Abstract
DNA damage in chromatin comes in many forms, including single base lesions that induce base excision repair (BER). We (and others) have shown that the structural location of DNA lesions within nucleosomes greatly influences their accessibility to repair enzymes. Indeed, a difference in the location of uracil as small as one-half turn of the DNA backbone on the histone surface can result in a 10-fold difference in the time course of its removal in vitro. In addition, the cell has evolved several interdependent processes capable of enhancing the accessibility of excision repair enzymes to DNA lesions in nucleosomes, including post-translational modification of histones, ATP-dependent chromatin remodeling and interchange of histone variants in nucleosomes. In this review, we focus on different factors that affect accessibility of BER enzymes to nucleosomal DNA.
Keywords: chromatin, nucleosome dynamics, rotational setting, glycosylase, AP endonuclease, Polymerase β, ligase, histone acetylation, histone variants, chromatin remodeling
Introduction
Eukaryotic DNA is organized in nuclei into a hierarchical scheme of folding and compaction of protein-DNA assemblies forming an intricate and dynamic structure called chromatin. The primary level of organization that regulates DNA accessibility in chromatin is the nucleosome core particle (NCP), consisting of approximately 147 bp of DNA wrapped in 1.65 turns around an octamer of histone proteins (1). This octameric complex is composed of the well-conserved core histones: histones H3-H4, which form a tetramer, flanked by two heterodimers of histones H2A and H2B (1). Positioning of the histone octamers along the genome impacts the ability of regulatory proteins to bind DNA (2). Histone-DNA interactions in the NCP are mainly electrostatic and include 14 contacts where the minor groove interacts primarily with arginine residues at regular ~10 bp intervals on each side of a pseudo-two-fold axis of symmetry termed the dyad (3).
In the central 60 bps these interactions are mediated by the H3-H4 tetramer and have been shown to be stronger than those made by the H2A-H2B dimers (4), creating intrinsic differences in susceptibility to structural disruptions within the NCP. The overall stability of the NCP can be regulated in a number of ways, including exchange of canonical histones with histone variants (5), DNA sequence (6) and the activity of ATP-dependent remodeling complexes (7,8). Posttranslational modifications of the core histones also play a key role in regulating changes in chromatin structure during transcription, replication, and DNA repair (9). In this review, we will present current knowledge on how these factors that are responsible for the dynamic state of chromatin influence accessibility to base excision repair enzymes.
1. Base Excision Repair Overview
BER plays an indispensable role in the faithful maintenance of genomic information. It repairs oxidized, alkylated and deaminated bases, single strand breaks, and AP sites that result from spontaneous or chemically induced hydrolysis (or even as a result of DNA glycosylase activities) (10,11). This pathway, in addition to its broad specificity, must be exquisitely sensitive with high substrate specificity, as most chemically modified bases only exhibit small structural changes in the DNA helix. Removal of lesions by BER may be accomplished via short-patch (SP) BER, where only a single nucleotide is excised, or long-patch (LP) BER, which excises 2-13 nucleotides [for recent reviews see (12,13)].
BER is a coordinated, stepwise process. In the first step, a specific DNA glycosylase searches for the lesion through a phenomenon known as “facilitated diffusion”. This three-dimensional (3D) search is followed by a one-dimensional (1D) scanning in which the glycosylase slides along the DNA backbone (14). Recognition of the lesion is facilitated by a combination of increased base breathing in the mispair and an induced-fit mechanism in the glycosylase’s active site where specific hydrogen bonds must be made before excision (15-18). Removal of the damaged base generates different types of products depending on whether the glycosylase is monofunctional or bifunctional (10,19). Monofunctional glycosylases, such as the E. coli and human uracil DNA glycosylases (UDGs), remove the base by cleaving the N-C1’ glycosydic bond between the target base and the sugar. The subsequent abasic site generated is processed by an apurinic/apyrimidinic (AP) endonuclease (APE1), which cleaves the DNA backbone 5′ to the abasic site, generating a 3′hydroxyl (OH) and a 5′-deoxyribose-5-phosphate (5′-dRp) (19).
