a). In 85 clinical ovarian cancer specimens, ten of the 11 HNF1 network genes showed overall hypomethylation in ovarian CCC specimens and six of these eleven genes showed significantly decreased methylation in CCC compared to non-CCC (HNF1A, p=0.0036; HNF1B, p=0.0041; C14orf105, p=0.0194; SERPINA6, p=0.0291; SRC, p=0.0004 and F2, p=0.0003; Figure 3b). When CCC was compared with SAC, eight genes were significantly hypomethylated in CCC (HNF1A, p=0.0007; HNF1B, p=0.0002; C14orf105, p=0.0073; MIA2, p=0.0480; SERPINA6, p=0.0071; SRC, p <0.0001; TM4SF4, p=0.0452 and F2, p <0.0001; Figure 3b and data not shown).
Fig. 3.
Quantitative methylation analyses of 11 HNF1 network genes (HNF1A, HNF1B, C14orf105, KIF12, MIA2, PAX8, SERPINA6, SGK2, SRC, TM4SF4 and F2) in cell lines by bisulfite pyrosequencing shows lower levels of methylation in CCC versus non-CCC, supporting the results from the Infinium BeadChip (A). For clinical specimens, six of eleven HNF1 network genes show significantly decreased methylation in CCC compared to non-CCC. When CCC is compared with SAC in clinical samples, eight genes (HNF1A, p=0.0007; HNF1B, p=0.0002; C14orf105, p=0.0073; SERPINA6, p=0.0071; SRC, p<0.0001 and F2, p<0.0001 included in data shown in 4B, top; MIA2, p=0.0480 and TM4SF4, p=0.0452 are shown below) are significantly hypomethylated in CCC relative to SAC (B). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001
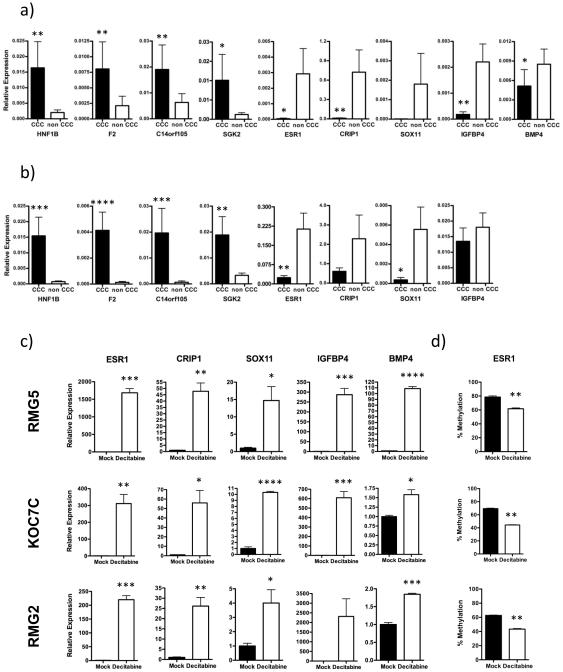
We selected four HNF1B network genes for validation studies using both cell lines and clinical specimens. Messenger RNA levels of HNF1B, F2, C14orf105 and SGK2 were significantly increased in N=13 CCC cell lines compared to N=23 non-CCC cell lines (p=0.0018, p=0.0057, p=0.0046 and p=0.0158, respectively; Figure 4a). Clinical samples also showed higher expression for all four genes comparing CCC (N=13) to non-CCC (N=36) specimens (HNF1B, p=0.0002; F2, p<0.0001; C14orf105, p=0.0002 and SGK2; p=0.0019; Figure 4b).
Fig. 4.
Validation of altered expression of HNF1 network and ERalpha pathway genes in CCC and reactivation and demethylation of ERalpha pathway genes with Decitabine treatment. Quantitative RT-PCR of HNF1 network genes (HNF1A, F2, C14orf105 and SGK2) and ERalpha pathway genes (ESR1, CRIP1, SOX11, IGFBP4 and BMP4) using cell lines (A) and clinical samples (B). The expression of ERalpha pathway genes (ESR1, CRIP1, SOX11, IGFBP4 and BMP4) is markedly induced by demethylating agent 5-aza-2'-deoxycytidine, (Decitabine) in three CCC cell lines (RMG-5, KOC-7C, RMG-2) (C). Methylation at the ESR1 promoter is decreased in response to Decitabine treatment (D). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
We used publicly available ovarian cancer expression microarray datasets representing clinical ovarian cancer samples (GSE6008 and GSE2109) to assess the expression levels of the genes we had identified as functionally regulated by methylation. Supervised hierarchical clustering was performed using only the genes regulated by DNA methylation listed in Supplementary Tables 5a and 5b. The CCC specimens were clearly divided from non-CCC samples based on the expression levels of these genes. The CCC-specific UM genes from our analysis were enriched among a distinct cluster of genes that are highly expressed in the CCC clinical specimens as compared to the presence of CCC-specific HM genes in this cluster (16 of 30 versus 0 of 378 in GSE6008, and 18 of 30 versus 1 of 378 in GSE2109; p <0.0001 for both; Supplementary Figures 4a and 4b).
