Abstract
The Barrier to Autointegration Factor (BAF or BANF1) is an abundant, highly conserved DNA binding protein. BAF is involved in multiple pathways including mitosis, nuclear assembly, viral infection, chromatin and gene regulation and the DNA damage response. BAF is also essential for early development in metazoans and relevant to human physiology; BANF1 mutations cause a progeroid syndrome, placing BAF within the laminopathy disease spectrum. This review summarizes previous knowledge about BAF in the context of recent discoveries about its protein partners, posttranslational regulation, dynamic subcellular localizations and roles in disease, innate immunity, transposable elements and genome integrity.
Introduction
The barrier to autointegration factor (BAF/BANF1) is a small (10 kDa), conserved and abundant DNA-binding protein. BAF is involved in multiple pathways including mitosis, post-mitotic nuclear assembly, intrinsic immunity against foreign DNA, transcription regulation, and the DNA damage response. BAF is a vital protein; complete loss of BAF is lethal during embryogenesis in Caenorhabditis elegans and Drosophila melanogaster [1,2]. However BAF also has intriguing roles in physiology, since BAF missense mutations appear sufficient to cause a human progeroid syndrome [3]. These and other new findings about BAF function and regulation are discussed below in the context of its known roles as an essential DNA-binding protein that also interacts with nuclear intermediate filament proteins (lamins), nuclear membrane proteins (`LEM-domain' proteins) and transcription regulators.
DNA binding properties of BAF
The unique DNA-binding properties of BAF are likely fundamental to its roles. Early studies showed that BAF forms homodimers, each subunit of which binds double-stranded DNA in a sequence-independent manner [1,4]. BAF can compact or loop DNA in vitro [1,5,6]. Rigorous biochemical and mutational studies combined with insights from atomic structures revealed an elegantly straightforward mechanism by which BAF interacts with DNA. Specifically, each BAF monomer has a helix-hairpin-helix DNA-binding domain, allowing BAF dimers to bind and `bridge' two strands of DNA [5,7] either intra-molecularly or inter-molecularly. BAF-DNA complexes formed in vitro are incredibly stable, with estimated dissociation constants in the low femtomolar range [6]. This poses the first conundrum, since BAF is not `glued' to DNA in living cells. Instead BAF is regulated by its partners in specific subcellular locations and by dynamic phosphorylation and dephosphorylation.
BAF-associated proteins
BAF is regulated at least in part by specific protein partners. Heterodimerization of BAF with BAF-L, a protein 40% identical to BAF but incapable of binding to DNA, is speculated to impair its DNA-bridging activity and potentially also its binding to other partners [8]. The best-characterized BAF partners are a family of proteins that share the ~40-residue LEM-domain fold, including LAP2, EMERIN, and MAN1 [9–12]. BAF homodimerization creates a binding cleft for one LEM domain, which contacts both BAF monomers [13]. BAF association with LEM-domain proteins can be enhanced by DNA or influenced by regions outside the LEM-domain [11,14–17]. Many LEM-domain proteins are anchored at the inner nuclear membrane and function with BAF and lamins as components of nuclear lamina structure ([18,19]; see Barton et al.; this issue). LEM-domain proteins and other BAF partners discussed here are summarized in Table I.
Table 1.
Summary of BAF protein partners
| BAF binding partners | Functions | Assays to analyze interaction with BAF1,2 | References |
|---|---|---|---|
| LEM-domain proteins | |||
| Emerin | Mitosis, integral component of nuclear inner membrane | Blot overlay assay, co-IP, microtiter binding assay, FRET, NMR, co-localization by IF | [11,14,21,38,40,67] |
| LAP2α | Mitosis, transcriptional regulator | Native gel shift assay, NMR, in vitro binding assay, co-IP, co-localization by IF | [13,15,77] |
| LAP2β | Mitosis, integral component of nuclear inner membrane, transcriptional repressor | Y2H, native gel shift assay, in vitro binding assay, NMR | [9,13,15] |
| MAN1 | Integral component of nuclear inner membrane | Microtiter binding assay | [10] |
| Lamin A | Mitosis, structural component of the nuclear envelope, signaling | Microtiter binding assay, FRET, AP-MS** | [40,67,68] |
| Prelamin A | Precursor form of lamin A | Co-IP, co-localization by IF | [28,32] |
| Progerin | Truncated form of farnesylated prelamin A | Co-IP, co-localization by IF | [28] |
| Lamin B | Structural component of the nuclear envelope | Subcellular co-fractionation | [78] |
| LEM2 | Integral component of nuclear inner membrane | Co-localization by IF | [79] |
| Ankle1/Lem3 | DNA damage response | In vitro pulldown assay | [73] |
| Nemp1 | Inner membrane nuclear protein in Xenopus, neural development | GST-pulldown assay, co-localization by IF | [80] |
| Transcriptional regulators | |||
| Crx * | Homeodomain transcription activator, organ morphogenesis | Y2H, in vitro pulldown, co-IP, IF, NMR | [65,66] |
| LAP2ζ | Regulator of LAP2β-mediated transcriptional repressor | Co-IP, co-localization by IF | [27] |
| Requiem | Transcription factor in myeloid cells, apoptosis | Co-IP, AP-MS | [68] |
| Sox2 | Embryonic stem cell differentiation | AP-MS (MudPIT) | [64] |
| Histones and histone regulators | |||
| H1.1 | Nucleosome | Blot overlay assay, microtiter binding assay, GST-pulldown | [70] |
