Abstract
Hypoxia, as a pervasive feature in the microenvironment of solid tumors, plays a significant role in cancer progression, metastasis, and ultimately clinical outcome. One key cellular consequence of hypoxic stress is the regulation of DNA repair pathways, which contributes to the genomic instability and mutator phenotype observed in human cancers. Tumor hypoxia can vary in severity and duration, ranging from acute fluctuating hypoxia arising from temporary blockages in the immature microvasculature, to chronic moderate hypoxia due to sparse vasculature, to complete anoxia at distances more than 150 μM from the nearest blood vessel. Paralleling the intra-tumor heterogeneity of hypoxia, the effects of hypoxia on DNA repair occur through diverse mechanisms. Acutely, hypoxia activates DNA damage signaling pathways, primarily via post-translational modifications. On a longer timescale, hypoxia leads to transcriptional and/or translational downregulation of most DNA repair pathways including DNA double-strand break repair, mismatch repair, and nucleotide excision repair. Furthermore, extended hypoxia can lead to long-term persistent silencing of certain DNA repair genes, including BRCA1 and MLH1, revealing a mechanism by which tumor suppressor genes can be inactivated. The discoveries of the hypoxic modulation of DNA repair pathways have highlighted many potential ways to target susceptibilities of hypoxic cancer cells. In this review, we will discuss the multifaceted hypoxic control of DNA repair at the transcriptional, post-transcriptional, and epigenetic levels, and we will offer perspective on the future of its clinical implications.
Keywords: Hypoxia, Tumor microenvironment, Genetic instability, Replication stress, DNA damage response, DNA repair, Gene regulation, Gene silencing, Post-translational modifications
1. Introduction
Cancer cells in solid tumors grow within a complex microenvironment, consisting of associated stromal and immune inflammatory cells, diverse extracellular signaling molecules, and the altered conditions of low oxygen, low pH, and low nutrient levels [1, 2]. These attributes of the microenvironment collectively enable the growth and proliferation of neoplastic cells. Hypoxia, or low oxygen content, is particularly prevalent in the tumor microenvironment. It arises as rapid cell proliferation outpaces the development of sufficient organized vasculature, leading to regions of tumors with sparse vessels as well as to structural and functional abnormalities of the new microvasculature [3]. Hypoxic or anoxic regions are found in 50–60% of locally advanced solid tumors and have been demonstrated in a wide range of malignancies, including breast, cervical, head and neck, prostate, rectal, pancreatic, lung, brain, and soft tissue cancers (reviewed in [4–6]). Though hypoxia is pervasive within tumors, it is temporally and spatially heterogeneous. Transient disruptions in perfusion can induce acute hypoxia on a timescale of minutes to hours while oxygen diffusion limitations can lead to more chronic hypoxia over hours to days [7]. In addition, cycles of hypoxia and reoxygenation can occur due to dynamic changes in microvessel perfusion and lead to the generation of reactive oxygen species [7].
In solid human cancers, the presence and level of hypoxia generally correlate with features of aggressive tumors, but also appear to have independent clinical significance (reviewed in [4, 5]). Direct oxygen level measurements in tumors have shown hypoxia to be an independent negative prognostic factor for disease-free and overall survival in cervical cancer, head and neck cancer, and soft tissue sarcomas [4]. Endogenous and exogenous markers of hypoxia also show prognostic significance for poorer patient outcome in many tumor types [4, 5]. In addition, hypoxic gene expression signatures serve as adverse prognostic markers in breast cancer, head and neck cancer, and glioblastoma [5]. Poorer outcomes in patients with hypoxic compared to normoxic tumors are associated with both progressive locoregional disease and increased risk of metastasis [4, 8].
Lack of oxygen increases cellular resistance to radiotherapy via a decrease in fixation of free radical DNA damage as well as resistance to some chemotherapy due to poor drug delivery or restrained cell proliferation [6]. However, cellular adaptions induced by hypoxia also directly promote tumor progression and metastasis through changes in gene expression, genomic changes, and clonal selection [6]. The hypoxia-inducible factors (HIFs), heterodimeric transcription factors consisting of a hypoxia-regulated α-subunit and a constitutively expressed β-subunit, mediate many of the cellular effects of hypoxia [9, 10]. Hypoxia-induced stabilization of the most broadly expressed α-subunit, HIF-1α, leads to direct transcriptional activation by the HIF-1α/β dimer of genes involved in cell growth, migration, energy metabolism, and angiogenic signaling. Additional signaling pathways, including the mTOR, NF-κB, and unfolded protein response pathways, are affected by hypoxia and mediate additional changes in transcription and translation [11, 12].
One key cellular event induced by hypoxia that contributes to tumor progression is genetic instability, itself an enabling feature of cancer (reviewed in [13]). The tumor microenvironment, and hypoxia in particular, have been shown to lead to both large-scale chromosomal aberrations and small-scale DNA mutations. Cells grown as tumors in vivo acquire increased levels of genomic rearrangements and higher levels of point mutations and small deletions in reporter genes compared with cells grown in cell culture [14–18]. In vitro, hypoxic stress leads to similar genomic rearrangements, comparable elevations in mutation frequency, DNA over-replication with gene amplification, and fragile site induction [16, 18–23]. Importantly, in vitro hypoxic exposure of fibrosarcoma and melanoma cells not only generated genomic instability, but also led to increased metastatic efficiency in mice [20]. The current evidence thus strongly supports a link between hypoxia, genomic instability, and tumorigenesis.
