Abstract
Enhancement of the fluorescent output of surface-based fluorescence assays by performing them upon nanostructured photonic crystal (PC) surfaces has been demonstrated to increase signal intensities by >8000×. Using the multiplicative effects of optical resonant coupling to the PC in increasing the electric field intensity experienced by fluorescent labels (“enhanced excitation”) and the spatially biased funneling of fluorophore emissions through coupling to PC resonances (“enhanced extraction”), PC enhanced fluorescence (PCEF) can be adapted to reduce the limits of detection of disease biomarker assays, and to reduce the size and cost of high sensitivity detection instrumentation. In this work, we demonstrate the first silicon-based PCEF detection platform for multiplexed biomarker assay. The sensor in this platform is a silicon-based PC structure, comprised of a SiO2 grating that is overcoated with a thin film of high refractive index TiO2 and is produced in a semiconductor foundry for low cost, uniform, and reproducible manufacturing. The compact detection instrument that completes this platform was designed to efficiently couples fluorescence excitation from a semiconductor laser to the resonant optical modes of the PC, resulting in elevated electric field strength that is highly concentrated within the region <100 nm from the PC surface. This instrument utilizes a cylindrically focused line to scan a microarray in <1 minute. To demonstrate the capabilities of this sensor-detector platform, microspot fluorescent sandwich immunoassays using secondary antibodies labeled with Cy5 for two cancer biomarkers (TNF-α and IL-3) were performed. Biomarkers were detected at concentrations as low as 0.1 pM. In a fluorescent microarray for detection of a breast cancer miRNA biomarker miR-21, the miRNA was detectable at a concentration of 0.6 pM.
Introduction
Circulating blood contains a diverse set of cellular and molecular elements that can be detected and quantified to indicate the presence of cancers, allergies, heart disease, and neurodegenerative disease 1–10. The detection of bloodborne biomarkers has value not only for disease detection, but also for informing the prescription choice for personalized therapies and in the monitoring of these treatments. For example, biomarker levels can help assist the development of novel molecular-targeted therapeutic strategies, identify patients who are likely to benefit from a specific targeted treatment, as well as provide molecular endpoints to predict and monitor treatment efficacy 11–13. Thus, the ultimate goal of such research is to reduce mortality rates through early disease detection and by administering novel targeted drug therapy in conjunction with a multiplexed, sensitive, rapid, and inexpensive biomarker-based monitoring system.
In the search for cancer biomarkers, the quantitative analysis of products of cancer cells, the tumor microenvironment, the host’s response, and the interaction between these three components has yielded several potential candidates. Circulating protein markers are currently in clinical use for the diagnoses of ovarian, pancreatic, colon, and prostate cancers 14–16. Exosomal microRNAs (miRNA), which are 18–24 bases long double stranded noncoding RNA that regulate expression through control over mRNA and protein translation, is another class of biomarker molecules that has been keenly studied as their expression is altered in disease states, notably in cancers 17–19 . A special consideration for detecting cancer biomarkers is that tumors initially develop from a small population of defective cells, and hence it is highly desirable to be able to detect the presence of the smallest number of tumor cells (i.e. early intervention) when a patient’s clinical outcomes and prognosis are still favorable. Although reports of several bioanalytical techniques for cancer biomarker detection exist, an unmet, critical limitation is the reliable and accurate detection of cancer biomarkers mainly due to (a) insufficient sensitivity of the assay and (b) insufficient dynamic range needed to detect biomarkers anywhere from the low ng/mL to the low pg/mL range 20. Furthermore, because single biomarkers often have inadequate predictive value, reliable cancer detection and monitoring platforms typically must include a biomarker panel 11, 14, 21–25. Multiplexed biomarker detection increases the predictive power through statistical and biochemical means to reduce false positive and false negative results 21, 23, 26, 27.
To maximize the applicability and accessibility of a biomarker detection platform, the ability to perform sensing noninvasively is highly desirable, particularly if it can utilize a sample comprised of a drop of blood obtained through a pin-prick. The adoption of biomarker tests will be further accelerated by methods that can be performed with minimal sample preparation and technical expertise, potentially enabling testing to be performed in close proximity to the patient. Such a portable platform would help reduce costs, minimize sample degradation, provide on-spot diagnosis thus alleviating patient stress, and finally guide the course therapy especially when timely adjustments in treatment are critical. Detection instruments used in a point-of-care setting must be inexpensive, compact, and rugged.
With the above design considerations in mind, we present, for the first time, the ability to use a silicon-based photonic crystal (PC) surface to achieve pg/ml-level sensitivity for multiplexed cancer biomarker detection (soluble proteins and miRNA) using photonic crystal enhanced fluorescence (PCEF) to amplify the output of surface-based fluorescent assays. Our efforts focus on two core elements of the detection platform - the PC surface and the detection instrument – and we demonstrate the capabilities of this platoform using miniaturized biological assays. The PC surfaces are designed to provide optical resonances for efficient coupling to the excitation laser and efficient extraction of fluorescence emission on silicon substrates using SiO2 and TiO2 materials selected to provide negligibly low levels of autofluorescence, thus enabling weak fluorescent signals generated by low concentration analytes to be easily observed. Furthermore, the Si-based PC allows the sensor to be inexpensively, uniformly, and reproducibly manufactured in a semiconductor foundry. The detection instrument was specifically designed so that all the light delivered by a miniature solid state laser can be coupled to PC resonant modes by taking advantage of a unique feature of the PC photonic band structure. PC enhancement enables the use of inexpensive components to detect otherwise weak fluorescent signals, resulting in a compact and inexpensive line scanning instrument.
To demonstrate the capabilities of this system, microspot fluorescent immunoassays for two breast cancer biomarkers (TNFα and IL-3) were performed. In order to minimize the usage of capture molecules while simultaneously minimizing the size of the array, capture protein and oligonucleotide spots were printed using dip-pen nanolithography (DPN) and electrohydrodynamic jetting (e-jet) technologies. Using only 10 µl volumes of detection samples, consistent with detection from a droplet of fluid, the biomarkers were detected at concentrations as low as 0.1 pM. Finally, in a fluorescent microarray for detection of a breast cancer miRNA biomarker (miR-21), the miRNA was detectable at a concentration of 0.6 pM. Our long-term goal is to demonstrate a system that can be applied broadly for multiplexed soluble biomarker analysis particularly for diseases in which no accurate imaging modality exists, where imaging would be cost-prohibitive as an initial screen, or for situations in which noninvasive and frequent biomarker monitoring would be beneficial.
Methods
Device design, fabrication, and surface characterization
The photonic crystal enhanced fluorescence (PCEF) surface is a nanostructured grating patterned in a thermally grown, low refractive index silicon oxide (SiO2) layer atop a silicon substrate. The sensor is completed with a conformal top layer of high index titanium oxide (TiO2). This periodic arrangement of the high (TiO2) and low (SiO2) index layers results in the characteristic narrow band resonance peak and local electric field enhancement.