| H3 | Nucleosome | Blot overlay assay, microtiter binding assay, GST-pulldown, AP-MS | [68,70] |
| H4 | Nucleosome | In vitro pulldown assay | [69] |
| RBBP4 | Histone chaperone | Co-IP, AP-MS | [68] |
| SET/I2PP2A | Mitosis, nucleosome assembly, gene expression | Co-IP, AP-MS | [68,69] |
| G9A | Histone methylation | Co-IP, AP-MS | [69] |
| DNA damage repair proteins | |||
| PARP1 | DNA damage response, gene expression | Co-IP, AP-MS | [68] |
| DDB1, DDB2 | DNA damage response, protein degradation | Co-IP, AP-MS | [68] |
| CUL4 | Protein ubiquitination | Co-IP, AP-MS | [68] |
| Kinases | |||
| VRK1 | Mitosis, histone phosphorylation, protein phosphorylation | In vitro kinase assay | [23,24,39,46] |
| VRK2 | Protein phosphorylation, signaling | In vitro kinase assay | [23,46] |
| B1 | Vaccinia kinase required for viral DNA replication and gene expression | In vitro kinase assay | [23,29] |
| Phosphatases | |||
| PP2A | Mitosis, protein dephosphorylation, gene expression | In vitro phosphatase assay | [35] |
| PP4 | Protein dephosphorylation, DNA damage response | siRNA depletion of PP4 | [36] |
| Others | |||
| BAF-L | Potential regulator of BAF DNA binding | In vitro pulldown assay | [8] |
Interaction with BAF is mediated by DNA
Bold letters indicate assays performed using purified proteins suggesting direct interaction with BAF
Underlined letters indicate assays performed using sequence common to all isoforms of LAP2
Abbreviations: Co-IP (co-immunoprecipitation), Y2H (yeast two hybrid), IF (immunofluorescence), NMR (nuclear magnetic resonance), FRET (fluorescence resonance energy transfer), AP-MS (affinity purification followed by mass spectrometry).
Dynamic subcellular localization
BAF can concentrate near the inner nuclear membrane but is also detected in the cytoplasm; its subcellular localization can vary in different cell types or at different stages of the cell cycle [20]. Seminal fluorescence photobleaching studies revealed separate nucleoplasmic and cytoplasmic pools of BAF, each of which had high diffusional mobility [21]. This dynamic mobility can be explained by several mechanisms including phosphorylation, which has a major role in regulating its localization and activity [22–25]. For example a phospho-mimetic mutation causes BAF to localize in the cytosol; BAF also localizes in the cytosol when transiently co-overexpressed with its kinase, VRK1 [23,26]. BAF partners also influence its distribution. For example BAF accumulates in the cytoplasm of cells that overexpress Lap2ζ (a cytosolic LEM-domain protein; [27]), but accumulates in the nucleus of cells that overexpress the lamin A precursor [28]. Viral infections can lead to interesting changes in the subcellular distribution of BAF. For example, infection with a B1 kinase-deficient vaccinia virus (discussed in more detail below) caused BAF to relocalize at sites of viral DNA accumulation in the cytoplasm, while no change in localization was found during infection with wild-type vaccinia [29]. By contrast in cells infected with herpes simplex virus type-1 (HSV-1) BAF localizes to the nucleus, where HSV-1 viral DNA replicates [30]. Interestingly, stresses including heat shock, caloric restriction and food deprivation reduce BAF-1 mobility in C. elegans, apparently stabilizing its association with the nuclear lamina [31]. Finally, cells from certain laminopathy patients show increased BAF accumulation in the nucleus [32]. These findings suggest BAF localization and mobility are controlled by multiple pathways that directly target either BAF, or its partners.
Posttranslational modification, especially phosphorylation, has key roles in regulating BAF function. BAF is phosphorylated by a conserved family of Ser/Thr kinases, named vaccinia-related kinases (VRKs), and by a closely-related vaccinia virus kinase, named B1 [23,24,33,34]. BAF is specifically phosphorylated on residues Ser-4 (the major VRK phosphorylation site), Thr-2 and Thr-3 [22,23]. Phosphorylation inhibits BAF binding to DNA, and can reduce its ability to homodimerize or bind LEM-domain proteins [22–24,26]. Phosphorylation also favors BAF localization in the cytoplasm [22–24,26], consistent with disrupted binding to DNA and LEM-domain proteins. BAF is dephosphorylated by at least two Ser/Thr phosphatases, named PP2A and PP4 [35,36], whose roles are best understood in the context of mitosis, discussed below.
BAF and nuclear reassembly during mitosis
BAF has essential roles during mitosis. Loss of BAF in either C. elegans or D. melanogaster leads to embryonic lethality with mitotic phenotypes that include anaphase chromosome bridges and aberrant nuclear envelope morphology [1,2,24,37]. Later in mitosis, BAF associations with DNA and nuclear membrane proteins are critical to recruit nuclear membranes to chromosomes [24,37,38]. Similar roles for BAF are reported during karyosome formation (the clustering of meiotic chromosomes after recombination) in oocytes of D. melanogaster [39]. Proper control of BAF association with other proteins, and with DNA, is critical at multiple stages of mitosis [40]. For example, phosphorylation by VRK1 early in mitosis triggers BAF release from chromatin and from LEM-domain proteins [24,25]. If BAF is not phosphorylated (e.g., due to loss of VRK1, or expression of an unphosphorylatable BAF mutant), nuclear disassembly is disrupted [24,25]. Thus BAF, like other nuclear proteins, is controlled by the wave of phosphorylation that drives nuclear disassembly.
