Abstract
Objective
To investigate the genetic cause of nonobstructive azoospermia (NOA) in a consanguineous Turkish family through homozygosity mapping followed by targeted exon/whole-exome sequencing to identify genetic variations.
Design
Whole-exome sequencing
Setting
Research laboratory
Patient(s)
We sequenced the exomes of two siblings in a consanguineous family with NOA.
Intervention(s)
All variants passing filter criteria were validated with Sanger sequencing to confirm familial segregation and absence in the control population.
Main Outcome Measure
Discovery of a mutation that could potentially cause NOA
Results
A novel non-synonymous mutation in neuronal PAS 2 domain (NPAS2) was identified in a consanguineous family from Turkey. This mutation in exon 14 (chr2: 101592000 C>G) of NPAS2 is likely a disease-causing mutation as it is predicted to be damaging, is a novel variant, and segregates with the disease. Family segregation of the variants showed the presence of homozygous mutation in the three brothers with NOA and heterozygous mutation in mother, one brother and one sister who were both fertile. The mutation is not found in the single nucleotide polymorphism (SNP) database, the 1000 Genomes Project, Baylor College of Medicine cohort of 500 Turkish patients (not a population specific polymorphism) or matching 50 fertile controls.
Conclusions
Using WES, we identified a novel homozygous mutation in NPAS2 as a likely disease-causing variant in a Turkish family diagnosed with NOA. Our data reinforce the clinical role of WES in the molecular diagnosis of highly heterogeneous genetic diseases which conventional genetic approaches have previously failed to conclude a molecular diagnosis.
Keywords: male infertility, circadian rhythm, spermatogenesis, genome, consanguineous
INTRODUCTION
Infertility affects ~ 10% of couples, with about half of this related to male factors. Men with infertility may present with either low sperm count or decreased sperm motility but more often exhibit a combination of both defects. The most severe phenotype of male infertility is nonobstructive azoospermia (NOA). NOA arises because of a lack of sperm in the ejaculate secondary to a production defect in the testis. NOA can be due to chromosomal aberrations such as 47, XXY (Klinefelter syndrome) and Y-chromosomal microdeletions. NOA secondary to these genetic diseases is associated with severe testicular atrophy, making genotype-phenotype correlations relatively straightforward in ~7% of men experiencing infertility.
However, the most common cause of male infertility remains unknown. Not knowing what is causing failure to reproduce can often be frustrating to couples. To date, there are approximately 500 “known” disease-associated genes for male infertility. A large proportion of cases, however, do not display characteristic phenotypic abnormalities and, as a result, we rely on molecular tests for definitive diagnoses. Recently, many molecular diagnoses have been guided by gene-disease association discoveries. Clinical whole-exome sequencing (WES) for identification of mutations leading to Mendelian disease has become increasingly implemented in clinical molecular diagnosis (1-3). WES using next-generation-sequencing(4) has proven to be a powerful tool for identifying mutations in genes known to underlie Mendelian disease, as well as for the discovery of novel disease-associated loci(5).
We applied WES to investigate the genetic cause of infertility due to NOA in a consanguineous Turkish family through homozygosity mapping followed by targeted exon/whole-exome sequencing to identify possible genetic variants that could be associated with NOA.
PATIENTS AND METHODS
Patients
Two siblings with male infertility were referred to Bahçeci Fulya IVF Center in Turkey. Both men with a history of infertility were diagnosed with NOA because of the absence of sperm in the ejaculate along with small testes and elevated serum follicle stimulating hormone (Table 1). This study was approved by the Institutional Review Board at Baylor College of Medicine, and informed consent was obtained from all subjects prior to enrolment in the project. Participants provided venous blood samples, and genomic DNA was extracted from blood according to the manufacturer’s protocol (Puregene Blood Kit, QIAGEN Sciences, Germantown, MD).