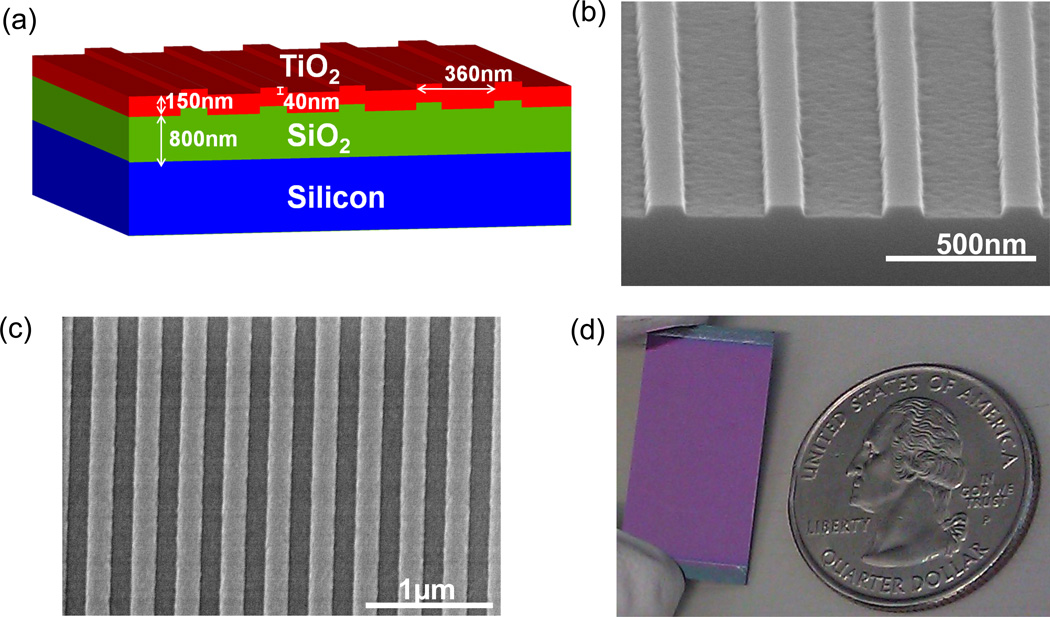
A commercially available simulation tool for rigorous coupled wave analysis (DIFFRACTMOD, RSoft) was used to aid in the optimal design of a PCEF device that provides maximal electric field enhancement for a TM mode at λ=632.8 nm. Simulation results dictated the use of a structure with a period of 360 nm, a duty cycle of 36%, a grating depth of 40 nm, and a TiO2 thickness of 120 nm. A cross-sectional view of the device schematic is presented in Figure 1(a).
Figure 1.
(a) Schematic of the silicon PC device design. (b) SEM cross-sectional view of grating pattern in SiO2 layer before TiO2 coating. Measured grating line width of 131nm and grating depth of 37.7nm. (c) SEM top view of grating after TiO2 coating. Measured grating period of 366 nm.
The first prototype devices were fabricated in a university cleanroom setting using a nano-imprint lithography process that has been reported previously 28. However, a commercial vendor (SVTC Inc., Austin TX) was contracted for the large scale fabrication of these devices over the area of 8” diameter silicon wafers. A thermal oxide (thickness of 800 nm) was grown on each Si wafer and a deep UV photolithography process (193 nm, ArF-line) was used to create the grating pattern in the oxide layer. For the lithography step, a 4X binary reticle (6” × 6” × 0.25 mm, quartz) with a critical dimension tolerance of +/− 0.05µm and uniformity of +/− 0.04 µm was used. Precisely timed reactive ion etching steps were then used to transfer the grating pattern from the resist layer into the SiO2 layer. Scanning electron micrographs showing the surface characteristics of these PCs are presented in Figure 1(b, c). The wafer was diced to produce 1” × 0.5” chips that were then coated with TiO2 (using a PVD sputtering process) to complete the device; a photograph of a completed device is presented in Figure 1(d). Additional images of a completed wafer and its surface characteristics are presented in Supplementary Figure S1.
Device optical characterization
The success of PCEF is critically dependent on the ability of the detection instrument to effectively couple light into a PC. The degree of coupling plays a critical role in the enhancement factor achievable by a PC-instrument combination 29. The process of characterizing a PC involves the measurement of the wavelength spectra associated with the PC. This was done to ensure suitable positioning of the resonance reflection peak for the PC. The angle spectrum of the PC was then measured at a fixed laser wavelength. The percentage of transmitted light at the resonance condition was indicative of the coupling efficiency of the PC with the instrument. For this purpose, an angle-tuned reflection system was developed that allowed for the measurement of the wavelength spectrum. The schematic of this setup is given in the supplemental section, Figure S2. The setup consists of a tungsten halogen lamp (white light), which is coupled to an optical fiber (Ocean Optics Inc.) with a 50 µm core. The output of the fiber is collimated using an achromatic lens. The white light is polarized using a linear polarizer (Thorlabs Inc.) and projected onto the photonic crystal device that is held in a customized holder. The setup design takes advantage of the 1-D PC’s relative insensitivity to angle change in the θ-direction. The PC holder is oriented at a fixed angle of θ = 5°. The detector is placed at distance of 150 mm from the base of the PC holder and tilted up by 2θ = 10°. This orientation allows for the detection of the reflected spectra at normal incidence. The detector height is lower than the height of the illumination beam and is composed of another collimator, which is coupled to a 50 µm core fiber. The other end of the fiber is coupled to a spectrometer (Ocean Optics Inc.). Utilizing commercial software (Spectra suite, Ocean Optics Inc.), we are able to measure the reflected spectrum. In order to measure the reflected wavelength spectra off normal, we mount the PC holder on a rotation stage. This stage is then mounted on a 180 mm custom arm that contains the detector mount at the other end. The arm is then sandwiched by another rotation stage. This is depicted in Supplemental Figure S2. In order to measure the reflected wavelength spectrum at an angle of incidence ϕ, we rotate the top rotation stage by ϕ clockwise and the bottom stage by 2ϕ counterclockwise. This allows the detector to collect the reflected wavelength spectra at every angle. Each spectrum is normalized using a gold reflection mirror.
LD-700 assay for fluorescence enhancement characterization
To characterize the fluorescence enhancement afforded by the silicon PCs, a fluorescent dye, LD-700 (Exciton, Inc., Dayton OH; excitation peak = 647 nm, emission peak = 673 nm) was used as previously reported 30. The dye was mixed with SU-8 which served as the carrier and this solution was spun coated onto the device. SU-8 2000.5 was mixed with an SU-8 thinner (Microchem Corporation) at a ratio of 1:20 (by volume). The stock LD-700 dye solution was diluted in methanol to a concentration of 100nM (53.8 ng/mL). The LD-700 was then mixed with the thinned SU-8 solution in 1:1 ratio (by volume) and the resulting solution was spun coated on the silicon PC and control glass substrates (Spincoater P6700, 5000 rpm, 30 seconds).
Device surface functionalization
The PC devices were functionalized using a vapor-phase epoxysilane process. The epoxysilane chemistry was chosen for its low background fluorescence 31 and high binding capacity to capture antibodies 32. The devices were first cleaned by sonication in 2” petri dishes of acetone, isopropanol, and deionozied (DI) water for 2 minutes each. The devices were then dried in a stream of N2 and then treated in an oxygen plasma system (Diener, Pico) for 10 minutes (power of 100 W, pressure of 0.75 mTorr). The backside of each device was then adhered to the inside of a screw top lid of a 2” glass container. At the base of the container, 100 µL of (3-Glycidoxypropyl) trimethoxysilane (GPTS, Sigma Aldrich, Saint Louis, MO) was placed and the screw top lid was securely placed over the dish. After securely tightening the lids, each dish with a device adhered to its lid was placed in a vacuum oven for an overnight incubation at a temperature of 80 °C and a pressure of 30 Torr. The devices were then detached from the lids and sonicated in 2” petri dishes of toluene, methanol, and DI water for 2 minutes each and dried under a stream of N2. Standard glass microscope slides that served as controls were also silanized using the same protocol but with appropriately sized glassware.
SA-Cy5 microspot assay for fluorescence enhancement characterization
To evaluate fluorescence enhancement, cyanine-5 (Cy5) conjugated streptavidin (GE Healthcare) at a concentration of 100 µg/mL in phosphate buffered saline (PBS) with 60% v/v glycerol was printed onto the substrates for a total of 14 replicate spots with a diameter of 45±6 µm. Protein spots were printed using an electrohydrodynamic jetting (e-jet) system previously introduced here 33, 34 as a non-contact method for high resolution printing. The print head consists of a syringe acting as an ink reservoir that is then connected to an Au-Pd coated (thickness of ~10 nm), glass, luer micropipette nozzle (World Precision Instruments) with an inner diameter of 5µm. Through a combination of capillary force and an appropriately selected back pressure applied to the fluid, a pendant meniscus is formed at the tip of the nozzle. A voltage is then applied between the nozzle tip and substrate to draw the meniscus into a Taylor cone and jetting from this cone creates printed features on the substrate. By fixing the separation distance between the nozzle and the substrate(s) and the applied back pressure, an initial jetting voltage is determined. To control the size and spacing of the printed droplets, a pulsed width modulated signal was used to print high droplet density images by specifying the jetting frequency and duty cycle. A pulsed-voltage mode with a square-wave function was used to create lines of droplets to form arrays for the SA-Cy5 and miRNA experiments. This tool also has the option of being operated in a Computer Numerical Controlled (CNC) mode so intricate images can be converted to a G-Code and run automatically. The Rubin’s vase image presented in this work is printed using this CNC mode with specified input parameters of line spacing and stage speed. After printing, the substrates were incubated overnight in a humid chamber and then rinsed thrice in a 0.05% Tween solution of PBS followed by a final set of three rinses in ultrapure deionizied water. The substrates were then dried under a stream of nitrogen and the fluorescence data was acquired soon thereafter.