Conversely, BAF is dephosphorylated in late mitosis by two phosphatases, PP2A and PP4 [35,36]. Studies in both C. elegans and HeLa cells indicate that PP2A is targeted to BAF by a LEM-domain protein named Ankle2 (also known as LEM4) [35]. Ankle2/LEM4 is required for BAF dephosphorylation; in its absence, BAF remains hyperphosphorylated throughout mitosis [35]. Ankle2/LEM4 also further enhances BAF dephosphorylation by associating with VRK1 and inhibiting its catalytic activity [35]. During interphase, Ankle2/LEM4 is detectable at the nuclear envelope, but the bulk of this protein is ER-localized [35], reducing its access to VRK1, which localizes predominantly in the nucleus [33]. Thus Ankle2/LEM4 may only encounter VRK1 (and thereby promote BAF dephosphorylation) later in mitosis when cytoplasmic and nuclear components are fully mixed, and provide a cue for nuclear reassembly as proposed by Asencio et al. [35]. PP4 is also a major regulator of BAF dephosphorylation and BAF localization during late mitosis in HEK293 cells [36]. Specifically, depletion of either the catalytic or regulatory subunit of PP4 led to an increase in BAF phosphorylation and a nuclear envelope invagination phenotype [36]. This phenotype might be only partially attributed to BAF, but raises the possibility that dynamic phosphorylation and dephosphorylation of BAF maintains nuclear envelope integrity.
BAF as an effector of intrinsic immunity
BAF actively protects the genome by intercepting foreign DNA. Ironically this protective function is exploited by retroviruses. BAF was first identified as a cellular factor that associates with the Moloney murine leukemia virus preintegration complex (PIC), and promotes integration into exogenous target DNA by blocking `suicidal' auto-integration within salt-stripped PICs [41,42]. BAF is also an essential host component of the HIV-1 PIC [43,44]. Elegant structural and functional analysis revealed that BAF helps compact retroviral DNA within the PIC [4,45]. Phosphorylation of BAF interferes with these activities, disrupting the PIC [46]. The big question— why BAF helps retroviruses— was recently answered by Izsvak and colleagues [47], who discovered that BAF assembles on transposon DNA in the nucleus and thereby protects at least two endogenous mobile genetic elements, Sleeping Beauty and piggyBac, from suicidal autointegration, much like it functions on retroviral PICs. Since mobile elements have profoundly influenced genome evolution (reviewed in [48–50]), we speculate that BAF has also contributed to metazoan development by modulating DNA transposition.
However BAF is a potent weapon against other viruses, including the poxvirus vaccinia. Vaccinia expresses its own replication and transcriptional machinery, and therefore completes its entire lifecycle in the cytoplasm of infected cells [51,52]. Vaccinia encodes a Ser/Thr kinase named B1; this kinase is essential for viral DNA replication and gene expression [53–57], and also phosphorylates BAF to block its DNA binding activity [23,29]. Without this inhibition, BAF co-localizes with viral DNA and interferes with both genome replication [29,58] and transcription [59], sharply reducing the number of virus progeny [58].
Antipoxvirus defense by BAF may be augmented by the protein phosphatase PP2A, which appears to actively counteract B1-induced BAF phosphorylation in the cytoplasm of vaccinia-infected cells [26]. The subcellular localizations and timing of these competing enzymes, and BAF, are interesting questions. Is this defensive role performed by BAF already in the cytoplasm, or do cells phosphorylate nuclear BAF to drive it into the cytoplasm, where the poxvirus DNA lurks? In either case, PP2A-mediated BAF dephosphorylation, potentially near sites of accumulating viral DNA, might be required to activate its antipoxvirus activity.
In addition to poxviruses, BAF may also defend against Herpes Simplex Virus type 1 (HSV-1), a DNA virus that replicates and transcribes its genes within the nucleus of infected cells. In cells that express an unphosphorylatable BAF mutant (normal DNA-binding activity; restricted to the nucleus), viral DNA replication and viral protein expression are both reduced [30]. Further studies of the BAF defense against HSV-1 may provide new insights into its roles in innate immunity on its `home turf': the nucleus.
The antiviral effector function of BAF requires its ability to bind DNA and homodimerize [26,58], strongly suggesting BAF must crossbridge DNA to impair viral replication. Bacterial proteins such as H-NS, with well-established roles in silencing foreign nucleic acid and regulating transposition events in prokaryotes [60], also crossbridge DNA. BAF and H-NS share no sequence homology, emphasizing the importance of DNA bridging proteins as intrinsic immune effectors against invading DNA or endogenous transposons in both prokaryotes and eukaryotes [30].