Validation of Hypermethylation and Decreased Expression of Estrogen Receptor Pathway Genes in CCC
Representative ER pathway genes including ESR1, BMP4, DKK1, SOX11, SNCG and MOSC1 were investigated for their methylation status in cell lines using MS-PCR. Twelve of 14 (86%) CCC cell lines exhibited methylation of the ESR1 promoter whereas 17 of 32 (53%) non-CCC cell lines showed methylation of this gene (p=0.0487). Similarly, more of the CCC versus non-CCC cell lines showed methylation at the BMP4 and DKK1 promoter regions (BMP4: 12 of 14 CCCs versus 8 of 32 non-CCCs, p=0.0002; DKK1: 10 of 14 CCCs versus 9 of 32 non-CCCs, p=0.0094; Table 2, top and Supplementary Figure 5a-f). For SOX11 and SNCG, all CCC cell lines exhibited methylation while there were several non-CCC unmethylated cell lines (SOX11: 13 of 13 CCCs versus 18 of 23 non-CCCs, p=0.1363; SNCG: 13 of 13 CCCs versus 21 of 24 non-CCCs, p=0.5382). The MOCS1 gene showed evidence of methylation in all cell lines analyzed. We also analyzed these six genes in clinical specimens, and three of the six (SOX11, BMP4 and MOCS1) were methylated more often in CCC than non-CCC, with SOX11 showing a significant difference (p=0.0116) and BMP4 and MOCS1 approaching significance (p=0.0645 and 0.1784, respectively; Table 2, bottom and Supplementary Figure 6a-f).
Table 2.
MS-PCR validation for six ERalpha network genesa in cell lines (top) and in clinical tissues (bottom)
| Cell lines | |||
|---|---|---|---|
| Genes | CCC | non-CCC | p value |
| ESR1 | 12/14 (86%) | 17/32 (53%) | 0.0487 |
| BMP4 | 12/14 (86%) | 8/32 (25%) | 0.0002 |
| DKK1 | 10/14 (71%) | 9/32 (28%) | 0.0094 |
| SOX11 | 13/13 (100%) | 18/23 (78%) | 0.1363 |
| SNCG | 13/13 (100%) | 21/24 (89%) | 0.5382 |
| MOCS1 | 13/13 (100%) | 24/24 (100%) | - |
| Clinical tissues | |||
| Genes | CCC | non-CCC | p value |
| ESR1 | 7/12 (58%) | 50/71 (70%) | 0.5034 |
| BMP4 | 9/13 (69%) | 27/71 (38%) | 0.0645 |
| DKK1 | 2/13 (15%) | 13/70 (19%) | 1.0000 |
| SOX11 | 7/7 (100%) | 28/60 (47%) | 0.0116 |
| SNCG | 6/7 (86%) | 56/60 (93%) | 0.4345 |
| MOCS1 | 7/7 (100%) | 41/60 (68%) | 0.1784 |
The number showing methylation (numerator) is shown relative to the total number analyzed (denominator)
Transcript levels for five ER pathway genes (ESR1, CRIP1, SOX11, IGFBP4 and BMP4) were decreased in CCC cell lines as compared to non-CCC cell lines, all of which were significant except for SOX11 (p=0.0112, p=0.0046, p=0.2108, p=0.0038 and p=0.0472, respectively; Figure 4a). Among the clinical specimens, four genes showed lower average expression levels in CCC compared to non-CCC, with a significant difference for two (ESR1, p=0.0014; CRIP1, p=0.1853; SOX11, p=0.0476; and IGFBP4, p =0.991; Figure 4b). When compared with SAC, IGFBP4 mRNA levels did not differ in CCC (p=0.9521), but expression of ESR1, CRIP1, SOX11 in CCC was significantly decreased in CCC (p=0.0022, p=0.0476 and p=0.0017, respectively).