Fluorescent sandwich immunoassay
The protein microarrays were produced using a desktop nanofabrication system, NLP2000, based on DPN technology (NanoInk Inc., Skokie, IL, USA). Prior to printing, the tips, DPN Probes type M-ED Side M-2 with 12 A frame cantilevers (NanoInk Inc., Skokie, IL, USA), with a pitch between each pen of 66 µm, were plasma cleaned for 40 second at low RF-value, using a gas mixture of Oxygen/Argon (21% / 79%) at 200 mTorr using a Plasma Cleaner (PDC-32G) (Harrick Plasma, Ithaca, NY, USA). Two type of cytokines interleukin 3 (IL-3) and tumor necrosis factor (TNF-α) (R&D Systems Inc., Minneapolis, MN) capture antibodies were printed on epoxysilane modified PC slides and Nexterion 1” × 3” Slide E (Schott AG, Maintz, Germany) control glass microscope slides. The printing was performed under a controlled environment using an environmental chamber (ambient temperature and 30% relative humidity). The antibodies (~5.0 mg/mL) where diluted in proprietary printing buffer (NanoInk Inc., Skokie, IL, USA); the printing buffer is formulated to keep the protein moisturized and to preserve their active and folded states. Four pens out of 12 pens were used to print two cytokines, a positive control (goat-anti Rabbit IgG) at 2.0 mg/mL and a negative control (Normal Rabbit IgG labeled with Alexa-Fluor-555) at 3.2 mg/mL. Each PC holds 10 subarrays and the glass slides hold 48 subarrays in a 4 × 12 format; each subarray contains 4 sets of 4 replicate spots per antibody for a total of 16 spots. Spot diameters were measured to be 15±3.7µm. The printed substrates were incubated in a sealed box with a desiccant for two days at 4 °C. Next, the slides were placed in a 48-well format slide module assembly (NanoInk Inc., Skokie, IL, USA), where each well could hold up to 12 µL. The arrays were blocked with casein blocking buffer (BioRad, Hercules, CA) for 1h. All incubations were performed at room temperature. The arrays were then washed three times with 0.01% (v/v) tween 20 in PBS (PBST) and then each well was incubated with a 10 µL mixture of different antigens concentration in casein buffer for three hours. This was followed by three rinses in PBST after which the glass slides was incubated in a bulk dish with 2 ml mixture of 1 µg/mL biotinylated detection antibodies while the PC substrates were incubated with only 10 µL of the biotinylated detection antibody mixture in each well of the slide module assembly for 1 hour. The PC substrates and glass slides were then washed three times with PBST, followed by the incubation with a 1 µg/mL solution of Alexa-Fluor-647 conjugated streptavidin (Invitrogen) for 30 minutes. Finally, the devices were washed 5 times with PBST and followed by a quick dip in DI water (3 sec.) to remove the salt, spin dried, and then scanned. Antigen standard curves were generated by using a 3-fold dilution scheme for a total of 7 concentrations. The starting concentrations for IL-3 and TNFα were 16.6 ng/mL and 2 ng/mL, respectively.
Fluorescent miRNA detection
In this experiment the miR-21 probe-target sequence was assayed in a microspot format. The capture oligonucleotide sequence (5’-TCA-ACA-TCA-GTC-TGA-TAA-GCT-A-3’, purchased from IDT DNA Technologies, Coralville, IA) was modified to have a 6-carbon chain amine modification at the 5’ end. This probe sequence was printed at a concentration of 50uM in a printing buffer of autoclaved, Milli-Q water (resistivity of 18.2 MΩ.cm−1) with 80% v/v of glycerol (Sigma-aldrich, Saint Louis, MO). A polydimethylsiloxane-based (PDMS; Sylguard 184, Dow Corning, Midland, MI) 8-well mold was used to create isolated, 4mm diameter wells on the printed substrates. The mold was prepared from a ~1 mm thick film of cured PDMS, rinsed with IPA and DI water, dried under N2 and firmly placed over the substrate. Through Van-der-Waal’s forces, the mold remained adhered to the substrate for the course of the experiment.
In each well, two rows of 8 spots were created for a total of 16 replicate spots per well. Printing was performed in ambient temperature and humidity conditions using the e-jet tool. Upon printing, the substrates were placed in a petri dish with a moist kimwipe at its base to keep the petri dish humid. The dish was sealed with parafilm and incubated overnight. The substrates were then rinsed in a wash buffer of DI water with 0.2% v/v of sodium dodecyl sulfate (SDS; purchas Sigma Aldrich, Saint Louis, MO), followed by two additional rinses in DI water, and then dried under a stream of N2. The target miRNA sequence (5’-UAG-CUU-AUC-AGA-CUG-AUG-UUG-A-3’; IDT DNA Technologies, Coralville, IA) was labeled with Cyanine-5 at the 5’ end. Dilutions of the target sequence was prepared in a buffer of 5X saline sodium citrate buffer (SSC, containing 75 mM sodium citrate and 750 mM sodium chloride) containing 10% v/v of formamide and 0.1% v/v of SDS (all reagents were molecular biology grade and were purchased from Sigma Aldrich, Saint Louis, MO). Seven concentrations of the target miRNA sequence were assayed over a concentration range of 10 nM – 0.6 pM; data was obtained from six replicate spots per concentration. For the hybridization step, the substrate was placed in a sealable, rubber chamber and 10 µL of each miRNA dilution was added to a unique well on the substrate. The chamber was sealed and left overnight in a water bath at a temperature of 42 °C. After the incubation period, all wells were aspirated, the PDMS mold was detached, and the substrates were rinsed in the following three buffers - (1X SSC containing 0.2% v/v of SDS), (0.2X SSC containing 0.2% v/v of SDS), and finally (0.1X SSC). The substrates were dried under a stream of N2 and imaged immediately thereafter.
Confocal laser scanning
Control glass substrates were scanned with a commercially available confocal laser microarray scanner (Tecan LS Reloaded). This scanner was fitted with a λ=632.8 nm, 5 mW laser for Cy5 excitation and a Cy5 emission filter (bandpass, λ=670–715 nm). The incident light was TM polarized and made incident on the substrates at an angle of 0°. Scans were obtained at a pixel resolution of 4um and the photomultiplier tube (PMT) gain was adjusted such that the largest fluorescence intensities did not saturate the PMT.