BAF in gene expression and epigenetic regulation
BAF can influence gene expression either positively or negatively, but its mechanisms of action remain unknown. For example, depletion of BAF in mouse embryonic stem cells (ESC) reduces the mRNA levels of ESC markers (Sox2, Oct4, Nanog), while increasing that of mesoderm and trophectoderm markers [61], suggesting BAF helps maintain the pluripotent state. In the L-HaCaT psoriasis cell model, BAF depletion up-regulated the expression of S100A9 and c-Jun, suggesting BAF normally suppresses inflammation [62]. In C. elegans, BAF is known to repress a gene (eff-1) involved in somatic cell fusion [63]. Interestingly BAF also represses viral (vaccinia) gene expression in the cytoplasm via mechanisms that remain to be explored [59].
BAF's roles in transcriptional regulation are major open questions, and might involve interactions with a variety of players including transcription factors, chromatin modifiers and nuclear lamina components (Table 1). For example, BAF associates with Sox2 in ESCs [64] and is proposed to help form or regulate Sox2 complexes, which maintain pluripotency [61]. BAF associates indirectly (via DNA) with transcription factor Crx in the mouse retina, and with other paired rule homeodomain proteins in vitro [65,66]. The transcriptional repressor, germ cell-less (GCL) competes with BAF for binding to the LEM-domain protein emerin [67], whereas the apoptotic regulator Requiem co-immunoprecipitates with BAF and emerin [68]. Clearly we are missing important pieces of these puzzles.
Direct BAF binding to histones H1.1, H3 and H4 has many implications, including potential roles in chromatin remodeling during transcription [69,70]. Indeed, BAF overexpression influences histone posttranslational modifications, globally reducing histone acetylation and increasing or decreasing many specific histone marks [69]. BAF depletion affects heterochromatin and transcriptional repression in C. elegans, highlighting its importance for gene expression across species [71]. However, much remains to be learned about these epigenetic roles. BAF does not appear to directly interact with HATs or HDAC1 [69]; instead it might block regulators, or recruit specific chromatin modifiers such as G9A, the DNA methyltransferase Dnmt3 [69,72] or RBBP4, all of which can interact with BAF and are components of chromatin remodeling complexes [68]. These interactions may be key to understanding how BAF relates to its generally repressive nuclear lamina partners at the nuclear periphery, and may provide novel insights into genome regulation.
BAF and the DNA damage response
Proteomic analysis of the BAF interactome identified over 70 potential partners, many of which are involved in DNA damage responses, such as PARP1, DDB1, DDB2 and CUL4 [68]. UV exposure altered BAF or emerin association with DDB1, DDB2 and CUL4A; this suggested BAF interactions are differentially regulated in response to UV damage [68], but did not illuminate the nature of its role. Further evidence linking BAF to the DNA damage response comes from the study of a previously uncharacterized LEM-domain protein, Ankle1 (C. elegans LEM-3), which is unique among LEM-domain proteins in possessing endonuclease activity conferred by a conserved GIY-YIG nuclease domain [73,74]. Its proposed role in the DNA damage response is based on its identification as a C. elegans mutation that increased sensitivity to ionizing radiation [74]. BAF appears to be required for LEM-3 functions, as the introduction of a baf-1 mutation in a lem-3 null background increased embryonic lethality even in the absence of DNA damage [74]. Interestingly Ankle1 is a soluble protein that localizes predominantly in the cytoplasm of mammalian cells, but cycles in and out of the nucleus [73]. These dynamics suggest subcellular localization may regulate Ankle1 interactions with BAF and DNA, raising new questions about how these proteins contribute to the DNA damage response [73].
BAF and Néstor-Guillermo Progeria Syndrome (NGPS)
NGPS is a rare `accelerated aging' syndrome with many clinical features similar to Hutchinson-Gilford Progeria Syndrome (HGPS), caused by dominant mutations in the lamin A precursor [3,75]. However, NGPS patients lack the cardiovascular pathology characteristic of HGPS, and have lifespans well beyond their 20s [3,75]. Whole genome and exome sequencing of two NGPS patients revealed both were homozygous for an Ala12-to-Thr12 (A12T) missense mutation in BANF1 [3]. Fibroblasts from these individuals showed aberrant nuclear morphology that could be rescued by transient expression of BANF1, arguing that NGPS is caused by recessive loss of functional BAF [3]. Further work suggested the BAF A12T mutant protein is stable, folds correctly and co-immunoprecipitates with lamin, emerin and histone H3 [76]. However the A12T mutation weakened BAF binding to DNA and, when overexpressed, caused aberrant nuclear morphology [76]. Thus, weakened DNA binding by BAF is likely a major determinant of NGPS. Future studies of this mutation may offer crucial biological insight into the many roles of BAF, potentially including roles that are DNA-independent, and its interactions with lamin A and other proteins disrupted in progeria.
Conclusions and Future Perspectives
We are only beginning to scratch the surface of BAF's involvement in mitosis, nuclear structure, chromatin regulation and myriad other roles including the interception of foreign DNA and the protection of endogenous transposable elements. The discovery of a BAF mutation in NGPS patients highlights its relevance to human physiology and disease. Further characterization of BAF will continue to yield novel insights into events governing nuclear structure and organization, gene expression, and genome integrity.
