Abstract
The degeneration of substantia nigra (SN) dopamine (DA) neurons in sporadic Parkinson’s disease (PD) is characterized by disturbed gene expression networks. Micro(mi)RNAs are post-transcriptional regulators of gene expression and we recently provided evidence that these molecules may play a functional role in the pathogenesis of PD. Here, we document a comprehensive analysis of miRNAs in SN DA neurons and PD, including sex differences. Our data show that miRNAs are dysregulated in disease-affected neurons and differentially expressed between male and female samples with a trend of more up-regulated miRNAs in males and more down-regulated miRNAs in females. Unbiased Ingenuity Pathway Analysis (IPA) revealed a network of miRNA/target-gene associations that is consistent with dysfunctional gene and signaling pathways in PD pathology. Our study provides evidence for a general association of miRNAs with the cellular function and identity of SN DA neurons, and with deregulated gene expression networks and signaling pathways related to PD pathogenesis that may be sex-specific.
Keywords: miRNAs, Dopamine Neurons, Parkinson’s Disease, Laser Capture Microdissection, IPA
1. Introduction
Sporadic Parkinson’s disease (PD) is associated with the progressive loss of substantia nigra (SN) dopamine (DA) neurons (Braak and Del Tredici, 2008), and the exact molecular mechanisms of this cell loss are still unknown. Several factors have been implicated in the pathogenesis of PD, such as genetic predisposition and environmental factors, which affect key signaling pathways in the function of DA neurons (Schapira and Jenner, 2011; Wirdefeldt et al., 2011). On the molecular level we, and others, have demonstrated that the SN DA neurons from sporadic PD patients display dysregulated gene expression networks that are related to major signaling pathways in PD pathogenesis (Cantuti-Castelvetri et al., 2007; Elstner et al., 2011; Simunovic et al., 2009; Simunovic et al., 2010).
Recently we have extended our studies to also determine the miRNA expression profiles of these neurons. miRNAs are short non-coding RNAs that regulate gene expression on the pre- or post-transcriptional level (Bartel, 2009), and evidence suggests that these molecules are involved in the pathology of PD (Heman-Ackah et al., 2013; Ma et al., 2013; Sonntag, 2010; Wong and Nass, 2012). We found that human SN DA neurons have a distinctive miRNA expression profile that is dysregulated in PD, and functional analysis of miR-126, which was upregulated in PD DA neurons, unraveled an association of this miRNA with Insulin/IGF-1/PI3K signaling, a pathway that has been implicated in PD (Kim et al., 2014a; Kim et al., 2014h). These data suggested that miRNAs may have functional roles in DA neurons and in the pathogenesis of PD.
In the current study, we conducted a comprehensive analysis of miRNA expression profiles in DA neurons and PD, including sex differences, and their associations with gene expression networks. We show that miRNAs are dysregulated in disease-affected neurons and differentially expressed between males and females. Correlation of up- or down-regulated miRNAs with upstream regulators that are dysregulated in PD demonstrate a network of miRNA/gene-target associations that is linked to dysfunctional genes and signaling pathways in PD pathology.
2. Results
2.1. PD DA neurons have dysregulated miRNA expression profiles
To assess differential miRNA expression profiles between PD and control DA neurons, we determined average FC for each miRNA with CT values < 35 using three independent methods based on either endogenous control sno-RNA RNU44, global mean, or ABIqPCR normalization (D’Haene et al., 2012; Deo et al., 2011; Mestdagh et al., 2009). Because our sample population consisted of 5 males and 3 females in each control or PD patients group (Kim et al., 2014a; Simunovic et al., 2009; Simunovic et al., 2010), we determined miRNA profiles for all samples and females or males separately. Altogether, 159 miRNAs had CT values < 35, and consistent with our previous observation (Kim et al., 2014a), DA neurons from PD patients had dysregulated miRNA expression profiles with patterns of miRNA changes that show a trend of more up-regulation in the male and more down-regulation in the female group (Fig. 1, Supplementary Table S1).
Fig. 1.
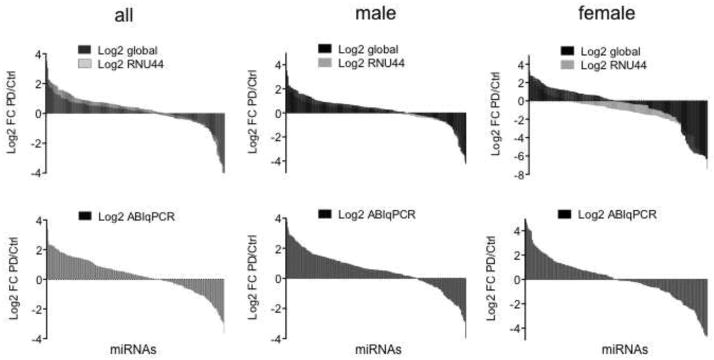
miRNA expression profiles in LMD DA neurons. Average Log2 FC of miRNA expression between controls and PD samples with CT values < 35 after data analysis with three independent methods using the endogenous control RNU44, global mean, or ABIqPCR for normalization (D’Haene et al., 2012; Deo et al., 2011; Mestdagh et al., 2009).
2.2. PD DA neurons have dysregulated cellular pathways and upstream regulators
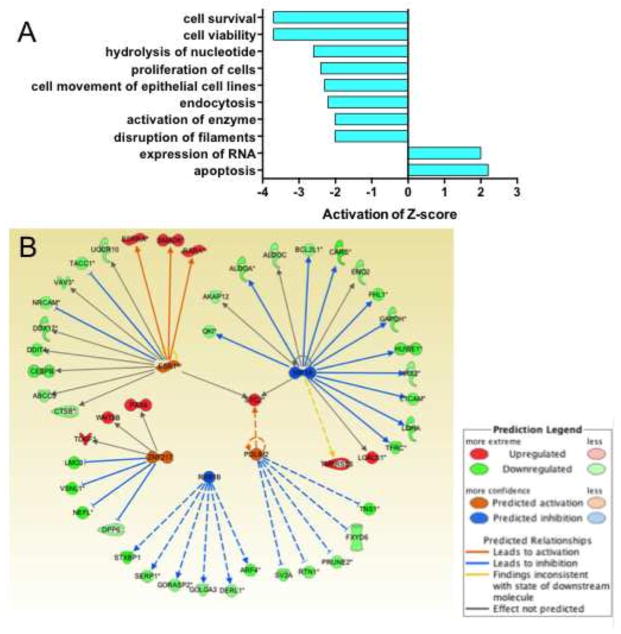
To identify PD-associated genes and signaling pathways, we first used IPA Core Analyses to determine up- or down-regulated pathways (IPA Biofunctions) or target genes (IPA Upstream Regulators) using our published mRNA arrays (Simunovic et al., 2009; Simunovic et al., 2010). Consistent with our previous observations, we identified dysregulated cellular pathways in PD DA neurons that are, among others, related to apoptosis, dysruption of filaments, cell proliferation, cell viability, and survival (Fig. 2A). These analyses identified 47 gene-targets of upstream regulators, from which 23 genes also contributed to the IPA Biofunctions, and an additional six dysregulated genes that were predicted effectors (absolute Z-scores > 2) based on their associations with the miRNA target-genes (Fig. 2B and Supplementary Table S2).