Bifunctional glycosylases (e.g., OGG1) remove the damaged base in a similar mechanism as monofunctional glycosylases, but because they possess intrinsic AP lyase activity, they subsequently cleave the DNA backbone (20). However, this cleaved substrate is not suitable for polymerase activity, and must be processed by the endonuclease APE1 or polynucleotide kinase/phosphatase (PNKP) in mammals to generate the 3′OH (20,21). In both subpathways, the 3′OH is used to fill the gap through template-directed synthesis by Pol β in SP-BER, or by one of several polymerases [Pol δ, Pol ε, and Pol β (22)] in LP-BER. LP-BER requires the assistance of additional scaffold proteins to stimulate polymerase binding and activity (23). After DNA synthesis, Pol β also removes the 5′dRp, generating a nick that is sealed by DNA ligase I, or the DNA ligase III/XRCC1 complex (19). Pol δ and Pol ε do not possess lyase activity and require the assistance of flap endonuclease I (FEN I) to cleave the 5′- flap structure, which is followed by restoration of the phosphodiester backbone through ligation as above (24).
It is important to note that failure to complete BER can be more detrimental to the cell than the initial base modifications themselves as the BER intermediates impede replication and can signal apoptosis (25). AP sites are generated in mammalian cells at a rate of ~10,000/cell/day due to depurination alone (26), and as they are a cytotoxic intermediates of BER, a common feature of most DNA glycosylases is tight binding to AP sites (10). In fact, studies from reconstituted BER reactions in vitro have shown that enzymes in this pathway function via a handoff mechanism (25,27). This mechanism is not only beneficial in protecting the cell from deleterious effects of BER abortive intermediates but also in aiding substrate recognition (25).
2. Factors Influencing DNA Accessibility in Chromatin at the Nucleosome Level
The eukaryotic cell possesses a number of interdependent mechanisms capable of changing chromatin compaction to modulate DNA accessibility. To gain a better understanding of how these activities influence excision repair, we first address how they shape the chromatin landscape at the nucleosome level. This is followed by a discussion on their involvement during base excision repair within individual nucleosomes.
2.1. Role of DNA Sequence on Nucleosome Positioning and Stability
Determination of nucleosome positioning in vivo was inspired by the findings showing different DNA sequences containing either AA/TT/TA dinucleotides spaced every 10 bp, or the GGGCCC motif, are intrinsically bent (6). Sequences with such motifs might be preferred as the bending of DNA around the histone octamer is energetically favored (6). Genomic DNA sequence can also affect DNA accessibility by promoting specific positioning of nucleosomes and generating nucleosomes with a range of stabilities (6). Segal et al. mapped nucleosome positions in the genome of Saccharomyces cerevisiae and developed a probabilistic model that indicates sequence preferences for AA/TT/TA dinucleotides that oscillate in phase with each other [and out of phase (5 base pairs) with GC dinucleotides] for nucleosome localization (28). Similar results supporting this finding had previously been found by Satchwell et al. using nucleosomal DNA from chicken erythrocyte core particles (29). Though such isolated nucleosomes can be positioned in vivo by factors other than DNA sequence, a study performed by Kaplan et al. (30) verified the contribution of DNA sequence in directing nucleosome placement by measuring the genome-wide occupancy of histone octamers from chicken erythrocytes assembled onto purified yeast genomic DNA. Overall these studies highlighted that DNA sequence preferences of nucleosomes are important determinants of nucleosome organization in vivo (31), and, as discussed below, such sequences effectively dictate the range of repair protein accessibility throughout the genome.
In addition to directing genomic nucleosome placement, DNA sequence is also a strong determinant of nucleosome stability (6). This concept of nucleosome stability and DNA sequence has been exploited for the generation of synthetic DNA sequences with a strong positioning power (i.e. the Widom 601 positioning sequence) that bind to the histone octamer with high affinity. Use of these DNA sequences allows the in vitro production of homogenous populations of nucleosome core particles that maintain a single rotational and translational position (6). Rotational setting refers to the orientation of the DNA backbone and/or minor groove with respect to the histone octamer surface, where “in” rotational orientations are toward the histone octamer and “out” regions are more solvent exposed. On the other hand, translational setting is the location along the DNA relative to the NCP’s dyad center of symmetry.
Natural DNA sequences exhibit a vast breadth of nucleosome positioning strength, with affinities for histone octamer binding differing by as much as 5000-fold (32). Analysis of 35 NCP crystal structures suggests that DNA sequence can have a dramatic effect on the DNA duplex structure, inducing changes in minor groove width, which in turn affects the rotational setting, shifting the DNA residues in contact with the histones by as many as three base pairs (33). These changes have pronounced effects on accessibility to DNA lesions by BER enzymes given that their activity on nucleosomal DNA is greatly dependent on the rotational setting of the lesion (see below).