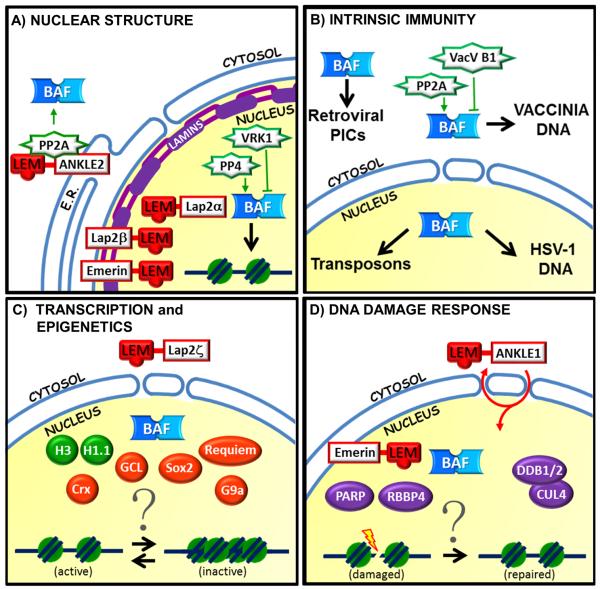
Figure 1.
Overview of BAF-associated proteins in different functional pathways. (A) Nuclear structure. BAF interacts dynamically with LEM-domain proteins, lamins and chromatin at the inner nuclear membrane and in the nucleoplasm. BAF also functions dynamically in the cytoplasm. BAF binding to key partners including DNA and LEM-domain proteins is inhibited by phosphorylation (by VRK1) and promoted by dephosphorylation (by PP2A and PP4). (B) Intrinsic Immunity: In the cytoplasm BAF intercepts and fights certain viruses by binding their DNA (the poxvirus vaccinia), but is acquired and exploited to protect retroviral DNA in preintegration complexes. BAF defense against vaccinia infection may be supported by PP2A-dependent dephosphorylation of BAF, competing against vaccinia B1 kinase-dependent inhibitory phosphorylation of BAF. Within the nucleus, BAF inhibits the replication and transcription of the DNA virus HSV-1 as part of the host innate immune defense. Remarkably BAF binds and protects endogenous transposons (mobile genetic elements), a role with implications for the evolution of eukaryotic genomes. (C) Transcription and Epigenetics: BAF influences transcription, and associates with proteins known to regulate transcription including LEM-domain proteins (emerin, MAN1, Lap2β not shown; Lap2ζ shown), transcription factors (Sox2, Crx, GCL, Requiem), specific histones (H3, H1.1) and histone modifiers (G9a). However the mechanisms of BAF involvement in chromatin regulation remain unknown. (D) DNA Damage Response. BAF is needed for a robust DNA damage response, and interacts with proteins that respond to or mediate the DNA damage response including Ankle1/LEM3, emerin, RBBP4, DDR1/2, CUL4 and PARP. However the mechanisms of BAF involvement are open questions.
Acknowledgements
Work in the Wiebe laboratory is supported by NIH grants (K22AI080941 and R56AI099062) and the Nebraska Center for Virology (P30GM10359). A.J. was also partially supported by a Ruth L. Kirschstein NRSA (T32 AI060547).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Furukawa K, Sugiyama S, Osouda S, Goto H, Inagaki M, Horigome T, Omata S, McConnell M, Fisher PA, Nishida Y. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Science. 2003;116:3811–3823. doi: 10.1242/jcs.00682. [DOI] [PubMed] [Google Scholar]
- •• [3].Puente XS, Quesada V, Osorio FG, Cabanillas R, Cadinanos J, Fraile JM, Ordonez GR, Puente DA, Gutierrez-Fernandez A, Fanjul-Fernandez M, et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am.J.Hum.Genet. 2011;88:650–656. doi: 10.1016/j.ajhg.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first reported disease-causing mutation in BANF1, identified in two patients with Néstor-Guillermo progeria syndrome.
- [4].Harris D, Engelman A. Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J.Biol.Chem. 2000;275:39671–39677. doi: 10.1074/jbc.M002626200. [DOI] [PubMed] [Google Scholar]
- [5].Bradley CM, Ronning DR, Ghirlando R, Craigie R, Dyda F. Structural basis for DNA bridging by barrier-to-autointegration factor. Nat.Struct.Mol.Biol. 2005;12:935–936. doi: 10.1038/nsmb989. [DOI] [PubMed] [Google Scholar]
- [6].Skoko D, Li M, Huang Y, Mizuuchi M, Cai M, Bradley CM, Pease PJ, Xiao B, Marko JF, Craigie R, Mizuuchi K. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc.Natl.Acad.Sci.U.S.A. 2009;106:16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Umland TC, Wei SQ, Craigie R, Davies DR. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry. 2000;39:9130–9138. doi: 10.1021/bi000572w. [DOI] [PubMed] [Google Scholar]
- [8].Tifft KE, Segura-Totten M, Lee KK, Wilson KL. Barrier-to-autointegration factor-like (BAF-L): A proposed regulator of BAF. Exp.Cell Res. 2006;312:478–487. doi: 10.1016/j.yexcr.2005.11.013. [DOI] [PubMed] [Google Scholar]
- [9].Furukawa K. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J.Cell.Sci. 1999;112(Pt 15):2485–2492. doi: 10.1242/jcs.112.15.2485. [DOI] [PubMed] [Google Scholar]
- [10].Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J.Biol.Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- [11].Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell. Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- [12].Laguri C, Gilquin B, Wolff N, Romi-Lebrun R, Courchay K, Callebaut I, Worman HJ, Zinn-Justin S. Structural characterization of the LEM motif common to three human inner nuclear membrane proteins. Structure. 2001;9:503–511. doi: 10.1016/s0969-2126(01)00611-6. [DOI] [PubMed] [Google Scholar]
- [13].Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 2001;20:4399–4407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cai M, Huang Y, Suh JY, Louis JM, Ghirlando R, Craigie R, Clore GM. Solution NMR structure of the barrier-to-autointegration factor-Emerin complex. J.Biol.Chem. 2007;282:14525–14535. doi: 10.1074/jbc.M700576200. [DOI] [PubMed] [Google Scholar]