Fig. 2.
IPA Biofunctions in DA neurons with activation/inhibition Z-scores. (A) Dysregulated signaling pathways in DA neurons that are associated with PD. Z-scores for each IPA Biofunction and Upstream Regulator were calculated in IPA’s Core Analysis based on a binomial distribution of up-regulated and down-regulated pathway genes (in IPA Biofunctions) or target genes (of IPA Upstream Regulators) on mRNA array data from LMD DA neurons (Simunovic et al., 2009; Simunovic et al., 2010). Genes with |FC| > 1.2 and t-test p values < 0.05 were selected from the experimental data as analysis-ready genes for the IPA Core Analysis. (B) Network associations of 47 IPA upstream regulators and 6 predicted effectors with activation/inhibition Z-scores > 2.0.
2.3. miRNAs are associated with dysregulated gene and signaling pathways in PD
To determine miRNA associations with the gene expression networks in PD, we used the list of globally normalized miRNAs in all samples consisting of 109 up- and 50 down-regulated miRNAs (Supplementary Table S1). When we correlated the upregulated miRNAs with the 47 targets of upstream regulators (Fig. 2B and Supplementary Table S2), 52 miRNAs were associated with 17 target genes (Fig. 3A, Table 1 and Supplementary Table S3). From these miRNAs, 12 had p values < 0.05 and p values from an additional 14 miRNAs were > 0.05 but < 0.1. From the 50 down-regulated miRNAs, we identified 4 targets of upstream regulators that correlated with 8 miRNAs, from which 2 were significantly down-regulated in PD (p < 0.05) and were associated with 2 target genes (Fig. 3B, Table 1 and Supplementary Table S3). Altogether, these data analyses revealed a network of 14 significantly (p < 0.05) dysregulated miRNAs in PD DA neurons that correlated with 16 PD-associated target genes (Table 1). Data mining of the literature showed that the gene-targets were associated with several aspects of PD pathogenesis and additional miRNAs that were not included in the IPA analysis (Table 2). As for sex differences, 8 of the 12 upregulated miRNAs with p values < 0.05 were also upregulated in males (miR-106a, -135a, -148a, -223, -26a, -28-5p, -335, -and -92a), while 3 were upregulated in females (let-7b, miR-106a, and -95) (Supplementary Table S3). Accordingly, 10 of the down-regulated target genes (IRS2, STXBP1, TFRC, FHL1, VAV3, DDX17, HUWE1, CEBPB, L1CAM, and NEFL) were associated with males, 4 with females (ABCC5, AKAP12, IRS2, and VAV3), and 1 (LMO3) with both.
Fig. 3.
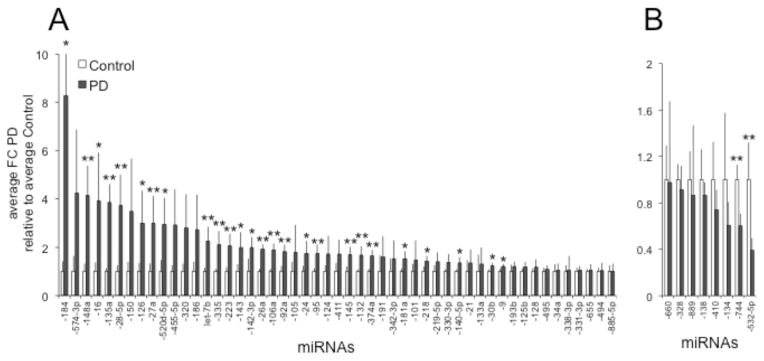
Dysregulated miRNAs with target gene associations. 52 up- (A) and 8 down-regulated (B) miRNAs that target 17 down- and 4 up-regulated target genes, respectively (see Table 1 and Supplementary Table S3). **: FC PD versus Control with p values < 0.05; *: FC PD versus Control with p values > 0.05 < 0.1.
Table 1.
Down-regulated target-genes associated with up-regulated miRNAs and up-regulated target-genes associated with down-regulated miRNAs. miRNAs with p values < 0.05 are marked in italic, bold, and gray, and with p > 0.05 but < 0.1 in italic and bold (for more details see Fig. 3 and Supplementary Table S1). The FCs of the identified target-genes in IPA were highly consistent with data from our previously published gene lists (*: Simunovic et al., 2010).
| Down-regulated target genes associated with up-regulated miRNAs | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Title | FC | FC* | miRNAs IPA analysis |
| ABCC5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | −1.41 | −2.08 | let-7b, -101, -125b, -494 |
| AKAP12 | A kinase (PRKA) anchor protein 12 | −1.22 | −1.54 | -145, -16, -186, -21, -219-5p, -338-3p, -455-5p, -95 |
| BCL2L1 | BCL2-like 1 | −1.23 | −1.45 | let-7b, -133a, -140-5p, -142-3p, -184, -331-3p, -342-3p, -495 |
| CARS | cysteinyl-tRNA synthetase | −4.05 | −5.00 | -30b |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | −1.87 | −1.96 | -150, -191, -374a, -665 |
| CTSB | cathepsin B | −1.36 | −1.51 | -126, -140-5p, -218, -24, -320, -411, -455-5p |
| DDX17 | DEAD (Asp-Glu-Ala-Asp) box helicase 17 | −1.82 | −2.63 | -105, -145, -26a, -335, -34a, -9 |
| FHL1 | four and a half LIM domains 1 | −1.91 | −2.86 | -105, -223, -574-3p |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase | −1.98 | −1.82 | -184, -26a |
| IRS2 | insulin receptor substrate 2 | −1.32 | −1.85 | let-7b, -135a, -145, -16, -181a, -193b, -30b, -338-3p, -92a |
| L1CAM | L1 cell adhesion molecule | −1.64 | −1.85 | -181a, -324-3p, -331-3p, -374a |
| LDHA | lactate dehydrogenase A | −1.96 | −2.86 | -338-3p, -34a |
| LMO3 | LIM domain only 3 (rhombotin-like 2) | −2.51 | −4.76 | -101, -106a, -124, -142-3p, -181a, -186, -218, -219-5p, -320, -338-3p |
| NEFL | neurofilament, light polypeptide | −2.60 | −2.00 | -30b, -330-3p, -885-5p, -9, -92a |
| STXBP1 | syntaxin binding protein 1 | −2.96 | −3.85 | -132, -133a, -143, -148a, -16, -218, -30b, -331-3p, -9 |
| TFRC | transferrin receptor | −1.89 | −3.13 | -124, -140-5p, -145, -148a, -181a, -28-5p, -338-3p, -520d-5p, -9 |
| VAV3 | vav 3 guanine nucleotide exchange factor | −1.6 | −2.00 | let-7b, -125b, -128, -193b, -218, -223, -27a, -30b, -338-3p, -520d- 5p, -655, -9 |
| Up-regulated target genes associated with down-regulated miRNAs | ||||
|---|---|---|---|---|
| Gene Symbol | Gene Title | FC | FC* | miRNAs IPA analysis |
| ESRRA | estrogen-related receptor alpha | 1.25 | 1.22 | -328 |
| RARA | retinoic acid receptor, alpha | 1.23 | 1.39 | -138, -744 |
| SMAD6 | SMAD family member 6 | 1.23 | 1.21 | -134, -410, -889 |
| TDGF1 | teratocarcinoma-derived growth factor 1 | 1.43 | 1.58 | -532-5p, -660 |
Table 2.