Several groups have shown that DNA sequence plays an important role in nucleosome dynamics and accessibility. For instance, it was shown that NCPs containing the 5S rDNA are more readily destabilized by salt and dilution compared to the 601 NCPs (34). Similarly, it was shown that differences in NCP stability occur in DNA positioning sequences around promoter regions of Mouse Mammary Tumor Virus, where the nucleosome at the promoter is inherently more dynamic/unstable compared to another nucleosome nearby, possibly for targeting transcriptional activators (35). Additionally, DNA sequence plays an important role in regulating transient dynamic states of DNA on the surface of the histone octamer in which the DNA transiently lifts off the surface of the histone octamer to allow regulatory proteins access to buried target sites (36-38). Although these ‘site exposure’ or ‘DNA unwrapping’ events were originally considered to occur only in vitro, in vivo results also point to the existence of such a mechanism. Bucceri et al. observed very rapid repair (time scale of seconds) of UV lesions by photolyase in nucleosomal regions, which can only be explained by DNA unwrapping events (39). Similarly, another study demonstrated that as many as ~40 base pairs may partially unwrap, allowing binding of transcription factors in yeast (40).
2.2. Interplay Between Histone Acetylation and ATP-dependent Chromatin Remodeling
The role of histone posttranslational modifications in shaping chromatin structure has been studied extensively as it relates to transcriptional regulation. Given the large number of modifications and the complexity of their combinatorial effects, this review will only focus on addressing the role of histone acetylation as it pertains to nucleosome structure and dynamics. Details on specific histone modifications and their effects on chromatin structure are covered elsewhere (41). Histone acetylation is regulated by the opposing action of histone acetyl transferases (HATs), which bind Acetyl-CoA and transfer acetyl groups to the ε-amino group of lysines, and histone deacetylases (HDACs) that remove those acetyl groups. Because histone-DNA interactions are largely electrostatic and acetylation neutralizes the charge of lysines, histone acetylation at residues located in the globular domain of histones can have the direct local effect of weakening these interactions. Importantly, the structural location of histone acetylation has been shown to be a strong determinant of nucleosome dynamics.
In general, acetylation of lysine residues within the globular domain of histone H3 impact nucleosome structural stability, with acetylation near the DNA entry/exit point regulating NCP unwrapping dynamics, and lysine acetylation near the central dyad primarily promoting NCP disassembly (42,43). Acetylation of lysine 56 on histone H3 (H3K56Ac) has been shown to increase unwrapping dynamics within one turn of the DNA ends (44,45). Lysine acetylation is also capable of regulating NCP stability and formation by changing histone-histone interactions. For example, acetylation at H4K91 disrupts octamer formation, as this residue is important in facilitating dimer/tetramer interactions via a salt bridge with glutamic acid residue in histone H2B (46). On the other hand, histone acetylation in the N-terminal tails, whose association with local DNA is more salt-sensitive (47), primarily influences chromatin structure indirectly by recruiting bromodomain-containing factors that recognize site-specific acetylated lysines. For example, the Rsc4 subunit of the ATP-dependent chromatin remodeler RSC (Remodels the Structure of Chromatin) contains tandem bromodomains (TBD) that specifically bind acetylated lysine 14 of histone H3 (H3K14Ac), located in the N-terminal tail of H3 (48,49). It was also reported that acetylation at H3K14 is important for DNA damage checkpoint activation by directly regulating chromatin structure through the activity of RSC in fission yeast (50). Indeed, hyperacetylation at H3K14 and/or H3K9 is induced following ultraviolet (UV) irradiation in yeast (51). At the nucleosome level, H3K14Ac is an essential mark for histone chaperone-mediated histone octamer eviction from promoter DNA, which is necessary for transcriptional activation (52).
2.3. Exchange of Canonical Histones with Specialized Histone Variants
In addition to the combinatorial effects of histone modifications, core histone themselves can be replaced by histone variants in a replication independent process mediated by chaperones (53). H2A.Bbd has been shown to destabilize the histone octamer and NCPs containing this variant exhibit a relaxed conformation compared to native NCPs (54,55). Because of the differences in histone-DNA interactions and the order of histone deposition along the DNA, the histone H3-H4 tetramer dominates positioning (56); however, this might not always be the case with nucleosome core particles entailing histone variants. Positioning of a histone octamer containing the histone variants H2A.Z and H3.3, when compared to the positioning of either variant associated with the other canonical histones, revealed that positioning was dominated by H2A.Z (57). The crystal structure of the nucleosome core particle containing H2A.Z revealed significant changes in the histone fold that lead to subtle destabilization of interactions between the (H2A.Z-H2B) dimer and (H3-H4)2 tetramer, enlarging the ‘acidic patch’ in histone H2A (58). Although the overall architecture of the NCP is not significantly affected (58), the stability of the histone octamer is likely changed, impacting accessibility to nucleosomal DNA.