Angle-tuned, objective-coupled fluorescence line scanner
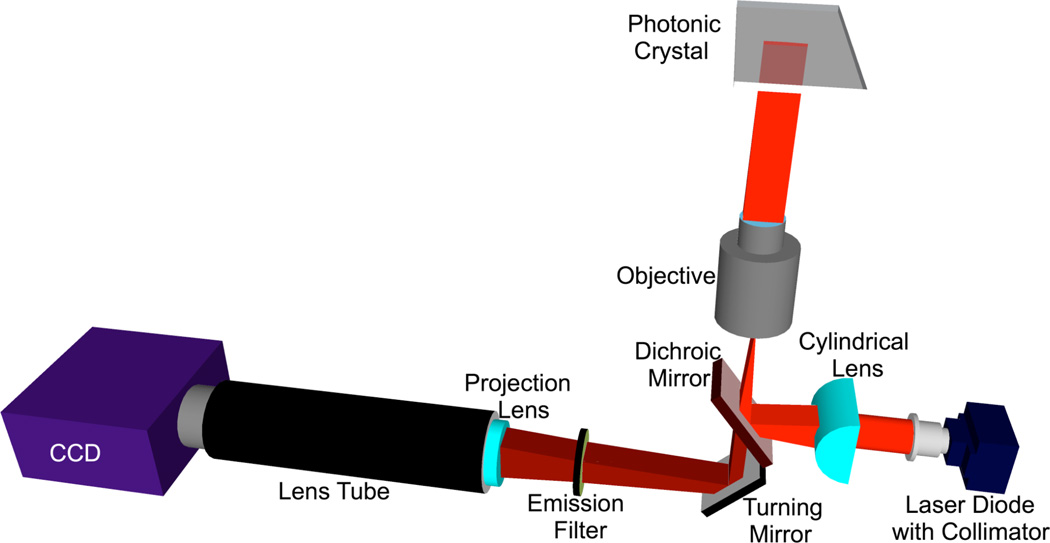
The schematic diagram of the objective-coupled line scanning (OCLS) instrument is illustrated in the Figure 2. The illumination of this system consists of a 70 mw solid-state laser (AlGaAs) at λ=637 nm, coupled to a polarization maintaining fiber, a half wave plate, a cylindrical lens, a long pass dichroic mirror, and a 10× objective (Olympus Plan N) of focal length 18 mm. The fiber tip is coupled to a fiber collimator giving a highly collimated output beam 3.4 mm in diameter. The output beam is then passed through a half-wave plate, which is used to rotate the polarization of the output beam to match with the PC-mode to be excited. The laser beam is then focused to a line by the cylindrical lens (f = 100 mm). The focused laser line is directed onto the back focal plane of the microscope objective via a dichroic mirror. The output of the objective is a 6 µm × 1mm line that is focused along the direction of the PC grating while remaining collimated in the direction perpendicular to the grating. The PC is placed on a motorized sample stage (MS2000, Applied Scientific Instruments) that is translated perpendicular to the laser line for a fast scan (750 lines/second). The fluorescence image is constructed by sequential scanning across the sensor in fixed increments. The PC, placed at the focal plane of the infinity-corrected 10× objective (f0 = 18 mm), interacts with a beam that is collimated in one plane but focused in the orthogonal plane. The assembly of the cylindrical lens, half-wave plate and fiber collimator is mounted on a two-dimensional motion stage. The stage is manually adjustable in one plane and automated in the other. The manual adjustment is utilized to fine-tune the focus of the beam onto the back focal plane of the objective lens. In order to achieve angle tuning, the line-focused beam is translated on the back focal plane of the objective, by tuning the position of the cylindrical lens-wave plate fiber collimator assembly. This fine stepping is achieved by utilizing a motorized linear stepping stage (Zaber LSM-25). The result is a change in the incident angle in the ϕ direction; here the focal length used would be that of the objective lens. The emitted fluorescence signal is collected by the objective and projected onto a CCD camera (Hamamatsu 9100C) by a tube lens (f=150 mm). A bandpass fluorescence filter is inserted between the objective and tube lens to block the excitation laser beam.
Figure 2.
Schematic of the objective-coupled, line scanning instrument used to acquire fluorescence data at the precise PC resonant angle. Equipped with a solid-state laser diode, this instrument illuminates with the PC with a beam of light that is focused in one plane for higher illumination density but collimated in the other plane for optimally coupling the incident light to the PC.
Image construction
A fluorescent image of the PC surface is obtained by adjustments of the incident angle of the illumination line upon a region of the PC adjacent to the microarray, and then translating the PC holding stage in increments of 2 µm past the assay region, gathering a fluorescent intensity image of the line for each motion increment. Using a custom-built C# based user interface, the assembly of the fluorescent image of each line into a two-dimensional image of fluorescent intensity is performed. The software provides a synchronous integration of the various components of the OCLS. It can be used to both capture the angle spectrum of a PC as well as perform fluorescence measurements. We utilized the information about the beam shape and size in order to set up an acquisition scheme. The user interface requires inputs of start and stop positions for defining a x,y scan range and an angle scan range. The fixed x,y scan step size of 2 µm is also the effective width of an individual pixel on the CCD; the width of the beam is measured to be 6 µm. Thus we oversample our images by a factor of 3 in the scan direction. The oversampling is done so that only the peak intensity at each pixel is used to generate the image. If we were to utilize a step size equal to the focus beam width, we would encounter a variability of ±25% in the scan direction as opposed to a variability ±6%. The tradeoff here is a 3× slower scan speed due to the shorter step size. The motivation for the new image-processing algorithm came as we recognized that by only selecting the peak intensity value, we effectively discard valuable data from the lower intensity pixels adjacent to the peak. The image processing algorithm works on the assumption that, as we step through a 6 µm region, we are illuminating the region with a different intensity beam. If we aggregate the fluorescence emitted for all three frames, our final intensity value for that pixel should be over 2× without increasing the laser power or integration time. Thus if we do need to oversample in steps to achieve the lower variability in the scan direction, we can utilize an algorithm designed to enhance the integration time as well. This algorithm is depicted in the supplemental Figure S3. Position 2 indicates the peak value that we would normally utilize and the processed peak indicates the aggregated intensity value we obtain. Owing to this processing algorithm we further noticed that we lowered the variability in the scan direction by 3×, due to the flat-top effect for a Gaussian profile. Theoretically, a Gaussian convolved with a Gaussian gives a flattop beam. This algorithm results in an optical convolution that allows reduction of variability and hence noise. For the arrays used in this work, a single scan is performed in 60 seconds.
Image analysis and quantification
Spot segmentation and intensity calculations of the constructed fluorescence images were performed using either ImageJ or Genepix Pro 6.1 (Molecular Devices). Net spot intensity was calculated as the local background subtracted spot intensity where the local background is an annular region around a given spot. Spot signal-to-noise ratio (SNR) was calculated as the local background subtracted spot intensity divided by the standard deviation of the local background.
Results and Discussion
Device optical characteristics
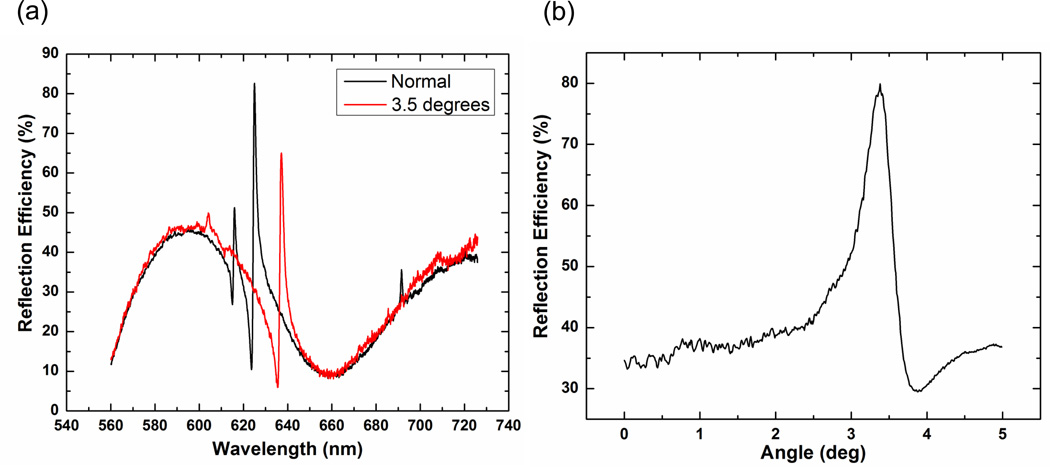
The broadband optical response of the silicon PC (Si-PC) as measured from a finished device is presented in Figure 3(a). Here the reflected spectra for a PC both at normal and at 3.5 degrees (resonance peak at the Cyanine-5 excitation wavelength of λ=637 nm) is presented; the peak has a full width at half maximum (FWHM) of 2 nm. The broad features observed in the reflection spectrum are a result of the thin film interference of the SiO2 and TiO2 thin films, while the high efficiency, narrow reflection peak results from the presence of the PC, and indicates the wavelength at which PC resonance is established. The optical response of a Si-PC over a range of illumination angles and a fixed illumination wavelength (λ=637 nm, in this case) was also obtained to characterize the device angular profile and accurately identify the device resonant angle (see Figure 3(b)) for a fixed wavelength.