IPA-identified target-genes, their biofunctions, and miRNAs associated with gene functions according to the published literature. miRNAs that are also dysregulated in PD DA neurons with p values < 0.1 are marked in italic and bold (for details see Fig. 3 and Supplementary Table S1).
| Gene Symbol | Selected Biological Functions | Associated miRNAs | Reference |
|---|---|---|---|
| ABCC5 | Multidrug-resistant proteins mediating ATP-dependent export of organic anions from cells; substrates include cyclic AMP and GMP, and nucleotide analogs; seems to be involved in Na+/H+ exchange and protection to glutamate-induced toxicity in pyramidal neurons and astrocytes. | miR-129-5p, -128 | (Nies et al., 2004; Wu et al., 2014; Zhu et al., 2011b) |
| AKAP12 | Key mediator of protein kinase A and C signaling regulating several signaling cascades that affect ERK2 activation; contributes to synaptic plasticity and memory function in the hippocampus by regulating β2-adrenergic receptor signaling; associated with neurodegeneration. | (Havekes et al., 2012; Poppinga et al., 2014) | |
| BCL2L1 | Plays a role in multiple aspects of cell death and has been directly linked to factors in DA cell function and PD pathogenesis, e.g., by inducing several midbrain DA-specific neuronal transcription factors and interacting with the PD-associated molecules PINK1 and DJ1. | let-7 group, miR-122, -133a, -98, -491 | (Arena et al., 2013; Courtois et al., 2010; Hertz et al., 2013; Kang et al., 2011; Michels et al., 2013; Ren et al., 2012; Seiz et al., 2012) |
| CEBPB | Associated with synaptic plasticity and memory, inflammation, neurogenesis, and possibly apoptosis; downstream factor of BDNF/TrkB signaling and transcriptional activity on the GLUT4 promoter. | miR-155, -378 | (Calella et al., 2007; Chen et al., 2013; Gerin et al., 2010; Pena-Altamira et al., 2014) |
| CTSB | Lysosomal protease that functions in α-Synuclein homeostasis in DA neurons contributing to its attenuation and aggregate formation. | miR-218 | (Crabtree et al., 2014; Tsujimura et al., 2014; Venkataraman et al., 2013) |
| DDX17 | Transcriptional activator involved in miRNA biogenesis, transcriptional regulation and RNA splicing; participates in the co-activation of the tumor suppressor p53 or the estrogen receptor α (ESR1). | (Dardenne et al., 2014; Fuller-Pace, 2013) | |
| ESRRA | Linked to ESR1 functions; meta-analyses showed that genetic polymorphism of ESR1 may be a risk factor for PD. | (Chung et al., 2011; Gao et al., 2014; Palacios et al., 1991) | |
| FHL1, LMO3 | Members of a group of nuclear and cytoplasmic scaffolding proteins that mediate the assembly of multiprotein complexes; LMO3 is highly expressed in the brain and seems to play a role in neurogenesis, neuroblastoma or astrocyte cell proliferation, and in the induction of apoptosis by inhibiting the tumor suppressor p53. | (Sang et al., 2014; Shathasivam et al., 2010) | |
| HUWE1 | Ubiquitin ligase implicated in neural stem cell differentiation, adult neurogenesis, and the DNA damage response pathway; suggested as a possible therapeutic target in PD. | (Salama, 2012; Zhou et al., 2014) | |
| IRS2 | Like IRS1, adapter molecule of the insulin or IGF-1 receptor; regulates insulin/IGF-1/PI3K and RAS/ERK1/2 signaling; dysfunctional insulin/IGF-1 signaling has been implicated in aging and neurodegeneration, including PD. | LIN28, let-7 group, miR-7-5p, -33b, -126, -128a, -135a, -200a, -145, -96 | (Agarwal et al., 2013; Bassil et al., 2014; Chakraborty et al., 2014; Crepin et al., 2014; Frost and Olson, 2011; Giles et al., 2013; Gurung et al., 2014; Rottiers et al., 2011; Wen et al., 2014; Zhu et al., 2011a) |
| L1CAM | Belongs to a group of extracellular adhesion molecules that are associated with many CNS diseases and implicated as therapeutic targets, including in PD. | miR-29a, -34a, -146a, -503 | (Berezin et al., 2014; Diedrich et al., 2008; Hulley et al., 1998) |
| NEFL | Plays a role in axonal transport and organizing the cytoarchitecture affecting mitochondrial morphology, and ER, endosome and lysosome distribution; mutations have been associated with Charcot-Marie-Tooth disease; mRNA is stabilized by miRNAs, which are downregulated in the spinal cord of ALS patients. | miR-b1336, -b2403 | (Gentil and Cooper, 2012; Ishtiaq et al., 2014) |
| RARA | Involved in Vitamin A/retinoic acid (RA) metabolism; key molecule in homeostatic synaptic plasticity and neuronal strength; factor in neuroprotection by promoting multiple anti-inflammatory processes, including in midbrain DA neurons; RA-mediated neuroprotection may be disturbed in PD; RA- and Toll-like receptor-associated signaling cascades appear to induce multiple miRNAs that play a role in inflammatory processes. | let-7 group, miR-125b, -132, -146a, -147, -155, -21, -223, -27b, -9, -98 | (Chen et al., 2014b; Li and Shi, 2013) |
| STXBP1 | Chaperone for syntaxin-1; controls the N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) complex; participates in exocytosis/membrane fusion, neurosecretion and neurotransmission. | miR-130a, -132, -212, -218, -322, -335 | (Cijsouw et al., 2014; Esguerra et al., 2011; Han et al., 2010; Lang et al., 2014) |
| TDGF1 | Co-receptor for transforming growth factor (TGF)-β family members; activates several signaling cascades, including ALK4/ALK7/SMAD, PI3K/AKT, MAPK, WNT/β-CATENIN, and NOTCH/CBF-1 pathways; plays a role in cell morphogenesis and lineage specification in development, and is associated with a variety of cancers. | miR-15a, -16 | (Chen et al., 2014a; Klauzinska et al., 2014) |
| TFRC | Mediates the extracellular transport of transferrin and regulates cellular iron metabolism; iron deficiency has been associated with brain dysfunction, including DA abnormalities in restless leg syndrome; disease-associated gene in a microarray and protein-protein- interaction databases in postmortem brain tissue samples from PD patients. | (Jellen et al., 2013; Rakshit et al., 2014) | |
| VAV3 | GDP/GTP exchange factor for Rho/Rac GTPases that regulate multiple cell functions; plays a role in cerebellar development, GABAergic axon guidance, and peripheral nerve regeneration; disease-associated gene in a microarray and protein-protein-interaction databases in postmortem brain tissue samples from PD patients. | (Keilhoff et al., 2012; Quevedo et al., 2010; Rakshit et al., 2014; Sauzeau et al., 2010) |
3. Discussion
Here we show that miRNA profiles are dysregulated in PD DA neurons with a trend of more up-regulated miRNAs in males and more down-regulated miRNAs in females. In addition, IPA-based data analysis revealed a network of miRNA/gene-target associations in these neurons that is consistent with dysfunctional gene and signaling pathways in PD pathology, and appears to be more prominent in males. Together, these data complement our previous observations that midbrain DA neurons exhibit unique expression networks that are dysregulated in PD, and may be sex-specific (Kim et al., 2014a; Simunovic et al., 2009; Simunovic et al., 2010).