3. Factors Affecting Nucleosome Accessibility To BER
In the context of excision repair, only a few of the factors that affect lesion accessibility in nucleosomes have been studied. We and others have reported that translational and rotational positioning of DNA lesions in NCPs influence the activity of different BER enzymes, primarily driven by enzyme-specific structural requirements to act on the lesion. The first step of BER, the detection of chemically modified bases by DNA glycosylases, entails a combination of non-specific 3D searching and 1D tracking along the sugar-phosphate backbone (59,60). Importantly, glycosylase interaction with a target lesion in nucleosomes requires transient DNA dissociation from the histone surface at regions where the DNA backbone faces the histones, whereas there is a higher probability of gaining access to outwardly-oriented lesions (61). This is supported by findings that uracils and oxidative DNA lesions exhibit preferential repair when facing away from the histone octamer (62-68). Because the DNA also exhibits rotational flexibility at normal temperatures (e.g., 37°C), inwardly oriented lesions are not completely refractory to repair, even near the dyad where there is little to no DNA accessibility contribution due to DNA unwrapping. Near the DNA ends, where DNA unwrapping plays an important role on DNA accessibility, the effect of lesion rotational orientation on glycosylase activity is diminished (63,66,68), and inwardly oriented DNA lesions are, in general, removed more efficiently by DNA glycosylases. Interestingly, Ye et al. also observed a correlation with minor groove width and the ability of the human DNA UDG glycosylase (UNG) to remove the uracil irrespective of rotational orientation at some sites (69). Given UNG’s mechanistic properties for catalysis (70,71), it is plausible that a narrow minor groove presents a steric impediment for insertion of a key leucine residue (Leu 272) required to prevent deoxyuridine from returning to the base stack (69). This supports the notion that enzyme-specific structural requirements (i.e. enzyme-induced distortion on their substrates) are key parameters for repair of nucleosomal DNA. Indeed, the apurinic/apyrimidinic endonuclease1 (APE1), which kinks the DNA only slightly more than UDG (25), exhibits similar impairment by the histone octamer during the repair of rotationally-positioned abasic sites and its analog, tetrahydrofuran (72). Additionally, AP sites, which are highly reactive and can bind with neighboring nucleobases and/or lysines, within NCPs near the dyad have been shown to crosslink preferentially with the N-terminal tail of histone H4 (73), and protein-DNA crosslinks at AP sites are dependent on their structural location and proximity to the histone N-terminal tails (74). Therefore, the local structure of an AP site in NCPs determines not only its probability of removal by APE1 but also its propensity to form protein-DNA crosslinks, underscoring the importance for its efficient repair in the context of chromatin.
In repair of the gapped BER intermediate, Nilsen et al. (75) found only partial inhibition of Pol β by the histone octamer, which was also dependent on the translational position of the DNA gap, in agreement with our recent findings (68). Although these results deviate from those of Beard et al. and Menoni et al. (54,62), who observed little to no Pol β activity on DNA gaps in any helical orientation near the dyad, the actual experiments are not directly comparable due to different DNA sequences used and the translational positioning of the gaps. Pol β induces a significant degree of distortion in the template strand, opposite the DNA gap (~90°), which may not be as easily accommodated near the dyad due to stronger histone-DNA interactions, and this effect may be DNA-sequence dependent. In fact, Odell et al. showed that, in general, greater enzyme concentration was required to process DNA lesions, including DNA gaps, in 601-NCPs compared to 5S-NCPs (65). The effect of rotational setting of the DNA gap on Pol β activity is similar to that observed for DNA glycosylases and APE1 when the DNA gap is located near the DNA ends (65,68). However, we recently showed that this is not the case near the dyad, where accessibility and bendability of the templating base for a gap-out substrate more significantly inhibits Pol β activity compared to an inwardly-oriented gap, where the templating strand is not as constrained for this high degree of distortion (68).
Unlike the first three steps of BER, which can be performed in the context of NCPs, the final ligation step of repair by DNA liagaseIII-XRCC1 is inhibited by the presence of histones, likely due to its requirement to completely encircle its substrate (65). Thus, ligation of nicked DNA intermediates, independent of rotational orientation, require disruption of histone-DNA contacts. As DNA-histone disruption is achieved more readily in the 5S-NCP compared to the 601-NCP (34), it appears that DNA sequence plays a key role for the ligation step.