The potential to reactivate expression of five genes, ESR1, CRIP1, SOX11, IGFBP4 and BMP4, was evaluated in three CCC cell lines (RMG-2, RMG-5, KOC-7C) using the DNA methyltransferase inhibitor, 5-aza-2'-deoxycytidine (Decitabine). There was a marked and significant increase in mRNA levels for all five genes in cells treated with Decitabine relative to the mock treated cells (Figure 4c). We examined the methylation status of ESR1 in these same cell lines and showed that in each case, methylation was significantly decreased following Decitabine treatment relative to mock treated cells (p≤0.01; Figure 4d). Altogether, these results support a functional relationship between DNA methylation and transcriptional activity that is deregulated in CCC.
Discussion
Recent genome-wide technologies have provided opportunities to develop profiles that can distinguish, identify and classify discrete disease subsets as well as predict outcome or the response to therapy in a variety of malignant diseases. Although ovarian cancer is a morphologically and biologically heterogeneous disease, these approaches have enabled classification of ovarian cancers into distinct subtypes 2, 31, 33, 34. However, there is a paucity of genome-wide methylation data to examine if epigenomic patterns discriminate histological subtypes of cancers, including ovarian cancer.
The comparison of methylation profiles between ovarian cancer and non-cancerous cells indicated that, of the 2,003 genes that showed differential methylation levels, as many as 93.4% showed hypermethylation in the cancer cell lines. Based on prior focused studies of promoter CpG island methylation and next generation sequencing, acquisition of methylation occurs more frequently than loss of methylation in almost all types of primary tumors as compared to their normal counterparts 35, 36. Because 26,956 (97.7%) of the 27,578 CpG sites are located at promoter regions on the Illumina Infinium HumanMethylation27 BeadChip, our results are comparable with those of published reports. The ratio of HM genes (65.5%) in primary cancer tissues was smaller than that in cell lines probably due to the inclusion of CpG sites for all known genes (485,577 CpG sites) in addition to the relative heterogeneity of cell types present within clinical specimens.
Consensus clustering identified a CCC-specific cluster and a cluster comprised of non-cancerous cells together with approximately half of the SAC both in cell lines and clinical samples. In the clinical specimens, MAC and some of the EAC formed sub-clusters with the CCC clinical specimens, indicating closer similarity to CCC as compared with the other tissues analyzed with respect to DNA methylation. These results suggest that CCC, MAC and some EAC acquire distinct methylation profiles, which may in part reflect differences in the carcinogenic process for the different histologic types of ovarian cancer. For example, high grade SAC and EAC are thought to arise de novo (type 2 ovarian cancer), whereas CCC and MAC are thought to arise from precursor benign cysts, indicating an adenoma-carcinoma sequence (type 1 ovarian cancer) 37. These differences in carcinogenic steps may be brought about by, or affect specific epigenetic patterns 38. Differences in genome-wide methylation status between histologic subtypes of ovarian cancer have been controversial. Although single genes (14-3-3 sigma also known as SFN, PYCARD, WT1 and HNF1B) have been reported as showing aberrant methylation in CCC 1, 13, 16, 21, comprehensive methylation analyses showed that differentially methylated genes in CCC overlap with SAC and EAC 14, 39. However, these studies were limited in their analysis to 1,505 CpG loci selected from 871 genes (GoldenGate Methylation Cancer Panel I) and analyzed a small number of CCC samples (only three and four specimens, respectively). Our study found altered methylation in CCC for three of four genes which are known as epigenetically regulated in CCC (14-3-3 sigma as SFN, PYCARD, WT1 and HNF1B) whereas the prior studies did not identify hypomethylation of HNF1B network genes. Furthermore, Houshdaran et al.14 identified hypermethylation of ESR1 and WT1 genes in CCC, and Yoon et al. 39 did not identify these genes. Our study is the first that has begun to resolve these limitations with the identification of specific methylation profiles that distinguish the histologic subtypes, especially ovarian CCC, suggesting that epigenetic regulation contributes to the ontogeny of epithelial ovarian cancer.