The data from our study provide a platform for the identification and characterization of functional miRNAs in DA neurons. The combined analysis of miRNA and mRNA profiles indicated that known features of miRNA functions which have been described in other cell contexts, are also present in human midbrain DA neurons. This includes the observation that multiple miRNAs act on the same target and pathway(s), while a single miRNA can be associated with multiple targets and pathways. Importantly, potential miRNA targets in the DA neurons are also targets in other cell types, consistent with the view that miRNAs are not necessarily “cell-specific”; rather, they are involved in the determination of many cellular phenotypes and, thus, may play a role in several disease entities, and in particular cancer (Du and Pertsemlidis, 2011; Sonntag, 2010; Sonntag et al., 2012). In fact, many of the targets and pathways that we identified in our study have been described in cancer biology as well. On the other hand, it is also possible that the unique composition of miRNAs (as now determined for midbrain DA neurons) defines a cellular phenotype and when disturbed, contributes to (cell-specific) disease pathogenesis.
Several studies have indicated associations of miRNAs with some aspects of PD pathology (summarized in (Heman-Ackah et al., 2013; Ma et al., 2013; Wong and Nass, 2012)). “PD-specific” miRNA/target associations of interest have been miR-7, miR-153, and miR-34b/c with α-Synuclein, (Doxakis, 2010; Junn et al., 2009; Kabaria et al., 2015), miR-433 with FGF20 (Wang et al., 2008), miR-133b with PITX3 (Kim et al., 2007), let-7a-5p, miR-184-5p, and miR-205 with LRRK2 (Cho et al., 2013; Gehrke et al., 2010), miR-132 with NURR1 (Yang et al., 2012) or AChE (Shaked et al., 2009), and miR-34b/c indirectly with PARKIN and DJ1 (Minones-Moyano et al., 2011), and decreased expression of miR-133b and miR-34b/c, or miR-205 was identified in dissected human postmortem midbrain, or cortical tissue from sporadic PD patients, respectively. Except for miR-132 and miR-184, which were upregulated (Log2 FC 0.75; p = 0.05, and Log2 FC 2.05; p = 0.89, respectively), and miR-433, which was slightly, but not significantly downregulated (Log2 FC -0.09; p = 0.4) in PD, our results did not provide strong support for a role of most of these miRNAs in DA neurons or PD: miR-133b, miR-34b/c, miR-153, and miR-205 were expressed below detection threshold and miR-7 was not present on the TaqMan® Human MicroRNA A Array v2.0.
Altogether, our data did not reveal strong indication for miRNAs targeting specific key factors that are thought to be major players in PD, e.g., members from the PARK gene group (Schiesling et al., 2008), which are markedly downregulated in the DA neurons (Simunovic et al., 2009; Simunovic et al., 2010). However, we identified a network of miRNA target-genes and predicted effectors (summarized in Tables 1 and 2) that is linked to dysfunctional signaling pathways related to metabolism, apoptosis, protein degradation, synaptic plasticity/transmission, and inflammation. Many of these genes and pathways have been implicated in PD pathology, but also overlap with other disease entities, and particular cancer. Identified effectors include the estrogen receptor α (ESR1), which is transcriptionally co-activated by DDX17 (Dardenne et al., 2014; Fuller-Pace, 2013), and genetic polymorphism of this gene may be a risk factor for PD (Chung et al., 2011; Gao et al., 2014; Palacios et al., 2010); RAS oncogene member RAB1B, which has been proposed to participate in vesicle transfer from ER to the Golgi apparatus in neuronal dendritic spines (Pierce et al., 2001), and PDZ and LIM domain 2 (PDLIM2), which is an ubiquitin E3 ligase and has been implemented in inflammatory processes, cell adhesion, apoptosis, and proliferation (Mankan et al., 2009). Intriguingly, these analyses also identified hypoxia inducible factor 1 alpha (HIF1A), which plays a role in ubiquitination, hypoxia and oxidative stress, and has been shown to be negatively regulated by different PD-associated molecules, including DJ-1 (Parsanejad et al., 2014), PINK1 (Lin et al., 2014), and Rotenone (Wu et al., 2010).
As for sex differences, the higher number of up-regulated miRNAs and associations with down-regulated gene-targets in the male population is somewhat consistent with our previous observation that males may have a more prominent downregulation of gene expression than females (Simunovic et al., 2010). In addition, some of the down-regulated gene targets in males could be linked to more “male-dominant” signaling pathways, including synaptic transmission (STXBP1, CEBPB), and apoptosis (DDX17, HUWEI1) (Simunovic et al., 2010), while other targets such as IRS2 and VAV3 that are related to metabolism and axon guidance or nerve regeneration, were associated with both males and females. Although these observations are derived from a limited sample number, they could suggest that some dysregulated miRNA/gene-target networks in PD pathology may be sex-specific.