The N-terminal histone tails themselves likely have direct and indirect influence on BER in nucleosomes. The N-terminal tails of histones might assist in orienting or fixing the DNA flap generated during LP-BER, supporting increased recognition and binding by FEN1 (76). In addition, histone tails influence translational positioning of nucleosomal DNA (77,78), though this effect is variable depending on DNA sequence (78). Consequently, while the presence of N-terminal tails interfere with DNA ligase I activity at DNA nicks near the dyad in the presence of linker DNA (79), removal of the histone tails in the context of a different DNA positioning sequence has no effect on the activity of UDG, APE1 and Pol β compared to intact NCPs (80).
Studies elucidating the roles of nucleosome positioning strength, NCP stability, and lesion position within nucleosomes on BER enzyme activities have provided a wealth of information for understanding BER in the context of chromatin. However, new studies are investigating the likely role for ATP-dependent chromatin remodeling in regulation of BER. It is clear that remodelers can play a role in providing enzyme access to target sequences within nucleosomes. Hara et al. (81) demonstrated with restriction enzyme activity assays that ySWI/SNF stimulates accessibility to DNA within the nucleosome core and has only a marginal effect on restriction sites near the ends of the DNA-histone interaction region. Indeed, in the context of DNA repair, ySWI/SNF stimulates removal of acetylaminofluorene-guanine adducts near the dyad axis by nucleotide excision repair in NCPs (81). These results are consistent with studies demonstrating that the strongest interactions between the SWI/SNF complex and nucleosomal DNA (using the 601 sequence) occur about two helical turns from the dyad axis, where the catalytic subunit SWI2/SNF2 interacts with DNA (82). Interestingly, this interaction was not strongly localized at specific regions in crosslinking studies performed with the weaker positioned 5S rDNA sequence (83), and it was proposed that the variability in translational positioning prevented exact positioning of the SWI/SNF complex (82).
Menoni and colleagues used the 601 sequence containing a single 8-oxoG 10 bp from the dyad axis to reconstitute NCPs with either ‘conventional’ or variant H2A.Bbd-containing histone octamers (54). It was found that the presence of ySWI/SNF stimulated BER of 8-oxoG in the conventional histone octamer but not in the variant-containing nucleosome (54), which exhibits a more open structure (as discussed previously). Nakanishi et al. confirmed that inhibition of Pol β by histones is alleviated by the use of ATP-dependent remodeling complexes, yISWI1 and yISWI2, (84). This stimulation by ATP-dependent remodelers might occur more strongly for damage located near the dyad as some ATP-dependent remodeling factors have been shown to stimulate accessibility more significantly at those sites (81). Indeed, we have found that Pol β is inhibited more strongly at APE1-nicked sites (or gaps) located closer to the dyad axis using a modified 601 sequence (68). It would be important to test how Pol β activity is affected in the presence of the ATP-dependent chromatin remodeling complex SWI/SNF as a function of translational position, as it is quite possible that this stronger inhibition of Pol β close to the dyad can be alleviated by its preferential binding and remodeling at those sites.
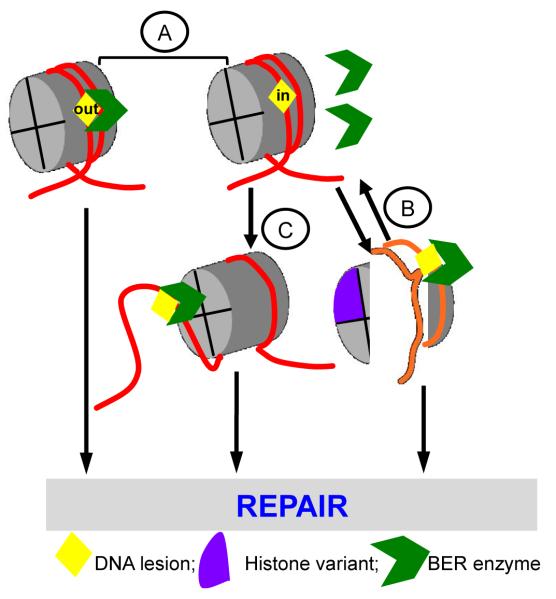
Taken together these findings illustrate how the inherently dynamic structure of the NCP alone is capable of regulating BER. This regulation is dependent on NCP composition (DNA sequence and histones) as well as enzyme structure and mode of action. Figure 1 illustrates that, in general, the repair of most lesions handled by BER is driven by their rotational orientation (A), where accessibility to inwardly-oriented lesions is aided by intrinsic nucleosome dynamics and stability (B) and (possibly) the action of ATP-dependent chromatin remodelers (C).
Figure 1.
Schematic illustration of the various modalities that influence DNA accessibility to BER enzymes in NCPs: (A) Rotational orientation, (B) Intrinsic nucleosome dynamics and stability, and (C) ATP-dependent chromatin remodeling.