Figure 3.
(a) Reflection spectra of a Si-PC illuminated with a broadband light source and captured at normal incidence (black) and at an incidence angle of 3.5 degrees (red). At an incidence angle of 3.5 degrees, the resonant peak is located at a wavelength of λ=637 nm. (b) Reflection spectra of a Si PC obtained when illuminated with a collimated solid state laser (at λ=637 nm) over a range of illumination angles. All data is normalized to the reflection from a gold mirror.
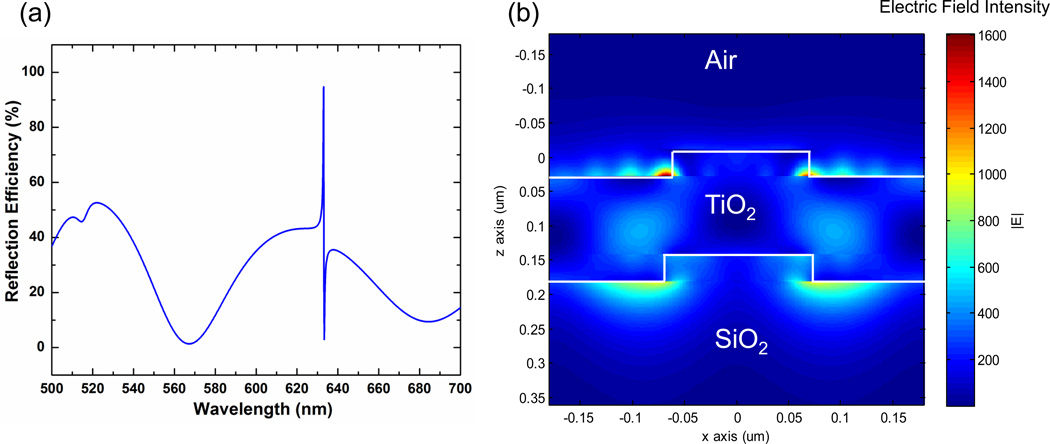
Finite Difference Time Domain (FDTD) computer simulations were used to aid in the design of the Si-PC structure to predict the resonant spectrum expected from such a device. FDTD also enables visualization of the electric field distribution on the PC surface at the resonant coupling condition. As shown in Figure 4, excellent agreement was observed between the simulation and measured broadband optical responses in key parameters such as the spectral location of the peak, the peak width, and its reflection efficiency. The model can be used to estimate the potentially available excitation enhancement provided by a PC structure where the incident excitation light is collimated and matched to the PC resonant coupling condition. We report a maximum electric field enhancement of 1767 times the incident electric field and an average electric field enhancement of 401 times and 135 times the incident electric field for 10 nm and 100 nm tall regions extending above the TiO2 layer into the device superstrate (air, in this case). Our design goal is to provide a narrow high reflection efficiency peak, as the potential enhancement is inversely proportional to the bandwidth of the resonance, as shown in previous work 28.
Figure 4.
(a) Reflection efficiency as a function of wavelength for the simulated Si-PC. When illuminated at normal incidence, the device resonance is located at 633nm. (b) Electric field intensity cross section plotted for one period of the device. The maximum field intensity is 1767 times the incident field intensity. When averaged over a 10 nm and 100 nm tall region above the top TiO2 layer, the electric field intensity is 401 times and 135 times the incident field intensity, respectively.
Silicon-based PCs provide substantial advantages compared to previously reported PCEF surfaces prepared on plastic or quartz substrates. First, these devices can be fabricated on a wafer scale with semiconductor process technology and is thus amenable to inexpensive, high volume manufacturing. Second, the SiO2 and TiO2 materials of the PC have negligibly low levels of autofluorescence, thus enabling weak fluorescent signals generated by low concentration analytes to be more easily observed. As demonstrated in this work, Si-PCs provide narrow bandwidth optical resonances, which have been shown to generate the greatest fluorescence enhancement factors when the excitation illumination matches the resonance wavelength and coupling angle.
Fluorescence enhancement characterization
A critical aspect of the Si-PC detection platform is the design of the detection instrument used for fluorescence excitation, and imaging of fluorescence emission. The goal of the detection instrument is to illuminate the PC at the exact wavelength/angle combination that satisfies the resonant condition. Because light focused to a point contains a wide range of incident angles, only a small fraction of the incident light will be resonant with the PC, and thus the enhancement effect provided by point-focused light is not capable of achieving the maximum available enhancement effect 29. One approach to overcome this problem is to illuminate a broad area of the PC with collimated light, which unfortunately results in substantially reduced excitation intensity compared to a focused beam 35. We take advantage of the unique optical properties of the linear PC grating structure, for which the resonant coupling condition only need be satisfied for incident angles oriented perpendicular to the grating lines. As described previously 36 a cylindrical lens may be used to provide light that is focused in the plane parallel to the grating, but completely collimated in the plane perpendicular to the grating, thus simultaneously achieving nearly 100% resonant coupling to the PC, and a high intensity focused line of illumination, with a line width of 6 µm. Initial bulk fluorescence enhancement measurements on the Si-PC as compared to an unpatterned glass control substrate were performed by spin coating an LD-800 (dye) doped layer of SU-8 (photoresist) onto cleaned devices. Fluorescence data on all substrates was acquired using the OCLS instrument at a fixed laser power of 3 mW. CCD exposure time was adjusted to maximize fluorescence output on each substrate and gathered measurements were later normalized based on the exposure setting to allow for comparisons between substrates. Fluorescence signal enhancement is defined as the ratio of the system dark noise subtracted maximal fluorescence intensity of the PC to a control glass substrate. When scanned on-resonance, a 96.7× factor in fluorescence enhancement was observed on the Si-PC as compared to its off-resonance condition, representing the gain supplied by the enhanced excitation effect.
Next, we characterized the device fluorescence enhancement performance in the context of a simple microarray assay where microspots of streptavidin labeled with Cy5 were deposited on a silane-functionalized PC. Such an experiment provides a measure of net fluorescence signal intensity enhancement as well as another important parameter of SNR enhancement. Enhancements in the SNR are especially meaningful as they more readily translate to lowering the limits of detection for a biological specimen because such gains indicate an enhancement in the fluorescence intensity that is greater than any associated increases in the background fluorescence intensity. An average fluorescence signal enhancement of 113.5× and an average SNR enhancement of 10.3× was observed when the Si PC resonance was excited.