It is important to keep in mind that results from the assessment of miRNA expression levels and the computational data analyses, as presented here, are often not absolute. First, although the three methods used for determining miRNA expression profiles (D’Haene et al., 2012; Deo et al., 2011; Mestdagh et al., 2009) revealed largely similar results for differentially expressed miRNAs between controls and PD, not all of these data had overlapping significant p values. This was not surprising since such discrepancies have been previously associated with these methods and could have been attributed to mathematical disparities in the methodologies, the restricted sample size, and variations between individual samples (D’Haene et al., 2012; Deo et al., 2011; Mestdagh et al., 2009). In particular, studies on human postmortem material are afflicted with substantial variability between subjects, and small sample numbers can influence data strength (discussed in (Simunovic et al., 2010)). Second, results on differential gene expression and miRNA/target associations also depend on the choice of the computational algorithms. IPA provides microRNA Target Filter functionality for combining miRNA and mRNA expression data in order to facilitate the identification of miRNA-target pairs and networks relevant to the phenotype of interest. IPA’s miRNA-target reference database is comprised of validated interactions from external databases including TarBase, miRecord and Targetscan, and findings from peer-reviewed literature. In addition, IPA provides a ranking scale for the strength of evidence for each identified miRNA-target pair: Observed (experimentally), High (predicted) and Moderate (predicted). All of the upstream regulators and miRNA target-genes identified by IPA were also present in our previously published gene lists based on ANOVA and SAM (Simunovic et al., 2009; Simunovic et al., 2010) indicating a high level of validity in the IPA analysis. However, further studies are necessary to validate miRNA expression and to analyze their functional relevance in both healthy and disease-affected human midbrain DA neurons.
Along this line, we selected two miRNAs of interest, miR-126 and miR-320, which target factors in insulin signaling. miR-320 was detected at high levels in pigmented neurons in the SN by in situ hybridization (Nelson et al., 2008), confirming the qRT-PCR data in the current study. As for functional relevance, we could show that miR-126 may play an important role in neurotoxicity and neuroprotection by regulating growth factor/PI3K/AKT and MAPK/ERK signaling (Kim et al., 2014a; Kim et al., 2014h), and preliminary data indicate similar neurotoxic functions for miR-320 as well (unpublished results). Together, these data suggest that these, and other miRNAs, may be functionally involved in aging and neurodegeneration, and therefore, could be therapeutic targets in neurological disorders.
4. Experimental procedure
4.1. Subjects and material collection
All data about subjects and information about Affymetrix-based array analysis are publicized at the National Brain Databank (http://national_databank.mclean.harvard.edu/brainbank) or have been published elsewhere (Kim et al., 2014a; Simunovic et al., 2009; Simunovic et al., 2010). Material collection, preparation, and data generation for the miRNA arrays were conducted according to previously published protocols (Benes et al., 2007; Pietersen et al., 2011; Simunovic et al., 2009; Simunovic et al., 2010). Briefly, frozen tissue blocks obtained from the Harvard Brain Tissue Resource Center containing SN from 8 control subjects and 8 patients with idiopathic PD (5 males and 3 females in each group, respectively) matched for age and postmortem interval (PMI), were cut using a Microm HM 560 CryoStar cryostat (8 μm), mounted on LEICA Frame Slides with a PET-membrane (1.4 μm), dehydrated, and either directly used for laser microdissection (LMD) with a LEICA AS LMD apparatus, or stored at −80 °C, prior to RNA extraction using the mirVANATM miRNA Isolation Kit (Ambion, Austin, TX).
4.2. miRNA profiling by Megaplex miRNA TaqMan® Arrays
miRNA profiling was performed as recently published (Kim et al., 2014a). Human MicroRNA TaqMan® Arrays A 2.0 (Life Technologies, Foster City, CA) were used for miRNA analysis. About 300 neurons were collected per sample and 20 ng of total RNA was reverse-transcribed using MegaplexTM RT Primers Human Pool A and TaqMan miRNA reverse transcription kit (Life Technologies, Foster City, CA) in a total of 7.5 μl volume. For pre-amplification, 2.5 μl of cDNA was pre-amplified using MegaplexTM PreAmp Primers Human Pool A and TaqMan PreAmp Master Mix (Life Technologies, Foster City, CA) in a 25 μl PCR reaction. Fourteen cycles of pre-amplification were carried out following the manufacturer’s protocol and the activated Taq polymerase inhibited at 99.9 °C for 10 min. The pre-amplified cDNA was diluted 4-fold with 0.1 X TE buffer (pH 8.0). Quantitative real-time PCR was performed using the Applied Biosystems 7900HT system and TaqMan® Universal PCR Master Mix with 9 μl diluted cDNA input per TaqMan array. miRNAs with cycle threshold (CT) values <35 were analyzed according to published protocols using RNU44 or global mean normalization, or ABI’s R package for quantitative real-time polymerase chain reaction (qRT-PCR) analysis (ABIqPCR) in Bioconductor (D’Haene et al., 2012; Deo et al., 2011; Mestdagh et al., 2009). Expression fold changes (FC) were calculated with 1 tail, type 2 t-test.
4.3. Microarray data processing
Raw signal intensity values were uploaded into Bioconductor (Gentleman et al., 2004), an open-source, R-based platform of software packages for processing and analysis of microarray data. Intensity values were read into Bioconductor, log2-transformed and quantile normalized using the lumi package (Du et al., 2008). FC and moderated t-test p values were calculated between patients and controls for all probes with the limma package in Bioconductor (Smyth, 2005).
4.4. Identification of dysregulated biological functions and upstream regulators in PD versus controls in Ingenuity Platform Analysis (IPA)
FC and t-test p values for all probes were uploaded to IPA and filtered to select Analysis Ready Genes based on the user’s criteria, in our case, genes with t-test p values < 0.05 and |FC| >1.2. Analysis Ready Genes were then used for the IPA Core Analysis. The Core Analysis includes algorithms to infer and score IPA upstream regulators and downstream biological effects based on the expression data for the Analysis Ready Genes, as described by Krämer et al. (Kramer et al., 2014). Downstream biological functions are scored by Fisher’s Exact Test, an enrichment analysis for functional gene sets in the Analysis Ready expression data compared to reference data (IPA Knowledge Base). The Core Analysis also calculates a Z-score for the predicted activation state (activated or suppressed) of IPA upstream regulators based on the FC direction (up-regulated or down-regulated) observed among known downstream targets.
4.5. Identification of dysregulated miRNAs targeting key genes in PD patients
miRNA qPCR data were uploaded to IPA. IPA’s Target Filter paired miRNAs with gene-targets among Analysis Ready genes from the expression data. According to IPA, “The microRNA Target Filter in IPA provides insights into the biological effects of microRNAs, using experimentally validated interactions from TarBase and miRecords, as well as predicted microRNA-mRNA interactions from TargetScan. Additionally, IPA includes a large number of microRNA-related findings from the peer-reviewed literature”.
Supplementary Material
Highlights.