BER enzyme structure and mode of binding to DNA strongly dictates their ability to recognize and remove DNA lesions in NCPs. DNA glycosylases and APE1 interact primarily with one strand of the DNA to remove their respective DNA lesions. Thus, their activity is significantly dependent on the rotational orientation of the DNA lesion (Figure 1A), and intrinsic nucleosome dynamics may play a major role in the repair of DNA gaps near DNA ends (Figure 1B). DNA glycosylase, APE1, and Pol β activity on lesions located near the central (dyad) region of NCPs, require the additional action of ATP-dependent chromatin remodeling (Figure 1C). Ligation of nicks in the DNA backbone by DNA ligases is, however, more independent of their rotational orientation, but still requires disruption of the NCP (65); whereas, upstream enzymes can function, albeit with reduced activity relative to free DNA, with minimal disruption to the NCP structure. Because these parameters that regulate accessibility to nucleosomal DNA are not mutually exclusive, the overall repair efficiency of a given lesion can be determined by the variable contribution of each parameter.
4. Future Directions in DNA Repair in Chromatin
Given that DNA damage and repair has been a major focus of anticancer therapy, in vitro studies with NCPs will assist in revealing the fundamental mechanisms that regulate chromatin structure during DNA repair for future clinical applications. Recently, these findings have become the foundation of innovative research on improved methodologies for cancer treatment taking into account different levels of chromatin structure. For example, histone deacetylase inhibitors (or HDACis), vorinostat and romidepsin, which are FDA approved, have shown promising results in the treatment of refractory cutaneous T-cell lymphomas (85). Despite these promising results, most HDACis lack specificity and their usefulness as single agents is yet to be determined. In the future, studies addressing the role of site-specific histone acetylation on nucleosome structure and DNA repair are needed to dissect the possible mechanism(s) of action of HDACis. This can also aid in the design of HDACis with greater target specificity, as little is known about the effect of site-specific histone acetylation in the DNA damage response.
Additionally, Poly (ADP-ribose) polymerase 1 (PARP-1) is a chromatin architectural protein that has been shown to be clinically relevant for the treatment of cancer. Although PARP-1 activity to protect single strand breaks and recruit BER enzymes is well established, recent findings point to a complex multifaceted molecular mechanism. PARP-1 exhibits alternative modes of binding to free DNA and nucleosomes with a preference for nucleosomes containing two linker DNA extensions (86). Moreover, PARP-1 forms covalent attachments to AP site intermediates in cells during alkylating agent-induced BER, and this is potentiated in the presence of a PARP-1 inhibitor (4-AN) (87). These findings are biologically and clinically significant given that AP sites and AP-incised sites near the dyad are not repaired efficiently, by APE1 and Pol β respectively, and thus are expected to persist in the absence of chromatin modifiers. However, the ability of PARP-1 to form these crosslinks in NCPs is yet to be determined. Studies addressing PARP-1 binding and PARylation activity in NCPs containing rotationally and translationally positioned DNA lesions, and how this influences BER, would be an important first step in delineating the molecular mechanisms involved in PARP-1 inhibition and predicting the outcome of single and/or combinatorial therapeutic approaches.
Understanding how repair is regulated by chromatin has also been improved by induction of particular types of lesions. Several groups have recognized the importance of determining the structural location of nucleosomal DNA damage induced by platinating agents and its influence on nucleosome structure and dynamics (88,89). Such investigations should assist in the development of improved drugs for targeting regions of DNA in chromatin that are less accessible for repair, which can limit the efficacy of these drugs in cancer cells.
Another area expected to gain momentum is the effect of histone variants and ATP-dependent remodeling complexes on BER in chromatin. Understanding the role for these chromatin ‘structural modulators’ in repair efficiency could provide a number of additional therapeutic targets and potential drug combinations for enhanced therapeutic efficacy and specificity. A better understanding of the fundamental process of DNA repair in chromatin is essential for establishing the platform for the translational bridging of such basic research with the clinic.
Finally, improving our understanding of the mechanism(s) of DNA damage and repair of environmentally relevant agents in chromatin will bring us closer to a clearer understanding of these processes in vivo. This improved understanding will help provide a more rational basis for regulatory policies associated with environmental exposure to DNA damaging agents.