Although many single genes have been reported as regulated by DNA methylation in ovarian cancer, the biological role of genome-wide methylation in ovarian carcinogenesis has not been elucidated, with questions remaining as to whether or not changes in methylation reflect cause or consequence. We considered the genes showing an inverse correlation between expression and methylation as functionally important genes that are regulated at least in part through epigenetic mechanisms. For our purposes, the data from cell lines was used to evaluate expression-methylation correlations and proved more powerful at revealing these relationships than the clinical samples, likely because of the heterogeneity in cell type composition of the clinical tissue specimens and because the DNA and RNA were extracted from different parts of the same piece of tissue. Therefore, in order to analyze how DNA methylation influences biological features of CCC, we identified genes for which transcription and methylation are coordinately related and that are CCC-specific. Interestingly, our pathway and categorical analyses showed that HNF1 transcription factor binding sites as well as HNF1B were synchronously hypomethylated in CCC. Furthermore, genes that function in the ERalpha network in CCC were among the 276 HM genes, including ESR1 and WT1. These findings suggest that hypomethylation and hypermethylation in CCC appear to target genes belonging to specific and highly relevant pathways in this particular histologic subtype of ovarian cancer.
Previously we reported that HNF1 transcription is activated in CCC relative to the other ovarian cancer histologic subtypes, and this activation is related to loss of DNA methylation at HNF1B 13, 21, 40. Our finding that activation of multiple components of the HNF1 pathway by synchronous hypomethylation occurs is a significant step forward in understanding this process and supports previous studies. HNF1B is involved in glucose homeostasis and may be responsible for the prominence of glycogen in CCC cells 41, 42 causing the cytoplasm to be clear in appearance, a defining feature of CCC. Our findings suggest that this morphologic feature of CCC is at least in part epigenetically regulated. Ovarian CCC is characterized by unique biology, including slow growth and resistance to chemotherapy and oxidative stress. Ovarian CCC also exhibits the molecular phonotypes of HNF1 pathway activation, PI3K pathway activation and MAPK activation, traits that have been collectively referred to as ‘ovarian CCC-likeness’ 4. Using GO term analysis, CCC-specific UM genes were enriched with genes related to oxidative stress, consistent with ‘ovarian CCC-likeness’. These results indicate that DNA methylation loss appears to target specific pathways and biological functions, with hypomethylation influencing pathways that help sculpt the characteristic biology of the disease.
ESR1, which is a key molecule in the ERalpha network targeted by hypermethylation in ovarian CCC, is known to be a target of methylation, and the absence of ERalpha expression in CCC has been reported 43-45. WT1, one of the genes regulated by ERalpha signaling has previously been shown to be inactivated by promoter methylation in ovarian CCC 46. These results support our findings and underscore the importance of epigenetic regulation of this pathway in CCC. Endometriotic cysts of the ovary are precursors of ovarian cancer, especially of the CCC and EAC histologic subtypes 4. Akahane et al. showed that reductions in ERalpha expression occurred with progression from endometriosis to atypical endometriosis in CCC specimens, whereas ERalpha expression increased with malignant transformation in EAC 47. Estrogen signaling through estrogen receptors has divergent outcomes in that it can induce activation of cell proliferation but also inhibit cell cycle progression 48, 49. Our results indicate that the ERalpha signaling-independent growth of CCC is largely attributable to aberrant epigenetic alterations in CCC. Although changes in CCC phenotype cannot be specifically assessed using demethylating agents, these findings support our hypothetical model for the malignant transformation of endometriosis, in which EAC is induced by estrogen and CCC is influenced by the unique microenvironment within endometriotic cysts, including persistent exposure to oxidative stress and chronic inflammation 4. Epigenetic changes can be the earliest initiation factor and complement single driver mutations in a human cancer 50, while our results suggest that coordinate deregulation of DNA methylation leads not only to tumor development but also defines the biological features of the malignancy.