We analyzed gene and miRNA expression profiles in postmortem dopamine neurons in PD
DA neurons in PD have dysregulated miRNA expression profiles that are sex-specific
miRNAs are associated with dysregulated target-genes in PD DA neurons
miRNAs are involved in dysregulated pathways that are linked to PD pathogenesis
Dysregulated miRNA/gene-target expression networks in PD may be sex-specific
Acknowledgments
We want to thank Dr. Bruce Cohen for additional financial support and intellectual and stimulating discussions, and Dr. Robert J. Rooney at Genome Explorations Inc. for additional ABIqPCR data analysis. This research was supported by a grant from the Massachusetts’ Alzheimer’s Disease Research Center and the Harvard NeuroDiscovery Center, and National Institute of Neurological Disorders and Stroke R21NS067335 to Kai C Sonntag.
List of abbreviations
- CT
Cycle threshold
- DA
Dopamine
- FC
Fold Change
- SN
Substantia nigra
- LMD
Laser Microdissection
- IPA
Ingenuity Pathway Analysis
Footnotes
Competing interests
The authors declare no potential conflict of interest.
Author’s contributions
K.C.S.: conception, design, data acquisition, analysis, interpretation, presentation and write-up for publication; C.B.: design, data acquisition, analysis, interpretation, presentation, and manuscript preparation; Y.W. and B.K.: Data acquisition; T.U.W.W. and L.K.I.: Manuscript preparation.
Supportive supplementary data can be found in the online version of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal P, Srivastava R, Srivastava AK, Ali S, Datta M. miR-135a targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle. Biochim Biophys Acta. 2013;1832:1294–303. doi: 10.1016/j.bbadis.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013;20:920–30. doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil F, Fernagut PO, Bezard E, Meissner WG. Insulin, IGF-1 and GLP-1 signaling in neurodegenerative disorders: Targets for disease modification? Prog Neurobiol. 2014;118:1–18. doi: 10.1016/j.pneurobio.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–9. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezin V, Walmod PS, Filippov M, Dityatev A. Targeting of ECM molecules and their metabolizing enzymes and receptors for the treatment of CNS diseases. Prog Brain Res. 2014;214:353–88. doi: 10.1016/B978-0-444-63486-3.00015-3. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Invited Article: Nervous system pathology in sporadic Parkinson disease. Neurology. 2008;70:1916–25. doi: 10.1212/01.wnl.0000312279.49272.9f. [DOI] [PubMed] [Google Scholar]
- Calella AM, Nerlov C, Lopez RG, Sciarretta C, von Bohlen und Halbach O, Bereshchenko O, Minichiello L. Neurotrophin/Trk receptor signaling mediates C/EBPalpha, -beta and NeuroD recruitment to immediate-early gene promoters in neuronal cells and requires C/EBPs to induce immediate-early gene transcription. Neural Dev. 2007;2:4. doi: 10.1186/1749-8104-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Keller-McGandy C, Bouzou B, Asteris G, Clark TW, Frosch MP, Standaert DG. Effects of gender on nigral gene expression and parkinson disease. Neurobiol Dis. 2007;26:606–14. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Doss CG, Bandyopadhyay S, Agoramoorthy G. Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip Rev RNA. 2014;5:697–712. doi: 10.1002/wrna.1240. [DOI] [PubMed] [Google Scholar]
- Chen F, Hou SK, Fan HJ, Liu YF. MiR-15a-16 represses Cripto and inhibits NSCLC cell progression. Mol Cell Biochem. 2014a;391:11–9. doi: 10.1007/s11010-014-1981-y. [DOI] [PubMed] [Google Scholar]
- Chen L, Lau AG, Sarti F. Synaptic retinoic acid signaling and homeostatic synaptic plasticity. Neuropharmacology. 2014b;78:3–12. doi: 10.1016/j.neuropharm.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Siegel F, Kipschull S, Haas B, Frohlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Liu G, Jin SM, Parisiadou L, Xie C, Yu J, Sun L, Ma B, Ding J, Vancraenenbroeck R, Lobbestael E, Baekelandt V, Taymans JM, He P, Troncoso JC, Shen Y, Cai H. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2013;22:608–20. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SJ, Armasu SM, Biernacka JM, Lesnick TG, Rider DN, Cunningham JM, Maraganore DM. Variants in estrogen-related genes and risk of Parkinson’s disease. Mov Disord. 2011;26:1234–42. doi: 10.1002/mds.23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cijsouw T, Weber JP, Broeke JH, Broek JA, Schut D, Kroon T, Saarloos I, Verhage M, Toonen RF. Munc18-1 redistributes in nerve terminals in an activity- and PKC-dependent manner. J Cell Biol. 2014;204:759–75. doi: 10.1083/jcb.201308026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois ET, Castillo CG, Seiz EG, Ramos M, Bueno C, Liste I, Martinez-Serrano A. In vitro and in vivo enhanced generation of human A9 dopamine neurons from neural stem cells by Bcl-XL. J Biol Chem. 2010;285:9881–97. doi: 10.1074/jbc.M109.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree D, Dodson M, Ouyang X, Boyer-Guittaut M, Liang Q, Ballestas ME, Fineberg N, Zhang J. Over-expression of an inactive mutant cathepsin D increases endogenous alpha-synuclein and cathepsin B activity in SH-SY5Y cells. J Neurochem. 2014;128:950–61. doi: 10.1111/jnc.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin D, Benomar Y, Riffault L, Amine H, Gertler A, Taouis M. The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol Cell Endocrinol. 2014;384:1–11. doi: 10.1016/j.mce.2013.12.016. [DOI] [PubMed] [Google Scholar]
- D’Haene B, Mestdagh P, Hellemans J, Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–72. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, Chakrama FZ, Samaan S, Desmet FO, Tranchevent LC, Dutertre M, Rimokh R, Bourgeois CF, Auboeuf D. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7:1900–13. doi: 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Deo A, Carlsson J, Lindlof A. How to choose a normalization strategy for miRNA quantitative real-time (qPCR) arrays. J Bioinform Comput Biol. 2011;9:795–812. doi: 10.1142/s0219720011005793. [DOI] [PubMed] [Google Scholar]
- Diedrich M, Mao L, Bernreuther C, Zabel C, Nebrich G, Kleene R, Klose J. Proteome analysis of ventral midbrain in MPTP-treated normal and L1cam transgenic mice. Proteomics. 2008;8:1266–75. doi: 10.1002/pmic.200700754. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–34. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Pertsemlidis A. Cancer and neurodegenerative disorders: pathogenic convergence through microRNA regulation. J Mol Cell Biol. 2011;3:176–80. doi: 10.1093/jmcb/mjq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Elstner M, Morris CM, Heim K, Bender A, Mehta D, Jaros E, Klopstock T, Meitinger T, Turnbull DM, Prokisch H. Expression analysis of dopaminergic neurons in Parkinson’s disease and aging links transcriptional dysregulation of energy metabolism to cell death. Acta Neuropathol. 2011;122:75–86. doi: 10.1007/s00401-011-0828-9. [DOI] [PubMed] [Google Scholar]
- Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6:e18613. doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–80. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829:756–63. doi: 10.1016/j.bbagrm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Gao Z, Fu HJ, Xue JJ, Wu ZX, Zhao LB. Genetic polymorphisms in VDR, ESR1 and ESR2 genes may contribute to susceptibility to Parkinson’s disease: a meta-analysis. Mol Biol Rep. 2014;41:4463–74. doi: 10.1007/s11033-014-3317-0. [DOI] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–41. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentil BJ, Cooper L. Molecular basis of axonal dysfunction and traffic impairments in CMT. Brain Res Bull. 2012;88:444–53. doi: 10.1016/j.brainresbull.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles KM, Brown RA, Epis MR, Kalinowski FC, Leedman PJ. miRNA-7-5p inhibits melanoma cell migration and invasion. Biochem Biophys Res Commun. 2013;430:706–10. doi: 10.1016/j.bbrc.2012.11.086. [DOI] [PubMed] [Google Scholar]
- Gurung B, Muhammad AB, Hua X. Menin is required for optimal processing of the microRNA let-7a. J Biol Chem. 2014;289:9902–8. doi: 10.1074/jbc.M113.520692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GA, Malintan NT, Collins BM, Meunier FA, Sugita S. Munc18-1 as a key regulator of neurosecretion. J Neurochem. 2010;115:1–10. doi: 10.1111/j.1471-4159.2010.06900.x. [DOI] [PubMed] [Google Scholar]
- Havekes R, Canton DA, Park AJ, Huang T, Nie T, Day JP, Guercio LA, Grimes Q, Luczak V, Gelman IH, Baillie GS, Scott JD, Abel T. Gravin orchestrates protein kinase A and beta2-adrenergic receptor signaling critical for synaptic plasticity and memory. J Neurosci. 2012;32:18137–49. doi: 10.1523/JNEUROSCI.3612-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heman-Ackah SM, Hallegger M, Rao MS, Wood MJ. RISC in PD: the impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Front Mol Neurosci. 2013;6:40. doi: 10.3389/fnmol.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM. A neo-substrate that amplifies catalytic activity of parkinson’s-disease-related kinase PINK1. Cell. 2013;154:737–47. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulley P, Schachner M, Lubbert H. L1 neural cell adhesion molecule is a survival factor for fetal dopaminergic neurons. J Neurosci Res. 1998;53:129–34. doi: 10.1002/(SICI)1097-4547(19980715)53:2<129::AID-JNR1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Ishtiaq M, Campos-Melo D, Volkening K, Strong MJ. Analysis of novel NEFL mRNA targeting microRNAs in amyotrophic lateral sclerosis. PLoS One. 2014;9:e85653. doi: 10.1371/journal.pone.0085653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellen LC, Lu L, Wang X, Unger EL, Earley CJ, Allen RP, Williams RW, Jones BC. Iron deficiency alters expression of dopamine-related genes in the ventral midbrain in mice. Neuroscience. 2013;252:13–23. doi: 10.1016/j.neuroscience.2013.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–7. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589:319–25. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilhoff G, Wiegand S, Fansa H. Vav deficiency impedes peripheral nerve regeneration in mice. Restor Neurol Neurosci. 2012;30:463–79. doi: 10.3233/RNN-2012-120230. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee Y, McKenna ND, Yi M, Simunovic F, Wang Y, Kong B, Rooney RJ, Seo H, Stephens RM, Sonntag KC. miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol Aging. 2014a;35:1712–21. doi: 10.1016/j.neurobiolaging.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Noh H, Lee Y, Jeon J, Shanmugavadivu A, McPhie DL, Kim KS, Cohen BM, Seo H, Sonntag KC. MiR-126 Regulates Growth Factor Activities and Vulnerability to Toxic Insult in Neurons. Mol Neurobiol. 2014h Nov 17; doi: 10.1007/s12035-014-8989-x. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauzinska M, Castro NP, Rangel MC, Spike BT, Gray PC, Bertolette D, Cuttitta F, Salomon D. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin Cancer Biol. 2014;29C:51–58. doi: 10.1016/j.semcancer.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–30. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang H, Ai Z, You Z, Wan Y, Guo W, Xiao J, Jin X. Characterization of miRNA218/322-Stxbp1 pathway in the process of insulin secretion. J Mol Endocrinol. 2014 doi: 10.1530/JME-14-0305. [DOI] [PubMed] [Google Scholar]
- Li Y, Shi X. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol Immunol. 2013;10:65–71. doi: 10.1038/cmi.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Wadlington NL, Chen L, Zhuang X, Brorson JR, Kang UJ. Loss of PINK1 attenuates HIF-1alpha induction by preventing 4E-BP1-dependent switch in protein translation under hypoxia. J Neurosci. 2014;34:3079–89. doi: 10.1523/JNEUROSCI.2286-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wei L, Wu F, Hu Z, Liu Z, Yuan W. Advances with microRNAs in Parkinson’s disease research. Drug Des Devel Ther. 2013;7:1103–13. doi: 10.2147/DDDT.S48500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Lawless MW, Gray SG, Kelleher D, McManus R. NF-kappaB regulation: the nuclear response. J Cell Mol Med. 2009;13:631–43. doi: 10.1111/j.1582-4934.2009.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels J, Kepp O, Senovilla L, Lissa D, Castedo M, Kroemer G, Galluzzi L. Functions of BCL-X L at the Interface between Cell Death and Metabolism. Int J Cell Biol. 2013;2013:705294. doi: 10.1155/2013/705294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–78. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–8. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies AT, Jedlitschky G, Konig J, Herold-Mende C, Steiner HH, Schmitt HP, Keppler D. Expression and immunolocalization of the multidrug resistance proteins, MRP1-MRP6 (ABCC1-ABCC6), in human brain. Neuroscience. 2004;129:349–60. doi: 10.1016/j.neuroscience.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Palacios JM, Mengod G, Vilaro MT, Ramm P. Recent trends in receptor analysis techniques and instrumentation. J Chem Neuroanat. 1991;4:343–53. doi: 10.1016/0891-0618(91)90042-b. [DOI] [PubMed] [Google Scholar]
- Palacios N, Weisskopf M, Simon K, Gao X, Schwarzschild M, Ascherio A. Polymorphisms of caffeine metabolism and estrogen receptor genes and risk of Parkinson’s disease in men and women. Parkinsonism Relat Disord. 2010;16:370–5. doi: 10.1016/j.parkreldis.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsanejad M, Zhang Y, Qu D, Irrcher I, Rousseaux MW, Aleyasin H, Kamkar F, Callaghan S, Slack RS, Mak TW, Lee S, Figeys D, Park DS. Regulation of the VHL/HIF-1 pathway by DJ-1. J Neurosci. 2014;34:8043–50. doi: 10.1523/JNEUROSCI.1244-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Altamira E, Polazzi E, Moretto E, Lauriola M, Monti B. The transcription factor CCAAT enhancer-binding protein beta protects rat cerebellar granule neurons from apoptosis through its transcription-activating isoforms. Eur J Neurosci. 2014;39:176–85. doi: 10.1111/ejn.12407. [DOI] [PubMed] [Google Scholar]