Acknowledgements
This work was supported by NIH grants ES004106 and ES002614 (to MJS), and ES020955 (to JMH) from the National Institute of Environmental Health Sciences (NIEHS). Contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
Abbreviations
- BER
Base Excision Repair
- NCP
Nucleosome Core Particle
- SP-BER
Short-patch Base Excision Repair
- LP-BER
Long-patch Base Excision Repair
- UDG
Uracil DNA Glycosylase
- AP
apurinic/apyrimidinic
- APE
AP Endonuclease
- PNKP
polynucleotide kinase/phosphatase
- FEN I
flap endonuclease I
- HATs
histone acetyl transferases
- HDACs
histone deacetylases
- RSC
Remodels the Structure of Chromatin
- TBD
tandem bromodomains
- UV
ultraviolet
- HDACis
histone deacetylase inhibitors
- PARP-1
Poly (ADP-ribose) polymerase 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West SM, Rohs R, Mann RS, Honig B. Electrostatic interactions between arginines and the minor groove in the nucleosome. J Biomol Struct Dyn. 2010;27:861–866. doi: 10.1080/07391102.2010.10508587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes JJ, Clark DJ, Wolffe AP. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausio J. Histone variants--the structure behind the function. Brief Funct Genomic Proteomic. 2006;5:228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- 6.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 7.Liu N, Hayes JJ. When push comes to shove: SWI/SNF uses a nucleosome to get rid of a nucleosome. Mol Cell. 2010;38:484–486. doi: 10.1016/j.molcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson CL. Chromatin remodeling: nucleosomes bulging at the seams. Curr Biol. 2002;12:R245–247. doi: 10.1016/s0960-9822(02)00782-0. [DOI] [PubMed] [Google Scholar]
- 9.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews. Molecular cell biology. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 10.Baute J, Depicker A. Base excision repair and its role in maintaining genome stability. Crit Rev Biochem Mol Biol. 2008;43:239–276. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- 11.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krokan HE, Bjoras M. Base excision repair. Cold Spring Harbor perspectives in biology. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zharkov DO, Mechetin GV, Nevinsky GA. Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat Res. 2010;685:11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci U S A. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Royen ME, Zotter A, Ibrahim SM, Geverts B, Houtsmuller AB. Nuclear proteins: finding and binding target sites in chromatin. Chromosome Res. 2011;19:83–98. doi: 10.1007/s10577-010-9172-5. [DOI] [PubMed] [Google Scholar]
- 19.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 22.Asagoshi K, Liu Y, Masaoka A, Lan L, Prasad R, Horton JK, Brown AR, Wang XH, Bdour HM, Sobol RW, Taylor JS, Yasui A, Wilson SH. DNA polymerase beta-dependent long patch base excision repair in living cells. DNA Repair (Amst) 2010;9:109–119. doi: 10.1016/j.dnarep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen WK, Kelley MR. Review of mammalian DNA repair and translational implications. J Pharmacol Exp Ther. 2000;295:1–9. [PubMed] [Google Scholar]
- 24.Nilsen H, Krokan HE. Base excision repair in a network of defence and tolerance. Carcinogenesis. 2001;22:987–998. doi: 10.1093/carcin/22.7.987. [DOI] [PubMed] [Google Scholar]
- 25.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 26.Nickoloff JA, Hoekstra MF. DNA Damage and Repair: Volume III: Advances from Phage to Humans. 1 ed Humana Press; 2001. [Google Scholar]
- 27.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan N, Moore I, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Hughes TR, Lieb JD, Widom J, Segal E. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol. 2010;17:918–920. doi: 10.1038/nsmb0810-918. author reply 920-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu F, Olson WK. DNA architecture, deformability, and nucleosome positioning. J Biomol Struct Dyn. 2010;27:725–739. doi: 10.1080/073911010010524943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gansen A, Toth K, Schwarz N, Langowski J. Structural variability of nucleosomes detected by single-pair Forster resonance energy transfer: histone acetylation, sequence variation, and salt effects. The journal of physical chemistry. B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 35.Kelbauskas L, Yodh J, Woodbury N, Lohr D. Intrinsic promoter nucleosome stability/dynamics variations support a novel targeting mechanism. Biochemistry. 2009;48:4217–4219. doi: 10.1021/bi900476t. [DOI] [PubMed] [Google Scholar]
- 36.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 37.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 38.Tims HS, Widom J. Stopped-flow fluorescence resonance energy transfer for analysis of nucleosome dynamics. Methods. 2007;41:296–303. doi: 10.1016/j.ymeth.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bucceri A, Kapitza K, Thoma F. Rapid accessibility of nucleosomal DNA in yeast on a second time scale. EMBO J. 2006;25:3123–3132. doi: 10.1038/sj.emboj.7601196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geraghty DS, Sucic HB, Chen J, Pederson DS. Evidence that partial unwrapping of DNA from nucleosomes facilitates the binding of heat shock factor following DNA replication in yeast. J Biol Chem. 1998;273:20463–20472. doi: 10.1074/jbc.273.32.20463. [DOI] [PubMed] [Google Scholar]