We previously reported suppression of TGFbeta pathway activity by DNA methylation in ovarian cancers, primarily of serous histology 26, while the present study demonstrates that there are distinct pathways influenced by hypomethylation and hypermethylation in ovarian CCC. Genetic and epigenetic disruption plays a fundamental role in cancer development. However, mutations in ARID1A and PIK3CA are the only genetic alterations that have been identified in in approximately 50% and 40% of CCC cases, respectively. Thus, gene mutations alone cannot distinguish histologic subtypes of ovarian cancer. Our results indicate that coordinate changes in DNA methylation occur that affect expression of genes belonging to specific and relevant pathways in ovarian CCC, which contrasts with the more isolated pattern of genetic mutations observed in this disease. In high grade SAC, TP53 mutations initiate carcinogenesis and also contribute to the rapid growth that characterizes this histologic subtype, a ‘genetically-based disposition’. On the other hand, ARID1A mutations lead to malignant transformation from endometriosis to CCC and the specific biological characteristics of CCC and ‘ovarian CCC-likeness’ are developed through alteration of DNA methylation, an ‘epigenetically-based disposition’. Furthermore, although changes in methylation levels induced by environmental stress such as oxidative stress and inflammation were not analyzed in this study, our current and prior results collectively suggest that CCC tumor development is brought about by microenvironment-mediated effects resulting from oxidative stress that lead to these CCC-specific methylation profiles 13. Inflammatory diseases such as ulcerative colitis and chronic gastritis resulting from Helicobacter pylori infection induce increased DNA methylation and are associated with increased cancer risk 51. Together, these findings suggest that an ‘epigenetically-based disposition’ may be involved in inflammation-induced carcinogenesis.
In summary, our genome-wide methylation analyses have revealed that ovarian CCC has a distinct methylation profile relative to the other histological subtypes of epithelial ovarian cancer. The CCC-specific methylation profile includes synchronous gain of promoter methylation for multiple genes in the ERalpha pathway and loss of promoter methylation for multiple genes in the HNF1 pathway. Further work will be required to more precisely delineate the functions of these two pathways in this disease, to develop these changes as epigenetic biomarkers and to explore new modalities of treatment for CCC, including epigenetic therapies.
Supplementary Material
Novelty and impact of the work.
We found that genome-wide DNA methylation profiles distinguish clear cell from other histologic subtypes of ovarian cancer. Hypomethylated genes in clear cell cancers include many containing HNF1 binding motifs and are involved in stress response, a characteristic of clear cell cancer. In contrast, ERalpha pathway genes with tumor suppressive functions are hypermethylated in clear cell cancer. These results demonstrate that coordinate epigenetic deregulation contributes to prominent features characterizing the ovarian clear cell carcinoma phenotype.
Acknowledgments
We thank Carole Grenier and Darby Kroyer for excellent technical assistance. This work was supported by the American Cancer Society and the Gail Parkins Ovarian Cancer Awareness Fund.
Footnotes
Note: The gene expression microarray data used in this manuscript is publicly available through the Gene Expression Omnibus (GEO) web site, accessions GSE25428, GSE29175, GSE6008 and GSE2109. Illumina Infinium 27k methylation beadchip data for the ovarian cell lines analyzed in the present study is available at http://dl.dropbox.com/u/6904769/50CellLines_b-value_submission.txt.zip.
Conflict of Interest Statement: None declared.
References
- 1.Barton CA, Hacker NF, Clark SJ, O'Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109:129–39. doi: 10.1016/j.ygyno.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Gomez-Raposo C, Mendiola M, Barriuso J, Hardisson D, Redondo A. Molecular characterization of ovarian cancer by gene-expression profiling. Gynecol Oncol. 2010;118:88–92. doi: 10.1016/j.ygyno.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Cho KR, Shih Ie M. Ovarian cancer. Annu Rev Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandai M, Yamaguchi K, Matsumura N, Baba T, Konishi I. Ovarian cancer in endometriosis: molecular biology, pathology, and clinical management. Int J Clin Oncol. 2009;14:383–91. doi: 10.1007/s10147-009-0935-y. [DOI] [PubMed] [Google Scholar]