- [15].Shumaker DK, Lee KK, Tanhehco YC, Craigie R, Wilson KL. LAP2 binds to BAF.DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 2001;20:1754–1764. doi: 10.1093/emboj/20.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Segura-Totten M, Kowalski AK, Craigie R, Wilson KL. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 2002;158:475–485. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berk JM, Simon DN, Jenkins-Houk CR, Westerbeck JW, Gronning-Wang LM, Carlson CR, Wilson KL. The molecular basis of emerin-emerin and emerin-BAF interactions. J. Cell. Sci. 2014;127:3956–3969. doi: 10.1242/jcs.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brachner A, Foisner R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem.Soc.Trans. 2011;39:1735–1741. doi: 10.1042/BST20110724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. International review of cytology. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- [20].Haraguchi T, Koujin T, Osakada H, Kojidani T, Mori C, Masuda H, Hiraoka Y. Nuclear localization of barrier-to-autointegration factor is correlated with progression of S phase in human cells. J.Cell.Sci. 2007;120:1967–77. doi: 10.1242/jcs.03461. [DOI] [PubMed] [Google Scholar]
- [21].Shimi T, Koujin T, Segura-Totten M, Wilson KL, Haraguchi T, Hiraoka Y. Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J.Struct.Biol. 2004;147:31–41. doi: 10.1016/j.jsb.2003.11.013. [DOI] [PubMed] [Google Scholar]
- [22].Bengtsson L, Wilson KL. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol.Biol.Cell. 2006;17:1154–1163. doi: 10.1091/mbc.E05-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N' terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol.Biol.Cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 2007;26:132–143. doi: 10.1038/sj.emboj.7601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[25].Molitor TP, Traktman P. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol.Biol.Cell. 2014 doi: 10.1091/mbc.E13-10-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study elegantly demonstrates that the VRK1-BAF signaling axis is essential for NE architecture and the dynamics of BAF-chromosome interaction during mitosis in mammalian cells.
- •[26].Jamin A, Wicklund A, Wiebe MS. Cell and Virus Mediated Regulation of the Barrier-to-Autointegration Factor's Phosphorylation State Controls its DNA Binding, Dimerization, Subcellular Localization, and Antipoxviral Activity. J.Virol. 2014;88:5342–5355. doi: 10.1128/JVI.00427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that phosphorylation and dephosphorylation of BAF are both important for its ability to protect the host cell against poxviruses, highlighting the involvement of host enzymes in regulating this intrinsic immune response.
- [27].Shaklai S, Somech R, Gal-Yam EN, Deshet-Unger N, Moshitch-Moshkovitz S, Hirschberg K, Amariglio N, Simon AJ, Rechavi G. LAP2zeta binds BAF and suppresses LAP2beta-mediated transcriptional repression. Eur.J.Cell Biol. 2008;87:267–278. doi: 10.1016/j.ejcb.2008.01.014. [DOI] [PubMed] [Google Scholar]
- [28].Capanni C, Cenni V, Haraguchi T, Squarzoni S, Schuchner S, Ogris E, Novelli G, Maraldi NM, Lattanzi G. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell.Cycle. 2010;9 doi: 10.4161/cc.9.13.12080. [DOI] [PubMed] [Google Scholar]
- [29].Wiebe MS, Traktman P. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell.Host Microbe. 2007;1:187–197. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jamin A, Thunuguntla P, Wicklund A, Jones C, Wiebe MS. Barrier to Autointegration Factor Becomes Dephosphorylated during HSV-1 Infection and Can Act as a Host Defense by Impairing Viral DNA Replication and Gene Expression. PLoS One. 2014;9:e100511. doi: 10.1371/journal.pone.0100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[31].Bar DZ, Davidovich M, Lamm AT, Zer H, Wilson KL, Gruenbaum Y. BAF-1 mobility is regulated by environmental stresses. Mol.Biol.Cell. 2014;25:1127–1136. doi: 10.1091/mbc.E13-08-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper showed that BAF localization, mobility and phosphorylation are altered by caloric restriction and heat shock in C. elegans, revealing for the first time that BAF is regulated by environmental stress.
- [32].Capanni C, Squarzoni S, Cenni V, D'Apice MR, Gambineri A, Novelli G, Wehnert M, Pasquali R, Maraldi NM, Lattanzi G. Familial partial lipodystrophy, mandibuloacral dysplasia and restrictive dermopathy feature barrier-to-autointegration factor (BAF) nuclear redistribution. Cell cycle (Georgetown, Tex.) 2012;11:3568–77. doi: 10.4161/cc.21869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nichols RJ, Traktman P. Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J.Biol.Chem. 2004;279:7934–7946. doi: 10.1074/jbc.M310813200. [DOI] [PubMed] [Google Scholar]
- [34].Nezu J, Oku A, Jones MH, Shimane M. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics. 1997;45:327–331. doi: 10.1006/geno.1997.4938. [DOI] [PubMed] [Google Scholar]
- ••[35].Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjanacz M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]; By screening C. elegans mutants with defects in nuclear morphology, the authors identified a novel gene, lem-4, that promotes BAF dephosphorylation by PP2A and inhibits BAF phosphorylation by the kinase VRK1. This study revealed that a LEM-domain protein is critical for regulating the dynamic phosphorylation of BAF.