Table 1. Major Clinical Features of Three Brothers with nonobstructive azoospermia from consanguineous Turkish family.
| Clinical Parameter | A1 | A2 | A3 |
|---|---|---|---|
| Age | 33 | 35 | 37 |
| Testis Volume | 12ml | 12 ml | 14 ml |
| FSH | 36 mIU/ml | 32 mIU/ml | 30 mIU/ml |
| Testosterone | 560 ng/dL | 880 ng/dL | 800 ng/dL |
| Karyotype | 46, XY | 46,XY | 46,XY |
| Y-Chromosome microdeletion | No deletion | No deletion | No deletion |
| Microdissection TESE | N/A | Sperm (+) | Sperm (−) |
Whole Exome Sequencing
A total of two affected individuals from one consanguineous family were sequenced and analyzed through the Baylor-Hopkins Center for Mendelian Genomics research program(6). Genomic DNA samples were prepared and processed according to protocols previously described (7). Briefly, DNA was prepared in Illumina paired-end libraries, and capture was performed using the in-house developed BCM Human Genome Sequencing Center (HGSC) Core design. DNA samples were sequenced utilizing the Illumina HiSeq 2000 platform. Data was processed through the HGSC-developed Mercury pipeline to produce variant call format files (.vcf) using the Atlas2 variant calling method (8). Variants were annotated using the “Cassandra”(9) annotation pipeline based on ANNOVAR(10). We compared the resulting annotated variants in pairs of affected individuals within the same family and rare variants in shared genes among all affected individuals. During the analyses of candidate variants/mutations, we compared variants to genome-wide data using external publicly available databases such as the 1000 Genomes Project (http://www.1000genomes.org) and other large-scale exome sequencing projects including the Exome variant server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (http://evs.gswashington.edu/EVS/), and our “in-house-generated” exomes (from 5,000 individuals, of whom ~ 500 are of Turkish origin) at the Baylor College of Medicine (BCM) and Johns Hopkins University Center for Mendelian Genomics
Validation of Mutations With Sanger Sequencing
To confirm the identified exome sequencing candidate variants by an orthologous method and to segregate these variants within the families, exon 14 of the neuronal PAS 2 domain (NPAS2) gene (Figure 1A) was amplified from genomic DNA using conventional end-point PCR and the following primer pair (F-5′ATTCTGGGGAGGCGTCGAT, R-5′ATAGCCAGGGCCATACATCC). Standard PCR was performed in a 20 μl reaction volume using Phusion High-Fidelity PCR Master Mix with HF Buffer (Thermo, Cat: # F-531S). The PCR protocol from the manufacturer was used (anneal Tm: 65.1C, extension time: 30s) and amplification products were electrophoresed on a 2% agarose gel. PCR products were purified by Genewiz and analyzed by standard Sanger di-deoxy nucleotide sequencing (Genewiz). We analyzed the possible pathogenic functional effects of the variants using three types of prediction programs (Mutation Taster http://www.mutationtaster.org, PolyPhen (11) and SIFT http://sift.jcvi.org). The tools predict the possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations.
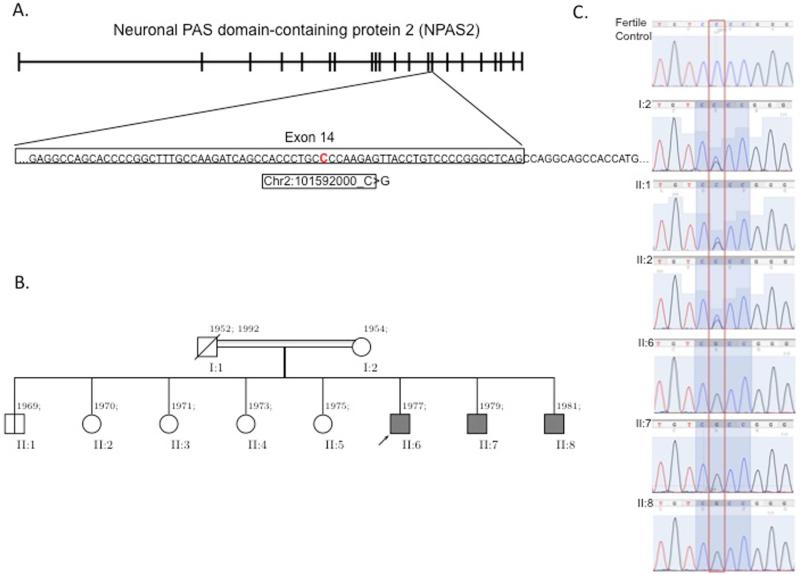
Figure 1.