- Pierce JP, Mayer T, McCarthy JB. Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol. 2001;11:351–5. doi: 10.1016/s0960-9822(01)00077-x. [DOI] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Macey L, Woo TU, Sonntag KC. Neuronal type-specific gene expression profiling and laser-capture microdissection. Methods Mol Biol. 2011;755:327–43. doi: 10.1007/978-1-61779-163-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppinga WJ, Munoz-Llancao P, Gonzalez-Billault C, Schmidt M. A-kinase anchoring proteins: cAMP compartmentalization in neurodegenerative and obstructive pulmonary diseases. Br J Pharmacol. 2014;171:5603–23. doi: 10.1111/bph.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevedo C, Sauzeau V, Menacho-Marquez M, Castro-Castro A, Bustelo XR. Vav3-deficient mice exhibit a transient delay in cerebellar development. Mol Biol Cell. 2010;21:1125–39. doi: 10.1091/mbc.E09-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit H, Rathi N, Roy D. Construction and analysis of the protein-protein interaction networks based on gene expression profiles of Parkinson’s disease. PLoS One. 2014;9:e103047. doi: 10.1371/journal.pone.0103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Fu K, Mu C, Zhen X, Wang G. L166P mutant DJ-1 promotes cell death by dissociating Bax from mitochondrial Bcl-XL. Mol Neurodegener. 2012;7:40. doi: 10.1186/1750-1326-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Naar AM. MicroRNAs in metabolism and metabolic diseases. Cold Spring Harb Symp Quant Biol. 2011;76:225–33. doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama MM. Huwe1: a possible role in PD. Med Hypotheses. 2012;79:419. doi: 10.1016/j.mehy.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Sang M, Ma L, Sang M, Zhou X, Gao W, Geng C. LIM-domain-only proteins: multifunctional nuclear transcription coregulators that interacts with diverse proteins. Mol Biol Rep. 2014;41:1067–73. doi: 10.1007/s11033-013-2952-1. [DOI] [PubMed] [Google Scholar]
- Sauzeau V, Horta-Junior JA, Riolobos AS, Fernandez G, Sevilla MA, Lopez DE, Montero MJ, Rico B, Bustelo XR. Vav3 is involved in GABAergic axon guidance events important for the proper function of brainstem neurons controlling cardiovascular, respiratory, and renal parameters. Mol Biol Cell. 2010;21:4251–63. doi: 10.1091/mbc.E10-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Jenner P. Etiology and pathogenesis of Parkinson’s disease. Mov Disord. 2011;26:1049–55. doi: 10.1002/mds.23732. [DOI] [PubMed] [Google Scholar]
- Schiesling C, Kieper N, Seidel K, Kruger R. Review: Familial Parkinson’s disease--genetics, clinical phenotype and neuropathology in relation to the common sporadic form of the disease. Neuropathol Appl Neurobiol. 2008;34:255–71. doi: 10.1111/j.1365-2990.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- Seiz EG, Ramos-Gomez M, Courtois ET, Tonnesen J, Kokaia M, Liste Noya I, Martinez-Serrano A. Human midbrain precursors activate the expected developmental genetic program and differentiate long-term to functional A9 dopamine neurons in vitro. Enhancement by Bcl-X(L) Exp Cell Res. 2012;318:2446–59. doi: 10.1016/j.yexcr.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–73. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Shathasivam T, Kislinger T, Gramolini AO. Genes, proteins and complexes: the multifaceted nature of FHL family proteins in diverse tissues. J Cell Mol Med. 2010;14:2702–20. doi: 10.1111/j.1582-4934.2010.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, Andersen SL, Stephens RM, Benes FM, Sonntag KC. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Stephens R, Sonntag KC. Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PLoS One. 2010;5:e8856. doi: 10.1371/journal.pone.0008856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. 2005. pp. 397–420. [Google Scholar]
- Sonntag KC. MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 2010;1338:48–57. doi: 10.1016/j.brainres.2010.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag KC, Woo TU, Krichevsky AM. Converging miRNA functions in diverse brain disorders: a case for miR-124 and miR-126. Exp Neurol. 2012;235:427–35. doi: 10.1016/j.expneurol.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura A, Taguchi K, Watanabe Y, Tatebe H, Tokuda T, Mizuno T, Tanaka M. Lysosomal enzyme cathepsin B enhances the aggregate forming activity of exogenous alpha-synuclein fibrils. Neurobiol Dis. 2014;73C:244–253. doi: 10.1016/j.nbd.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Birks DK, Balakrishnan I, Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD, Foreman NK, Vibhakar R. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem. 2013;288:1918–28. doi: 10.1074/jbc.M112.396762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Yang Y, Jin D, Sun J, Yu X, Yang Z. MiRNA-145 is involved in the development of resistin-induced insulin resistance in HepG2 cells. Biochem Biophys Res Commun. 2014;445:517–23. doi: 10.1016/j.bbrc.2014.02.034. [DOI] [PubMed] [Google Scholar]
- Wirdefeldt K, Adami HO, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson’s disease: a review of the evidence. Eur J Epidemiol. 2011;26(Suppl 1):S1–58. doi: 10.1007/s10654-011-9581-6. [DOI] [PubMed] [Google Scholar]
- Wong G, Nass R. miRNAs and their putative roles in the development and progression of Parkinson’s disease. Front Genet. 2012;3:315. doi: 10.3389/fgene.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y, Fan D. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget. 2014;5:11552–63. doi: 10.18632/oncotarget.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li X, Xie W, Jankovic J, Le W, Pan T. Neuroprotection of deferoxamine on rotenone-induced injury via accumulation of HIF-1 alpha and induction of autophagy in SH-SY5Y cells. Neurochem Int. 2010;57:198–205. doi: 10.1016/j.neuint.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Yang D, Li T, Wang Y, Tang Y, Cui H, Tang Y, Zhang X, Chen D, Shen N, Le W. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci. 2012;125:1673–82. doi: 10.1242/jcs.086421. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu Q, Mao M, Tong Y. Huwe1 as a therapeutic target for neural injury. Genet Mol Res. 2014;13:4320–5. doi: 10.4238/2014.June.9.18. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011a;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011b;17:7105–15. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.