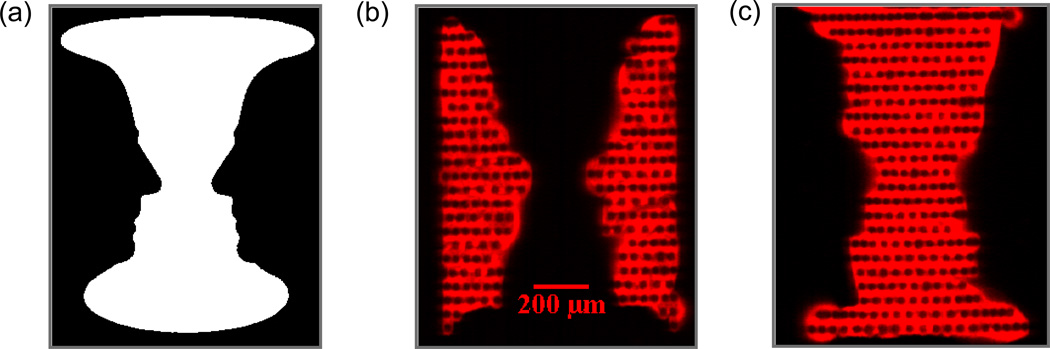
By utilizing the high resolution printing capability of the e-jet platform and the capability to acquire fluorescence data at precisely tuned angles of incidence with the OCLS system, the printing of an optical illusion known as Rubin’s Vase (Figure 5a) was performed on a Si-PC. This set of reversing figures consists of a face and a vase in the same image. We printed the two complementary figures using two different inks – Cy5-labeled streptavidin (purchased from GE Healthcare; printing concentration of 100 µg/mL, diluted in PBS with glycerol added at 80% v/v) and a Cy5-labeled oligonucleotide (purchased from Integrated DNA Technologies; sequence 5’-AT TTC CGC TGG TCG TCT GCA-3’, six-carbon amine modification on the 5’ end and Cy5 label on 3’ end; printing concentration of 1nM, diluted in sterile MilliQ water with glycerol added at 80% v/v). The e-jet deposited spots had a diameter of ~40–50 µm. Drying of the droplets after deposition resulted in surface adsorption of dye-labeled biomolecules around the perimeters of adjacent droplets, resulting in doughnut-shaped fluorescent spot morphology. Printing of spots into a dense array, thus results in a “chain link” appearance to the resulting image, where the regions between applied droplets are bright, and fluorophore is not observed in the droplet centroid. Adsorption of surface layers on the PC surface will shift the resonant coupling angle to greater values, proportional to the density of the adsorbed layer 35, 37. Because the protein-printed region has a greater surface density than the oligonucleotide-printed region, its resonant coupling angle is ~ 0.1 degrees higher. By tuning to each resonance angle, one for the oligonucleotide printed section and the other for the protein printed section, the face or vase components of this image can be observed without exciting the complementary component. (Figure 5b,c). This experiment demonstrates the importance of illuminating the PC so as to excite the resonance, as regions of the PC that are illuminated at an angle that does not match the resonance will experience substantially lower surface-bound excitation intensity, and will appear dark next to regions that are illuminated on-resonance. This experiment also serves to demonstrate the ability of the OCLS system to obtain uniform, high resolution images of fluorescent intensity.
Figure 5.
(a) The optical illusion, Rubin’s Vase, shown here as a gray-scale image consists of two reversing figures of a face and a vase. (b–c) Fluorescent images of the two reversing figures obtained by tuning to two distinct PC resonances. The two figures were printed with a Cy5 labeled oligonucleotide and a Cy5 labeled protein, respectively and each figure produced a separate shift in the PC resonance.
Fluorescent immunoassay and miRNA microarray
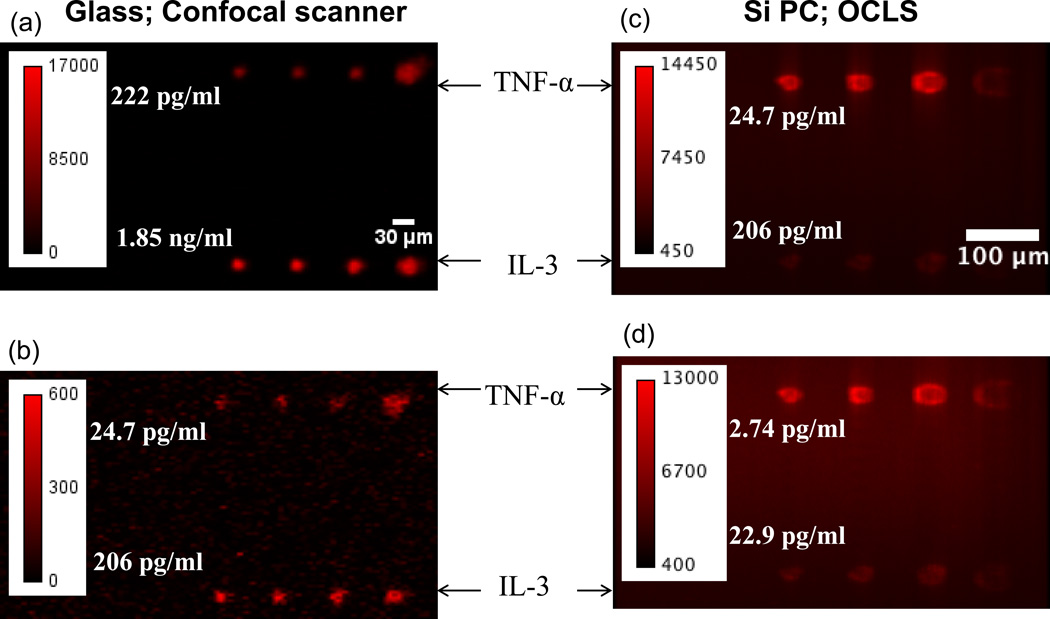
The performance of the Si-PC measured with the OCLS detection system was next studied in the context of a microspot-based fluorescent sandwich immunoassay. Si-PCs paired with glass control slides were partitioned into 8-sectors; in each sector four replicate microspots each of capture antibodies for IL-3 and TNF-α were printed using a dip-pen nanolithography system 38, 39. A mix of IL-3 and TNF-α was assayed in these sectors over a range of 7 concentrations in a 3-fold dilution series with a starting concentration of 17 ng/mL for IL-3 and 2.0 ng/mL for TNF-α. Fluorescence images for the Si PC were obtained with the OCLS and that for the glass control slide were obtained using a commercially available confocal microarray scanner (Tecan-LS, laser wavelength of 632.8 nm). It is worth noting that the power density available to a unit area for a glass slide scanned with the microarray scanner and a PC scanned with the OCLS are comparable. Figure 6 presents representative fluorescence images of microspots on the PC and glass surfaces at two sets of assayed concentrations.
Figure 6.
Representative fluorescence images of two subarrays on the glass slide (a–b) acquired using a commercial confocal microarray scanner. Two assayed concentrations of (a) 0.22 ng/mL for TNF-α and 1.9 ng/mL for IL-3 and (b) 25 pg/mL for TNF-α and 0.21 ng/mL for IL-3 are presented here. Fluorescent images of two subarrays on the Si PC (c-d) acquired using the OCLS at the following concentrations (a) 25 pg/mL for TNF-α and 0.21 ng/mL for IL-3 and (b) 2.7 pg/mL for TNF-α and 23 pg/mL for IL-3.
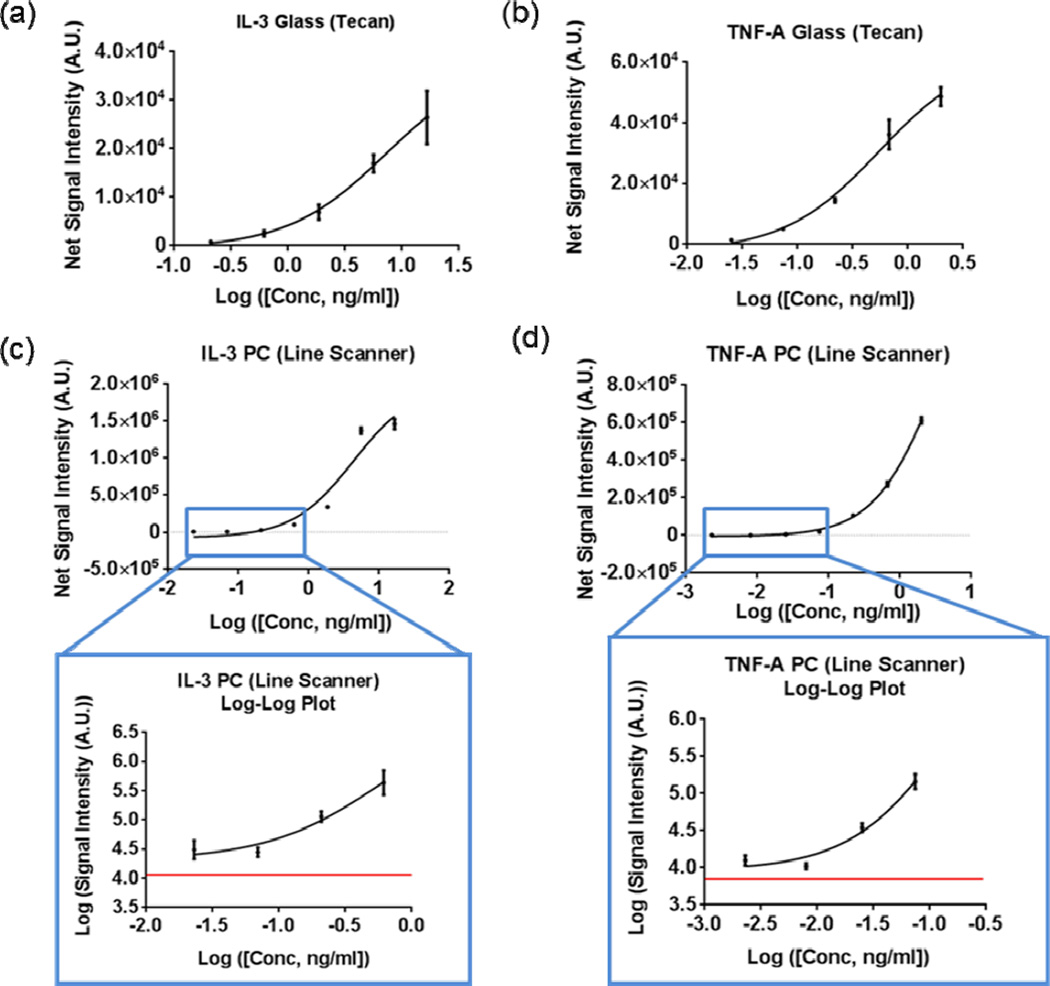
The dose-dependent response of each antigen assayed is presented in Figure 7. The lowest concentrations of TNF-α and IL-3 detected on the glass surface were 25 pg/mL and 0.21 ng/mL, respectively. In comparison, all seven assayed concentrations were detectable on the PC with the lowest concentrations being 2.7 pg/mL and 23 pg/mL for TNF-α and IL-3 with replicate averaged SNRs of 24.6 and 45.4, respectively. In Figures 7c–d, the red line represents the fluorescent intensity value measured in the regions directly adjacent to the microarray spots, thus establishing the local fluorescent background level. Negative controls performed by exposure of capture antibodies to a buffer-only sample resulted in no observable fluorescence signal above this background.