NPAS2 gene, pedigree of the family, and segregation study A. NPAS2 gene is located in chromosome 2 and consists of 21 exons. The specific mutation that the WES identified is in 14th exon with a C to a G leading to a nonsynonmous mutation. B. Pedigree of the family. Black filled boxes indicate affected individuals. C. Sanger chromatographs of the entire family for segregation analyses. Affected individuals have homozygous mutation whereas unaffected individuals are heterozygous or wild type, which is consistent with Mendelian recessive expectations
Immunohistochemistry
Bouin-fixed specimens from the testes of a fertile patient (who underwent orchiectomy for chronic orchialgia) was processed and embedded into a paraffin block. The slides prepared from this block were rehydrated, then were heated in boiling water bath for antigen retrieval (10 mmol/L citrate buffer, pH 6, 10 min). The sections were allowed to cool in citrate buffer for couple mins then washed twice in PBS for 5 mins, then incubated in 3% hydrogen peroxide in order to deactivate any endogenous peroxidase activity. After washing with PBS (2 × 5 min) the slides were incubated in 3% blocking solution (serum from the animal which the secondary antibody was made in) for 20 mins. Later, the sections were incubated in primary rabbit polyclonal anti-NPAS2 antibody (1:75 diluted, Abcam, ab137601) in 1% blocking solution overnight in 4 °C. Next, the slides were washed with PBS (2 × 5 min) and incubated 0.5% biotinylated secondary antibody (BA-1000, Vector Laboratories) for 40 mins. at room temperature. After being washed with PBS (2 × 5 min), the sections were incubated with a 2% tertiary antibody solution at room temperature to amplify the signal for 30 mins. Finally, after another wash with PBS (2 × 5mins) peroxidase activity was visualized using 0.05% diaminobenzidine (DAB; Sigma) in PBS, containing 0.01% hydrogen peroxide.
RESULTS
We studied two brothers with nonobstructive azoospermia (NOA) in a consanguineous family who met clinical criteria for NOA. The DNA from the two brothers (II:6 and II:7; Figure 1B) was isolated from blood samples and WES was performed at an average depth of coverage of greater than 120-fold, and more than 95% of the targeted bases were covered at 20-fold or greater. The results identified novel events, including single-nucleotide variants (SNVs) or small indel (insertion/deletion) mutations in the brothers, in four genes: NPAS2, IFT140, XIRP2 and M1AP. The sequence coordinates for the 4 genes based on human reference genome hg19 (UCSC Genome Browser) are as follows: homozygous c.C1363G (p.P455A) RefSeq ID: U77970 in NPAS2 (OMIM 603347; RefSeq NM_002518.3), homozygous c.3955_3960del (p.1319_1320del) in IFT140 (OMIM 266920; RefSeq NM_014714.3), homozygous c.T2664A (p.N855K) in XIRP2 (OMIM 609778; RefSeq NM_152381.5), and heterozygous c.1415_1416insTCC (p.P472delinsPP) in M1AP (RefSeq NM_138804.4);). We compared the minor allele frequencies between these 4 variants identified in these individuals to similar mutations in the NHLBI Exome Sequencing Project Exome Variant Server (EVS) and a local variant database at Baylor College of Medicine. We recruited some of the other family members (mother – I:2, fertile brother II:1, fertile sister II:2 and infertile brother II:8) and used their genomic DNA to perform segregation analysis (Figure 1B). We identified a novel non-synonymous mutation in NPAS2 as likely disease-causing candidate because of their occurrence in individuals without suspected infertility. This mutation in exon 14 (chr2: 101592000 C>G) of NPAS2 segregates in accordance with Mendelian expectations for an autosomal recessive trait. Familial segregation of the variants showed the presence of a homozygous mutation in all three brothers with NOA and a heterozygous mutation in the mother, one brother and one sister who were both fertile (Figure 1C).