- •[36].Zhuang X, Semenova E, Maric D, Craigie R. Dephosphorylation of barrier-to-autointegration-factor by protein phosphatase 4 and its role in cell mitosis. J. Biological Chemistry. 2013;289:1119–1127. doi: 10.1074/jbc.M113.492777. [DOI] [PMC free article] [PubMed] [Google Scholar]; This first evidence that BAF is regulated by PP4 provided unexpected insights into its control during mitosis.
- [37].Margalit A, Liu J, Fridkin A, Wilson KL, Gruenbaum Y. A lamin-dependent pathway that regulates nuclear organization, cell cycle progression and germ cell development. Novartis Found. Symp. 2005;264:231–240. [PubMed] [Google Scholar]
- [38].Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL, Hiraoka Y. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell. Sci. 2001;114:4575–4585. doi: 10.1242/jcs.114.24.4575. [DOI] [PubMed] [Google Scholar]
- [39].Lancaster OM, Cullen CF, Ohkura H. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J. Cell Biol. 2007;179:817–824. doi: 10.1083/jcb.200706067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A, Hiraoka Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J.Cell.Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- [41].Lee MS, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proceedings of the National Academy of Sciences. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proceedings of the National Academy of Sciences. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J.Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jacque JM, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- [45].Suzuki Y, Craigie R. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J.Virol. 2002;76:12376–12380. doi: 10.1128/JVI.76.23.12376-12380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Suzuki Y, Ogawa K, Koyanagi Y, Suzuki Y. Functional disruption of the moloney murine leukemia virus preintegration complex by vaccinia-related kinases. J.Biol.Chem. 2010;285:24032–24043. doi: 10.1074/jbc.M110.116640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••[47].Wang Y, Wang J, Devaraj A, Singh M, Jimenez Orgaz A, Chen JX, Selbach M, Ivics Z, Izsvak Z. Suicidal Autointegration of Sleeping Beauty and piggyBac Transposons in Eukaryotic Cells. PLoS Genet. 2014;10:e1004103. doi: 10.1371/journal.pgen.1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]; These authors discovered that BAF is a component of the Sleeping Beauty (SB) transposase complex and inhibits transposon autointegration, suggesting BAF is an important regulator of mobile genetic elements.
- [48].Chalopin D, Naville M, Plard F, Galiana D, Volff JN. Comparative Analysis of Transposable Elements Highlights Mobilome Diversity and Evolution in Vertebrates. Genome Biol. Evol. 2015 doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Konkel MK, Walker JA, Batzer MA. LINEs and SINEs of primate evolution. Evol. Anthropol. 2010;19:236–249. doi: 10.1002/evan.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am.J.Phys.Anthropol. 2007;(Suppl 45):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- [51].Condit RC, Moussatche N, Traktman P. Advances in Virus Research. Academic Press; 2006. In A Nutshell: Structure and Assembly of the Vaccinia Virion; pp. 31–124. [DOI] [PubMed] [Google Scholar]
- [52].Moss B. Poxvirus entry and membrane fusion. Virology. 2006;344:48–54. doi: 10.1016/j.virol.2005.09.037. [DOI] [PubMed] [Google Scholar]
- [53].Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–243. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- [54].Rempel RE, Anderson MK, Evans E, Traktman P. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J.Virol. 1990;64:574–583. doi: 10.1128/jvi.64.2.574-583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rempel RE, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J.Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kovacs GR, Vasilakis N, Moss B. Regulation of viral intermediate gene expression by the vaccinia virus B1 protein kinase. J.Virol. 2001;75:4048–4055. doi: 10.1128/JVI.75.9.4048-4055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lin S, Chen W, Broyles SS. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J.Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ibrahim N, Wicklund A, Wiebe MS. Molecular characterization of the host defense activity of the barrier to autointegration factor against vaccinia virus. J.Virol. 2011;85:11588–11600. doi: 10.1128/JVI.00641-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[59].Ibrahim N, Wicklund A, Jamin A, Wiebe MS. Barrier to autointegration factor (BAF) inhibits vaccinia virus intermediate transcription in the absence of the viral B1 kinase. Virology. 2013;444:363–373. doi: 10.1016/j.virol.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first evidence that BAF can repress transcription from a poxviral promoter in the cytoplasm. The poxviral B1 kinase counteracts this activity of BAF, suggesting a molecular struggle for control of the poxviral promoter.