Figure 7.
Dose response curves obtained on the glass slide for (a) IL-3 and (b) TNF-α. The five highest concentrations out of a total of seven assayed concentrations were detectable on the glass slide. All seven assayed concentrations of (c) IL-3 and (d) TNF-α were detectable on the PC. Fluorescent spot intensities on the PC for the four lowest assayed concentrations, as highlighted in the blue rectangle and magnified, are well above the local background value (red line). Data represents mean ± S.D. values of four replicate spots.
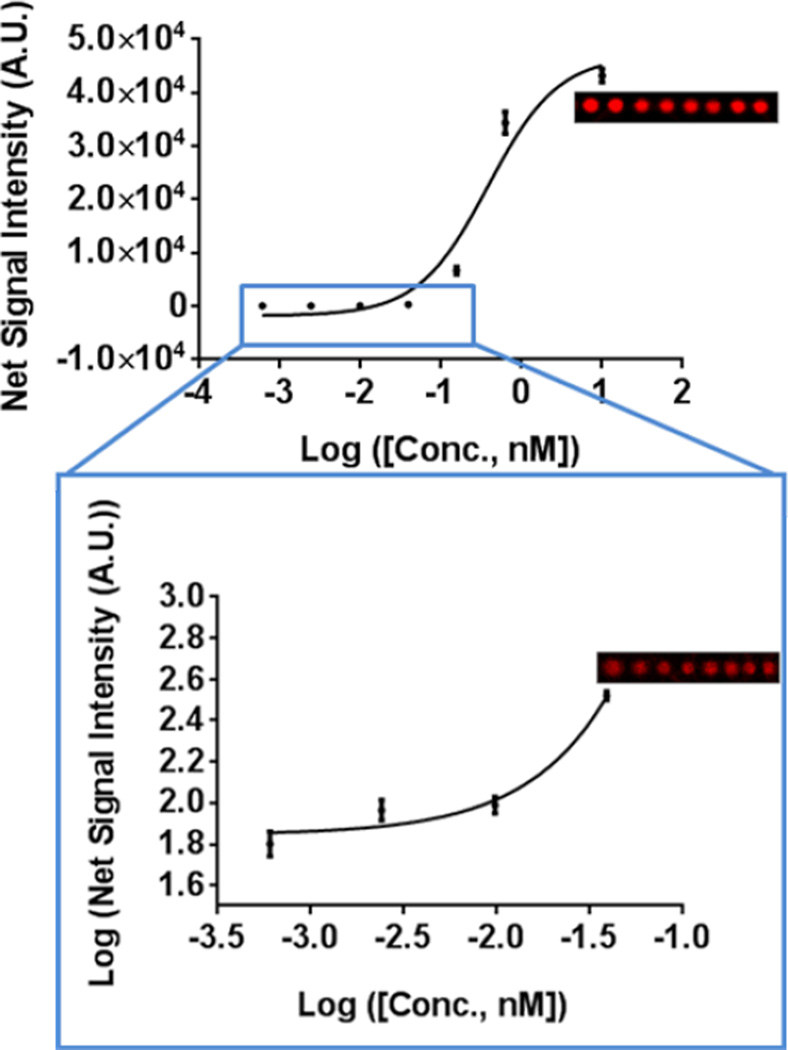
Finally, we characterized the performance of a miRNA microspot assay on the Si PC where we chose to assay miR-21, a miRNA sequence implicated in the progression of breast cancer 40. The miR-21 target miRNA sequence was assayed on the PC at the following seven concentrations – 10 nM, 2.5 nM, 0.16 pM, 39 pM, 9.8 pM, 2.4 pM, and 0.60pM. All seven concentrations yielded detectable signals with the lowest concentration of 0.60 pM having a replicate averaged spot SNR of 3.8. The dose response of the assayed set of concentrations is presented in Figure 8.
Figure 8.
Dose response curve depicting fluorescent spot intensities of miR-21 assayed on the PC over a concentration range of 1.0nM – 0.60pM. All seven assayed concentrations were detectable over the background fluorescence signal with the four lowest assayed concentrations highlighted in the blue rectangle and magnified. The local background is indicated as the red line. Representative fluorescence images of microspots at assayed miR-21 concentrations of 2.5nM and 39pM are presented alongside each graph. Data represents mean ± S.D. values of sixteen replicate spots.
While several promising optical 41, 42 and electronic label-free techniques 43–45 have been reported for biomarker detection, their development into point-of-care diagnostic tools are still in their infancy. Antibody arrays based on the traditional ELISA method remains as one of the most popular multiplexed platforms used for cancer biomarker analysis 23. The introduction of the protein ELISA microarray proved crucial for integrating antibodies to a panel of biomakers in a single array that is approximately 1 cm2 in size. Additionally, the protein ELISA microarray has improved sensitivity compared to a large number of traditional microwell plate ELISAs. Bead-based technologies, best exemplified by Luminex xMAP (Luminex Corp, Austin, TX), uses the basic "sandwich" assay format, but the capture antibody is coated onto the surface of a polystyrene bead rather than in a microwell plate. These beads are processed through the assay and separated for analysis via flow cytometry as they are spectrally unique and color coded into different sets that can be differentiated by the Luminex analyzer. The assay is capable of multiplex detection and has been commercially successful for a variety of in vitro diagnostics assays but has some critical limitations. The Luminex technology requires an expensive non-automated detection instrument, provides limited multiplexing capability, and lengthier assay protocols than surface-based fluorescent microarrays. These combined limitations make the Luminex platform less desirable for point-of-care diagnostics. Sandwich antibody microarrays on the other hand have been reported to optimally use 3 to 5 µl of sample to detect analytes at concentrations < 10 pg/ml, lack assay cross-reactivity, demonstrate the fastest binding kinetics, and provide lower limits of detection than bead-based assays due to the ability to wash away unbound material. Using a PC surface to enhance the fluorescence output from a biomarker microarray, it is our ultimate goal to automate this platform to enable even more rapid, high sensitivity detection for in vitro diagnostics. This platform will enable a more compact, inexpensive and practical instrument than current alternative approaches.