- 41.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J Biol Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J Mol Biol. 2011;408:187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangenot S, Leforestier A, Vachette P, Durand D, Livolant F. Salt-induced conformation and interaction changes of nucleosome core particles. Biophys J. 2002;82:345–356. doi: 10.1016/S0006-3495(02)75399-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 2004;23:1348–1359. doi: 10.1038/sj.emboj.7600143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, Yates JR, 3rd, Bahler J, O’Connell MJ, Jia S. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J Biol Chem. 2011;287:4386–4393. doi: 10.1074/jbc.M111.329417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci U S A. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luebben WR, Sharma N, Nyborg JK. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci U S A. 2010;107:19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. In and out: histone variant exchange in chromatin. Trends Biochem Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol Cell Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng C, Hayes JJ. Structures and interactions of the core histone tail domains. Biopolymers. 2003;68:539–546. doi: 10.1002/bip.10303. [DOI] [PubMed] [Google Scholar]
- 57.Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, Fukui K, Tomschik M, Ausio J, Zlatanova J. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–10857. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 58.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 59.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J Cell Physiol. 2012;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc Natl Acad Sci U S A. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. J Biol Chem. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proc Natl Acad Sci U S A. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Odell ID, Barbour JE, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol Cell Biol. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Mol Cell Biol. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1. DNA Repair (Amst) 2010;9:134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J Biol Chem. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye Y, Stahley MR, Xu J, Friedman JI, Sun Y, McKnight JN, Gray JJ, Bowman GD, Stivers JT. Enzymatic excision of uracil residues in nucleosomes depends on the local DNA structure and dynamics. Biochemistry. 2012;51:6028–6038. doi: 10.1021/bi3006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci U S A. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong I, Lundquist AJ, Bernards AS, Mosbaugh DW. Presteady-state analysis of a single catalytic turnover by Escherichia coli uracil-DNA glycosylase reveals a“pinch-pull-push” mechanism. J Biol Chem. 2002;277:19424–19432. doi: 10.1074/jbc.M201198200. [DOI] [PubMed] [Google Scholar]
- 72.Hinz JM. Impact of abasic site orientation within nucleosomes on human APE1 endonuclease activity. Mutation research. Fundamental and molecular mechanisms of mutagenesis. 2014;766-767:19–24. doi: 10.1016/j.mrfmmm.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sczepanski JT, Zhou C, Greenberg MM. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huggins CF, Chafin DR, Aoyagi S, Henricksen LA, Bambara RA, Hayes JJ. Flap endonuclease 1 efficiently cleaves base excision repair and DNA replication intermediates assembled into nucleosomes. Mol Cell. 2002;10:1201–1211. doi: 10.1016/s1097-2765(02)00736-0. [DOI] [PubMed] [Google Scholar]
- 77.Widlund HR, Vitolo JM, Thiriet C, Hayes JJ. DNA sequence-dependent contributions of core histone tails to nucleosome stability: differential effects of acetylation and proteolytic tail removal. Biochemistry. 2000;39:3835–3841. doi: 10.1021/bi991957l. [DOI] [PubMed] [Google Scholar]
- 78.Yang Z, Zheng C, Hayes JJ. The core histone tail domains contribute to sequence-dependent nucleosome positioning. J Biol Chem. 2007;282:7930–7938. doi: 10.1074/jbc.M610584200. [DOI] [PubMed] [Google Scholar]
- 79.Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beard BC, Stevenson JJ, Wilson SH, Smerdon MJ. Base excision repair in nucleosomes lacking histone tails. DNA Repair (Amst) 2005;4:203–209. doi: 10.1016/j.dnarep.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 81.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J Biol Chem. 2001;276:12636–12644. doi: 10.1074/jbc.m010470200. [DOI] [PubMed] [Google Scholar]
- 84.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res. 2007;35:4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. The Journal of clinical investigation. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark NJ, Kramer M, Muthurajan UM, Luger K. Alternative modes of binding of poly(ADP-ribose) polymerase 1 to free DNA and nucleosomes. The Journal of biological chemistry. 2012;287:32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prasad R, Horton JK, Chastain PD, 2nd, Gassman NR, Freudenthal BD, Hou EW, Wilson SH. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic acids research. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu B, Droge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat Chem Biol. 2008;4:110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 89.Todd RC, Lippard SJ. Consequences of cisplatin binding on nucleosome structure and dynamics. Chem Biol. 2010;17:1334–1343. doi: 10.1016/j.chembiol.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]