- [60].Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J.Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cox JL, Mallanna SK, Ormsbee BD, Desler M, Wiebe MS, Rizzino A. Banf1 is required to maintain the self-renewal of both mouse and human embryonic stem cells. J. Cell Sci. 2011;124:2654–2665. doi: 10.1242/jcs.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Takama H, Sugiura K, Ogawa Y, Muro Y, Akiyama M. Possible roles of barrier-to-autointegration factor 1 in regulation of keratinocyte differentiation and proliferation. J. Dermatol. Sci. 2013;71:100–106. doi: 10.1016/j.jdermsci.2013.04.007. [DOI] [PubMed] [Google Scholar]
- [63].Margalit A, Neufeld E, Feinstein N, Wilson KL, Podbilewicz B, Gruenbaum Y. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 2007;178:661–673. doi: 10.1083/jcb.200704049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28:1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang X, Xu S, Rivolta C, Li LY, Peng GH, Swain PK, Sung CH, Swaroop A, Berson EL, Dryja TP, Chen S. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J. Biol. Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- [66].Huang Y, Cai M, Clore GM, Craigie R. No interaction of barrier-to-autointegration factor (BAF) with HIV-1 MA, cone-rod homeobox (Crx) or MAN1-C in absence of DNA. PloS one. 2011;6:e25123. doi: 10.1371/journal.pone.0025123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional Repressor Germ Cell-less (GCL) and Barrier to Autointegration Factor (BAF) Compete for Binding to Emerin in Vitro. Journal of Biological Chemistry. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- [68].Montes de Oca R, Shoemaker CJ, Gucek M, Cole RN, Wilson KL. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS One. 2009;4:e7050. doi: 10.1371/journal.pone.0007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Montes de Oca R, Andreassen PR, Wilson KL. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus. 2011;2:580–590. doi: 10.4161/nucl.2.6.17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- [71].Towbin BD, Meister P, Pike BL, Gasser SM. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 2010;75:555–565. doi: 10.1101/sqb.2010.75.041. [DOI] [PubMed] [Google Scholar]
- [72].Rai K, Jafri IF, Chidester S, James SR, Karpf AR, Cairns BR, Jones DA. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J. Biol. Chem. 2010;285:4110–4121. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[73].Brachner A, Braun J, Ghodgaonkar M, Castor D, Zlopasa L, Ehrlich V, Jiricny J, Gotzmann J, Knasmuller S, Foisner R. The endonuclease Ankle1 requires its LEM and GIY-YIG motifs for DNA cleavage in vivo. J. Cell. Sci. 2012;125:1048–1057. doi: 10.1242/jcs.098392. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals the LEM-domain protein Ankle1 as a nuclease that cycles between the nucleus and cytoplasm. Ankle1 nuclease activity may be important for the DNA damage response.
- •[74].Dittrich CM, Kratz K, Sendoel A, Gruenbaum Y, Jiricny J, Hengartner MO. LEM-3 - A LEM domain containing nuclease involved in the DNA damage response in C. elegans. PLoS One. 2012;7:e24555. doi: 10.1371/journal.pone.0024555. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified C. elegans LEM-3 (Ankle1 in mammalian cells) in a screen for mutations that increased sensitivity to DNA damage. Lem-3 mutations increased sensitivity to ionizing radiation, UV-C light and chemical crosslinkers, yielding severe defects in chromosome segregation during mitosis. Mutations in BAF (baf-1) or the inner-nuclear-membrane LEM-domain proteins emerin or Lem2 also increased sensitivity to DNA damage.
- [75].Cabanillas R, Cadinanos J, Villameytide JA, Perez M, Longo J, Richard JM, Alvarez R, Duran NS, Illan R, Gonzalez DJ, Lopez-Otin C. Néstor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. A. 2011;155A:2617–2625. doi: 10.1002/ajmg.a.34249. [DOI] [PubMed] [Google Scholar]
- •[76].Paquet N, Box JK, Ashton NW, Suraweera A, Croft LV, Urquhart AJ, Bolderson E, Zhang SD, O'Byrne KJ, Richard DJ. Néstor-Guillermo Progeria Syndrome: a biochemical insight into Barrier-to-Autointegration Factor 1, alanine 12 threonine mutation. BMC Mol. Biol. 2014;15 doi: 10.1186/s12867-014-0027-z. 27-014-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; The NGPS-causing BAF A12T mutation weakened binding to DNA, but had no detectable affect on BAF association with three protein partners. These authors also disprove an earlier `protein instability' model: the apparently low level of endogenous BAF A12T protein was an artifact of epitope loss; other antibodies detect normal levels of mutant BAF protein.
- [77].Dechat T, Gajewski A, Korbei B, Gerlich D, Daigle N, Haraguchi T, Furukawa K, Ellenberg J, Foisner R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell. Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- [78].Berk JM, Maitra S, Dawdy AW, Shabanowitz J, Hunt DF, Wilson KL. O-Linked beta-N-acetylglucosamine (O-GlcNAc) regulates emerin binding to barrier to autointegration factor (BAF) in a chromatin- and lamin B-enriched “niche”. J. Biol. Chem. 2013;288:30192–30209. doi: 10.1074/jbc.M113.503060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Brachner A, Reipert S, Foisner R, Gotzmann J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell. Sci. 2005;118:5797–5810. doi: 10.1242/jcs.02701. [DOI] [PubMed] [Google Scholar]
- [80].Mamada H, Takahashi N, Taira M. Involvement of an inner nuclear membrane protein, Nemp1, in Xenopus neural development through an interaction with the chromatin protein BAF. Dev. Biol. 2009;327:497–507. doi: 10.1016/j.ydbio.2008.12.038. [DOI] [PubMed] [Google Scholar]