- 5.Skirnisdottir I, Seidal T, Karlsson MG, Sorbe B. Clinical and biological characteristics of clear cell carcinomas of the ovary in FIGO stages I-II. Int J Oncol. 2005;26:177–83. [PubMed] [Google Scholar]
- 6.Sugiyama T, Kamura T, Kigawa J, Terakawa N, Kikuchi Y, Kita T, Suzuki M, Sato I, Taguchi K. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer. 2000;88:2584–9. [PubMed] [Google Scholar]
- 7.Itamochi H, Kigawa J, Terakawa N. Mechanisms of chemoresistance and poor prognosis in ovarian clear cell carcinoma. Cancer Sci. 2008;99:653–8. doi: 10.1111/j.1349-7006.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JK, Teoh D, Hu JM, Shin JY, Osann K, Kapp DS. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol Oncol. 2008;109:370–6. doi: 10.1016/j.ygyno.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Ricciardelli C, Oehler MK. Diverse molecular pathways in ovarian cancer and their clinical significance. Maturitas. 2009;62:270–5. doi: 10.1016/j.maturitas.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr., Vogelstein B, Kinzler KW, Velculescu VE, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchiya A, Sakamoto M, Yasuda J, Chuma M, Ohta T, Ohki M, Yasugi T, Taketani Y, Hirohashi S. Expression profiling in ovarian clear cell carcinoma: identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. Am J Pathol. 2003;163:2503–12. doi: 10.1016/s0002-9440(10)63605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi K, Mandai M, Oura T, Matsumura N, Hamanishi J, Baba T, Matsui S, Murphy SK, Konishi I. Identification of an ovarian clear cell carcinoma gene signature that reflects inherent disease biology and the carcinogenic processes. Oncogene. 2010;29:1741–52. doi: 10.1038/onc.2009.470. [DOI] [PubMed] [Google Scholar]
- 14.Houshdaran S, Hawley S, Palmer C, Campan M, Olsen MN, Ventura AP, Knudsen BS, Drescher CW, Urban ND, Brown PO, Laird PW. DNA methylation profiles of ovarian epithelial carcinoma tumors and cell lines. PLoS One. 2010;5:e9359. doi: 10.1371/journal.pone.0009359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gargiulo G, Minucci S. Epigenomic profiling of cancer cells. Int J Biochem Cell Biol. 2009;41:127–35. doi: 10.1016/j.biocel.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Asadollahi R, Hyde CA, Zhong XY. Epigenetics of ovarian cancer: from the lab to the clinic. Gynecol Oncol. 2010;118:81–7. doi: 10.1016/j.ygyno.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, Li L, Wang Z, Wan JC, Davuluri RV, Karlan BY, Gifford G, et al. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788–94. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 18.Kamalakaran S, Kendall J, Zhao X, Tang C, Khan S, Ravi K, Auletta T, Riggs M, Wang Y, Helland A, Naume B, Dimitrova N, et al. Methylation detection oligonucleotide microarray analysis: a high-resolution method for detection of CpG island methylation. Nucleic Acids Res. 2009;37:e89. doi: 10.1093/nar/gkp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8:2246–52. [PubMed] [Google Scholar]
- 20.Ahluwalia A, Yan P, Hurteau JA, Bigsby RM, Jung SH, Huang TH, Nephew KP. DNA methylation and ovarian cancer. I. Analysis of CpG island hypermethylation in human ovarian cancer using differential methylation hybridization. Gynecol Oncol. 2001;82:261–8. doi: 10.1006/gyno.2001.6291. [DOI] [PubMed] [Google Scholar]
- 21.Terasawa K, Toyota M, Sagae S, Ogi K, Suzuki H, Sonoda T, Akino K, Maruyama R, Nishikawa N, Imai K, Shinomura Y, Saito T, et al. Epigenetic inactivation of TCF2 in ovarian cancer and various cancer cell lines. Br J Cancer. 2006;94:914–21. doi: 10.1038/sj.bjc.6602984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda T, Tashiro H, Katabuchi H, Begum M, Ohtake H, Kiyono T, Okamura H. Establishment of an immortalised human ovarian surface epithelial cell line without chromosomal instability. British Journal of Cancer. 2005;93:116–23. doi: 10.1038/sj.bjc.6602662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maines-Bandiera SL, Kruk PA, Auersperg N. Simian virus 40-transformed human ovarian surface epithelial cells escape normal growth controls but retain morphogenetic responses to extracellular matrix. Am J Obstet Gynecol. 1992;167:729–35. doi: 10.1016/s0002-9378(11)91579-8. [DOI] [PubMed] [Google Scholar]
- 24.Auersperg N, Maines-Bandiera SL, Dyck HG, Kruk PA. Characterization of cultured human ovarian surface epithelial cells: phenotypic plasticity and premalignant changes. Lab Invest. 1994;71:510–8. [PubMed] [Google Scholar]
- 25.Seiler M, Huang CC, Szalma S, Bhanot G. ConsensusCluster: a software tool for unsupervised cluster discovery in numerical data. OMICS. 2010;14:109–13. doi: 10.1089/omi.2009.0083. [DOI] [PubMed] [Google Scholar]