The observed nonsynonymous homozygous mutations in NPAS2 are not present in the 1000 Genomes database, or the EVS (about 6,500 individuals). To rule out the possibility that this mutation is not a potential polymorphism in the Turkish population and a benign variant, we compared our results to exome data from 500 Turkish patients sequenced at Baylor College of Medicine. We found a minor allele frequency of 0% and no individual was homozygous for this mutation. The absence of the homozygous mutation in NPAS2 (c.C1363G) in these databases suggests that a mutation at this locus is rare, and this, along with the genotype-phenotype familial segregation, and the absence of any other CNV detected by WES that fulfills these criteria is consistent with the possibility that this represents a potential pathogenic allele.
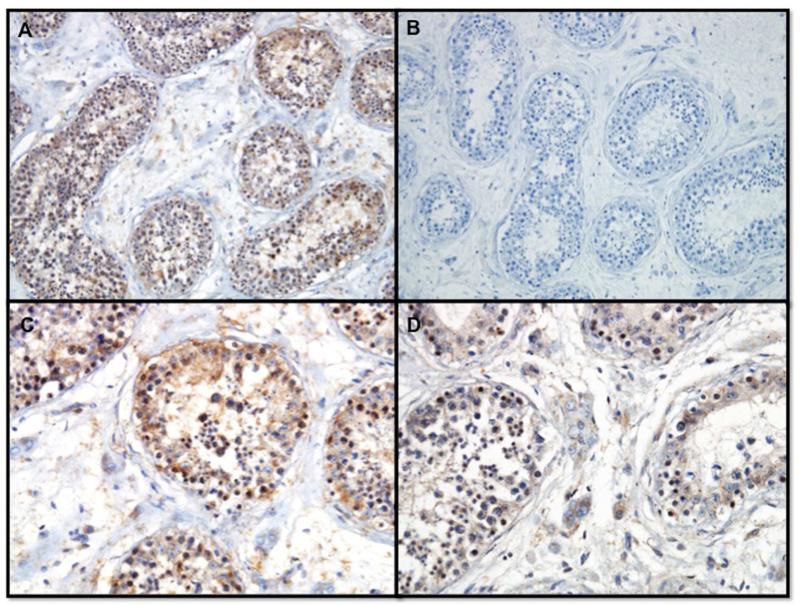
NPAS2 protein is expressed in the brain, more specifically the cerebral cortex, testis, and the prostate in both mouse and human (BioGPS, http://biogps.org/, Protein Atlas, http://www.proteinatlas.org/ENSG00000170485-NPAS2/tissue). This expression data is consistent with the role of NPAS2 function in circadian rhythm and fertility. The NPAS2 homozygous mutation is predicted as “benign” by Polyphen2, neutral by both SIFT and Mutation taster. The novel nonsynonymous mutation in NPAS2 (c.C1363G), results in a P455A amino acid change. By aligning human NPAS2 against the protein sequences of NPAS2 orthologs in rhesus monkey, mouse, and chicken, we found that the conserved amino acids are clustered into two regions, suggesting two functional units. HLH (Helix-loop-helix) has a DNA-binding basic region followed by two alpha-helices separated by a variable loop region located at codons 7-61, contained in conserved region 1. Two PAS domains located at codons 93 - 159 and 250 - 350, have been found to bind ligands, and to act as sensors for light and oxygen in signal transduction. To evaluate NPAS2 expression in human testis, immunostaining signals were evaluated by immunohistochemistry. NPAS2 was expressed in the germ cells within the tubules of the testes (Figure 2A and 2C) as well as in the Leydig cells in the interstitial space (Figure 2D). The negative control (no primary antibody, 2B) showed no staining.