Conclusion
In this work we have presented a miniaturized and inexpensive platform for the detection of soluble biomarkers. For the first time, PCs have been fabricated on a silicon substrate, using materials (SiO2 and TiO2) with very low levels of autofluorescence. By choosing such a device design, the detection of weak fluorescent signals generated at lower analyte concentrations can be more easily observed. These devices have been successfully designed and fabricated inexpensively at a semiconductor foundry. An objective-coupled line scanning detection instrument designed to efficiently couple incident light to PC resonant modes also enabling the detection of otherwise weak fluorescent signals is presented.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (GM086382A) the National Science Foundation (CBET 07-54122) and the Linda Su-Nan Chang Sah doctoral fellowship (awarded to SG). Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation. The authors thank the staff at the Micro and Nanotechnology Laboratory and colleagues from the Nano Sensors Group at the University of Illinois at Urbana-Champaign for their suggestions and helpful discussions.
Footnotes
Conflict of Interest Statement
BTC has founded a company with the goal of commercializing the PCEF technology. NA is employed by NanoInk, which commercially provides the DPN system used in this work.
Supplementary Information
† Electronic Supplementary Information (ESI) available: [Please see Supplemental_Information.pdf]. See DOI: 10.1039/b000000x/
References
- 1.Basil CF, Zhao Y, Zavaglia K, Jin P, Panelli MC, Voiculescu S, Mandruzzato S, Lee HM, Seliger B, Freedman RS. Cancer research. 2006;66:2953–2961. doi: 10.1158/0008-5472.CAN-05-3433. [DOI] [PubMed] [Google Scholar]
- 2.Zangar RC, Daly DS, White AM. Expert review of proteomics. 2006;3:37–44. doi: 10.1586/14789450.3.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava S. Molecular diagnosis & therapy. 2006;10:221–230. doi: 10.1007/BF03256460. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. The American journal of pathology. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masson S, Latini R, Anand IS, Vago T, Angelici L, Barlera S, Missov ED, Clerico A, Tognoni G, Cohn JN. Clinical chemistry. 2006;52:1528–1538. doi: 10.1373/clinchem.2006.069575. [DOI] [PubMed] [Google Scholar]
- 6.Felker GM, Petersen JW, Mark DB. Canadian Medical Association Journal. 2006;175:611–617. doi: 10.1503/cmaj.060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keating CD. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2263–2264. doi: 10.1073/pnas.0500024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hueber W, Robinson WH. Proteomics. 2006;6:4100–4105. doi: 10.1002/pmic.200600017. [DOI] [PubMed] [Google Scholar]
- 10.El-Agnaf OMA, Salem SA, Paleologou KE, Curran MD, Gibson MJ, Schlossmacher MG, Allsop D. The FASEB journal. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- 11.Hanash SM, Pitteri SJ, Faca VM. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 12.Weinstein IB, Joe AK. Nature Clinical Practice Oncology. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 13.Tourneau C, Le Faivre S, Siu LL. European journal of cancer (Oxford, England: 1990) 2007;43:2457. doi: 10.1016/j.ejca.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Kulasingam V, Diamandis EP. Nature Clinical Practice Oncology. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 15.Lilja H, Ulmert D, Vickers AJ. Nature Reviews Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 16.Smith RA, Cokkinides V, Brawley OW. CA: A Cancer Journal for Clinicians. 2008;58:161–179. doi: 10.3322/CA.2007.0017. [DOI] [PubMed] [Google Scholar]
- 17.Fleischhacker M, Schmidt B. Cancer Biomarkers. 2009;6:211–219. doi: 10.3233/CBM-2009-0131. [DOI] [PubMed] [Google Scholar]
- 18.Croce CM, Calin GA. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Cortez MA, Calin GA. 2009 [Google Scholar]
- 20.Rusling JF, Kumar CV, Gutkind JS, Patel V. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert MPA, Korc M, Malfertheiner P, Röcken C. Journal of proteome research. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
- 22.Weston AD, Hood L. Journal of proteome research. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 23.Tothill IE. Seminars in cell & developmental biology. Elsevier; 2009. pp. 55–62. [Google Scholar]
- 24.Stevens E, Liotta L, Kohn E. International Journal of Gynecological Cancer. 2003;13:133–139. doi: 10.1111/j.1525-1438.2003.13358.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson DS, Nock S. Angewandte Chemie International Edition. 2003;42:494–500. doi: 10.1002/anie.200390150. [DOI] [PubMed] [Google Scholar]
- 26.WAGNER PD, VERMA M, SRIVASTAVA S. Annals of the New York Academy of Sciences. 2006;1022:9–16. doi: 10.1196/annals.1318.003. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Clinical chemistry. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 28.Pokhriyal A, Lu M, Chaudhery V, Huang CS, Schulz S, Cunningham BT. Optics express. 2010;18:24793–24808. doi: 10.1364/OE.18.024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhery V, Lu M, Pokhriyal A, Schulz SC, Cunningham BT. Journal Sensors IEEE. 2012;12:1272–1279. [Google Scholar]
- 30.Estrada L, Martinez O, Brunstein M, Bouchoule S, Le-Gratiet L, Talneau A, Sagnes I, Monnier P, Levenson J, Yacomotti A. Optics express. 2010;18:3693–3699. doi: 10.1364/OE.18.003693. [DOI] [PubMed] [Google Scholar]
- 31.Dorvel B, Reddy B, Block I, Mathias P, Clare SE, Cunningham B, Bergstrom DE, Bashir R. Advanced Functional Materials. 2009;20:87–95. [Google Scholar]
- 32.Zhu H, Klemic JF, Chang S, Bertone P, Casamayor A, Klemic KG, Smith D, Gerstein M, Reed MA, Snyder M. Nature genetics. 2000;26:283–290. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- 33.Park JU, Hardy M, Kang SJ, Barton K, Adair K, kishore Mukhopadhyay D, Lee CY, Strano MS, Alleyne AG, Georgiadis JG. Nature materials. 2007;6:782–789. doi: 10.1038/nmat1974. [DOI] [PubMed] [Google Scholar]
- 34.Park JU, Lee JH, Paik U, Lu Y, Rogers JA. Nano letters. 2008;8:4210–4216. doi: 10.1021/nl801832v. [DOI] [PubMed] [Google Scholar]
- 35.Block ID, Mathias PC, Ganesh N, Jones SI, Dorvel BR, Chaudhery V, Vodkin LO, Bashir R, Cunningham BT. Optics express. 2009;17:13222–13235. doi: 10.1364/oe.17.013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhery V, Lu M, Huang C-S, Polans J, Tan R, Zangar RC, Cunningham BT. Optics Letters. 2012;37:2565–2567. doi: 10.1364/OL.37.002565. [DOI] [PubMed] [Google Scholar]
- 37.George S, Block I, Jones S, Mathias P, Chaudhery V, Vuttipittayamongkol P, Wu H, Vodkin L, Cunningham B. Analytical Chemistry-Columbus. 2010;82:8551. doi: 10.1021/ac101551c. [DOI] [PubMed] [Google Scholar]
- 38.Hong S, Zhu J, Mirkin CA. Science. 1999;286:523–525. doi: 10.1126/science.286.5439.523. [DOI] [PubMed] [Google Scholar]
- 39.Salaita K, Wang Y, Mirkin CA. Nature Nanotechnology. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 40.Si M, Zhu S, Wu H, Lu Z, Wu F, Mo Y. Oncogene. 2006;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 41.Washburn AL, Gunn LC, Bailey RC. ACS Publications; 2009. pp. 9499–9506. [Google Scholar]
- 42.Gohring JT, Dale PS, Fan X. Elsevier; pp. 226–230. [Google Scholar]
- 43.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nature Publishing Group; 2005. pp. 1294–1301. [DOI] [PubMed] [Google Scholar]
- 44.Dorvel BR, Reddy B, Jr, Go J, Duarte Guevara C, Salm E, Alam MA, Bashir R. ACS Publications; pp. 6150–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malhotra R, Patel V, Vaqué JP, Gutkind JS, Rusling JF. ACS Publications; pp. 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.