- 26.Matsumura N, Huang Z, Mori S, Baba T, Fujii S, Konishi I, Iversen ES, Berchuck A, Murphy SK. Epigenetic suppression of the TGF-beta pathway revealed by transcriptome profiling in ovarian cancer. Genome Res. 2010 doi: 10.1101/gr.108803.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newton MA, Quintana FA, den Boon JA, Sengupta S, Ahlquist P. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Annals of Applied Statistics. 2007;1:85–106. [Google Scholar]
- 30.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–19. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquez RT, Baggerly KA, Patterson AP, Liu J, Broaddus R, Frumovitz M, Atkinson EN, Smith DI, Hartmann L, Fishman D, Berchuck A, Whitaker R, et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin Cancer Res. 2005;11:6116–26. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 32.Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, Hanash S, Misek DE, Katabuchi H, Williams BO, Fearon ER, Cho KR. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11:321–33. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, Reed H, Ellenson LH, et al. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–9. [PubMed] [Google Scholar]
- 35.Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today. 2009;87:335–50. doi: 10.1002/bdrc.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11:137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurman RJ, Shih Ie M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer--shifting the paradigm. Human pathology. 2011;42:918–31. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott M, McCluggage WG. Current concepts in ovarian epithelial tumorigenesis: correlation between morphological and molecular data. Histol Histopathol. 2006;21:81–92. doi: 10.14670/HH-21.81. [DOI] [PubMed] [Google Scholar]
- 39.Yoon MS, Suh DS, Choi KU, Sol MY, Shin DH, Park WY, Lee JH, Jeong SM, Kim WG, Shin NR. High-throughput DNA hypermethylation profiling in different ovarian epithelial cancer subtypes using universal bead array. Oncol Rep. 2010;24:917–25. doi: 10.3892/or.2010.917. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura N, Mandai M, Okamoto T, Yamaguchi K, Yamamura S, Oura T, Baba T, Hamanishi J, Kang HS, Matsui S, Mori S, Murphy SK, et al. Sorafenib efficacy in ovarian clear cell carcinoma revealed by transcriptome profiling. Cancer Sci. 2010;101:2658–63. doi: 10.1111/j.1349-7006.2010.01736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J Am Soc Nephrol. 2000;11(Suppl 16):S140–3. [PubMed] [Google Scholar]
- 42.Kobel M, Kalloger SE, Carrick J, Huntsman D, Asad H, Oliva E, Ewanowich CA, Soslow RA, Gilks CB. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- 43.Smuc T, Hevir N, Ribic-Pucelj M, Husen B, Thole H, Rizner TL. Disturbed estrogen and progesterone action in ovarian endometriosis. Mol Cell Endocrinol. 2009;301:59–64. doi: 10.1016/j.mce.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 44.Fujimura M, Hidaka T, Kataoka K, Yamakawa Y, Akada S, Teranishi A, Saito S. Absence of estrogen receptor-alpha expression in human ovarian clear cell adenocarcinoma compared with ovarian serous, endometrioid, and mucinous adenocarcinoma. Am J Surg Pathol. 2001;25:667–72. doi: 10.1097/00000478-200105000-00016. [DOI] [PubMed] [Google Scholar]
- 45.Wiley A, Katsaros D, Chen H, Rigault de la Longrais IA, Beeghly A, Puopolo M, Singal R, Zhang Y, Amoako A, Zelterman D, Yu H. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107:299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 46.Kaneuchi M, Sasaki M, Tanaka Y, Shiina H, Yamada H, Yamamoto R, Sakuragi N, Enokida H, Verma M, Dahiya R. WT1 and WT1-AS genes are inactivated by promoter methylation in ovarian clear cell adenocarcinoma. Cancer. 2005;104:1924–30. doi: 10.1002/cncr.21397. [DOI] [PubMed] [Google Scholar]
- 47.Akahane T, Sekizawa A, Okuda T, Kushima M, Saito H, Okai T. Disappearance of steroid hormone dependency during malignant transformation of ovarian clear cell cancer. Int J Gynecol Pathol. 2005;24:369–76. doi: 10.1097/01.pgp.0000165173.90339.a2. [DOI] [PubMed] [Google Scholar]
- 48.Wright JW, Stouffer RL, Rodland KD. Estrogen inhibits cell cycle progression and retinoblastoma phosphorylation in rhesus ovarian surface epithelial cell culture. Mol Cell Endocrinol. 2003;208:1–10. doi: 10.1016/j.mce.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Cunat S, Hoffmann P, Pujol P. Estrogens and epithelial ovarian cancer. Gynecol Oncol. 2004;94:25–32. doi: 10.1016/j.ygyno.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.