Figure 2.
Immunohistochemistry demonstrating NPAS2 protein in germ cells from testis biopsy from a fertile man at low power - 200x (A) and higher power - 400x (C) and expression of NPAS2 protein in the Leydig cells (D). Lack of NPAS2 staining is demonstrated in negative control (B).
DISCUSSION
We identified a consanguineous family having three affected siblings with NOA, in whom we found a homozygous nonsynonymous mutation (c.C1363G; p.P455A, RefSeq ID: NM_002518.3; chr2:101592000C>G [hg19]) in NPAS2 in all three individuals. This is the first study to identify genetic variants in a family of men with NOA. Clinical evaluation of the three siblings demonstrated azoospermia due to a production defect. Small testes and elevated FSH levels, which are characteristic findings of NOA, were seen in all three siblings. Two of the siblings underwent microdissection testicular sperm extraction (TESE). One of them had successful sperm retrieval, clinical pregnancy and a live birth. The other sibling had no sperm at TESE.
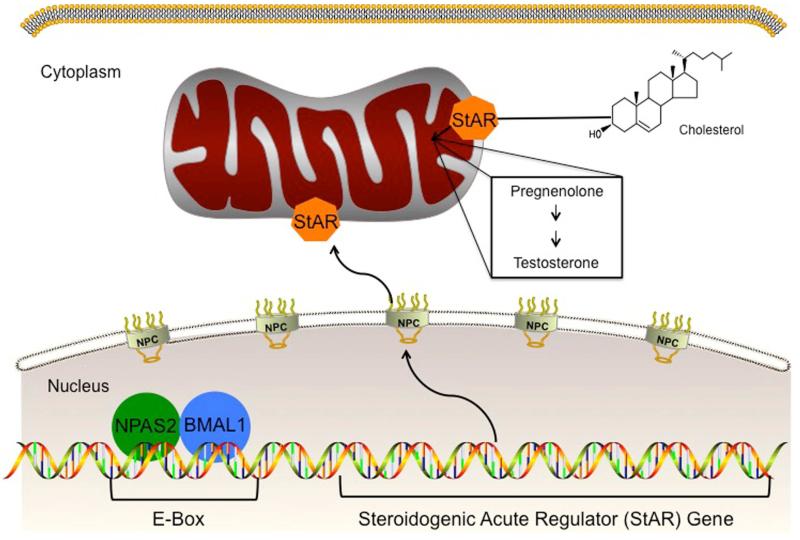
Several factors such as Klinefelter syndrome (ORPHA ID # 484) (KS) and Y-micro deletions (YMD) can lead to NOA. However, the underlying mechanism remains elusive in most cases. More than 500 genes have been associated with infertility (12). Using WES, we identified NPAS2 mutation in a family of men with NOA. Human NPAS2 encodes a protein of 824 amino acids. In vitro and in vivo studies have shown that NPAS2 interacts with a number of proteins including CLOCK and BMAL-1(13). Mice with disruption of two genes, CLOCK and neuronal PAS domain protein 2 NPAS2, lose their circadian rhythms (14). CLOCK/NPAS2 normally interact with BMAL1 and form a transcriptional complex (15). Male BMAL1 KO mice are infertile (16). In BMAL1 KO mice, expression of the steroidogenic acute regulatory protein (StAR) gene and protein, which regulates the rate-limiting step of steroidogenesis, was decreased in testes (Figure 3). It is posited that disruption in steroidogenesis leads to infertility. If BMAL1 functions as a circadian molecule to regulate StAR expression, then we would expect disruption of other circadian clock components to also affect StAR expression. For example, if NPAS2 partners with BMAL1 in Leydig cells to regulate StAR expression, we would expect that StAR expression would be low in mice with a disrupted NPAS2. Further studies are needed to evaluate the role of other circadian clock proteins such as CLOCK and NPAS2 on spermatogenesis and testosterone levels in in vivo and in vitro models.
Figure 3.
Role of NPAS2 in connection with CLOCK family member proteins. A hypothetical model demonstrates NPAS2 interacts with BMAL1 and functions by acting on promoter region of Steroidogenic Acute Regulatory Protein (StAR). StAR is essential for the rate-limiting step in the transport of free cholesterol from the cytoplasm into mitochondria. A series of enzymatic steps in the mitochondria of steroidogenic tissues convert cholesterol into all of the other steroid hormones such as testosterone.
This work represents one of the first comprehensive studies of a family of men with NOA by exome sequencing. Similar to other studies using WES technologies, known limitations should be considered. First, the coverage of the exome fraction was not complete; thus, it is possible that we missed disease-causing mutations. Second, we used stringent criteria to filter potential false-positives (0% frequency in general population), and as a consequence we may have missed true etiological variants that were not considered as being pathogenic. Third, we sequenced only 2 of the 3 affected probands, it is possible that we could have missed mutations present in the third proband. Fourth, the software used to predict the mutation predicted the NPAS2 mutation we identified as “benign or neutral”. For variants causing Mendelian disease, the reported sensitivity of these tools is between 0.68 and 0.86 (17-19). Thus, while variant prioritization tools often provide useful supporting evidence, they unfortunately classify between one third and one sixth of disease-causing variants as benign or neutral. Fifth, when we compared the variants present in the two brothers with NOA to the subjects included in the NHLBI database, we assumed that the subjects were fertile.
In conclusion, our findings, suggest NPAS2 as one of the potential causative genes for NOA. As seen in this case, rare variants in families with rare Mendelian phenotypes may provide novel insights into human biology. Further examples of NPAS2 variants and clinical NOA cases are needed to explore the function of NPAS2 and its interactions during spermatogenesis and gain insight into possible heritability of NOA.
Acknowledgments
Source of funding: RR is a K12 scholar supported by a Male Reproductive Health Research (MRHR) Career Development Physician-Scientist Award (Grant # HD073917-01) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Program to DJL, and in part by NIH grants to R.A.G. (NHGRI 5 U54 HG003273), J.R.L. (NHGRI RO1NS058529 and U54HG006542).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: J.R.L. has stock ownership in 23 and Me and Ion Torrent Systems, is a paid consultant for Regeneron, is on the Board of Directors for Lasergen and is a co-inventor on multiple United States and European patents related to molecular diagnostics. R.A.G. is interim Chief Scientific Officer of Baylor Medical Genetics Laboratory. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in the Medical Genetics Laboratory (MGL; http://www.bcm.edu/geneticlabs/). Other authors have no disclosures relevant to the manuscript.
References
- 1.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2014 doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainbridge MN, Wang M, Burgess DL, Kovar C, Rodesch MJ, D’Ascenzo M, et al. Whole exome capture in solution with 3 Gbp of data. Genome biology. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annual review of medicine. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. American journal of medical genetics Part A. 2012;158A:1523–5. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome medicine. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome research. 2010;20:273–80. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, et al. Whole-genome sequencing for optimized patient management. Science translational medicine. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nature medicine. 2008;14:1197–213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 14.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez JD, Hansen A, Ord T, Bebas P, Chappell PE, Giebultowicz JM, et al. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. Journal of biological rhythms. 2008;23:26–36. doi: 10.1177/0748730407311254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodzic A, Ristanovic M, Zorn B, Tulic C, Maver A, Novakovic I, et al. Genetic variation in circadian rhythm genes CLOCK and ARNTL as risk factor for male infertility. PloS one. 2013;8:e59220. doi: 10.1371/journal.pone.0059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11:361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 18.Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC genomics. 2013;14(Suppl 3):S7. doi: 10.1186/1471-2164-14-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genetic testing and molecular biomarkers. 2010;14:533–7. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]