Several studies have demonstrated that hypoxia, in the absence of reoxygenation, does not induce direct DNA damage [24–26]. Instead, hypoxia-induced genetic instability arises from the impact of hypoxia on DNA damage repair pathways [13]. Numerous mechanisms of DNA repair modulation by hypoxia have been reported, many of which depend upon the type or severity of hypoxia. Acute hypoxic stress rapidly stimulates changes in DNA repair pathways via post-translational modifications. On a slightly longer timescale, persistent hypoxia leads to transcriptional and/or translational downregulation of DNA repair proteins. More prolonged moderate hypoxia induces epigenetic regulation of DNA repair genes. Within this review, severe and moderate hypoxia will refer to conditions of ≤0.2% oxygen and 0.5%−2% oxygen, respectively. In the following sections, we will describe the diverse ways in which hypoxia impacts DNA repair function, classifying them according to post-translational, transcriptional, translational, and epigenetic mechanisms, and we will highlight areas for future research and with potential therapeutic promise.
2. Post-Translational Control of DNA Damage Signaling
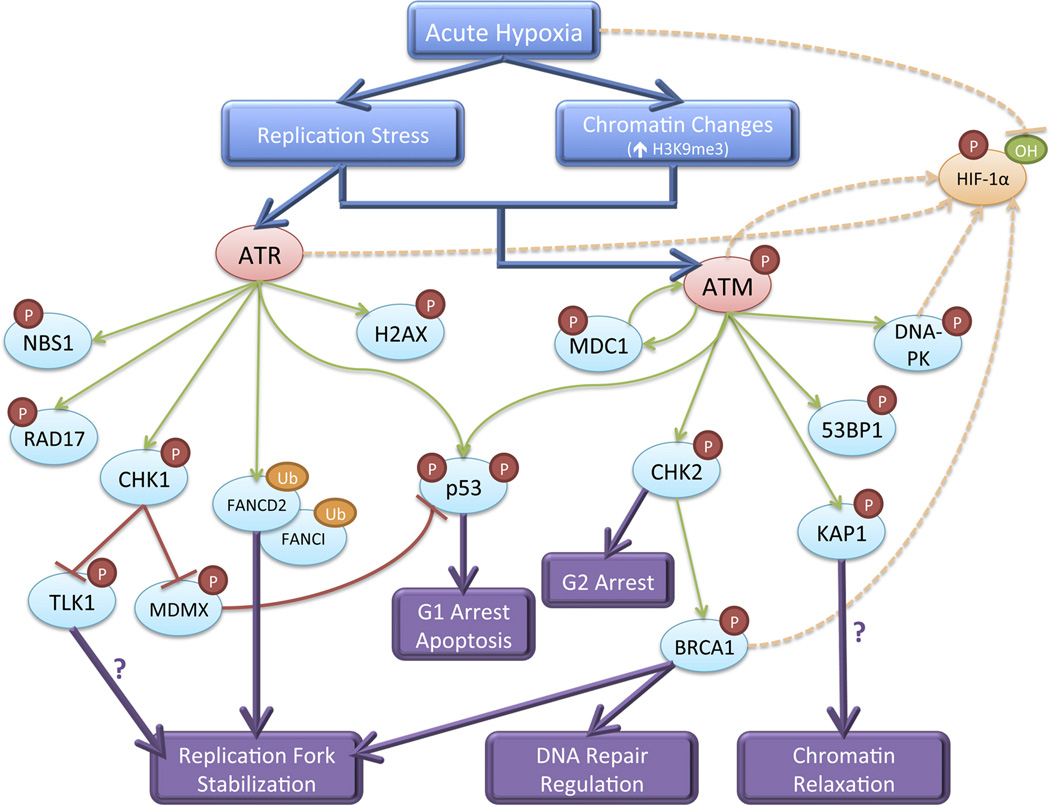
Post-translational protein modifications (PTMs) allow rapid control of protein functionality in response to cellular events or stressors. These covalent protein modifications, such as phosphorylation, hydroxylation, ubiquitination, or acetylation, can lead to changes in protein enzymatic activity, cellular localization, stability, and interactions with other proteins or DNA. Much of the cellular hypoxic response is initiated by changes in PTMs of HIF [10]. In parallel to HIF signaling, severe hypoxia rapidly induces a wide spectrum of PTMs of proteins involved in DNA damage response signaling and DNA repair, including components of both the ATR-CHK1 and ATM-CHK2 pathways [25, 27]. Given the absence of DNA damage under hypoxia, the main stimulus appears to be hypoxia-induced replication stress. Within six hours of severe hypoxic stress, replication initiation and elongation stall, giving rise to an accumulation of single-stranded DNA and RPA foci [28, 29]. It is generally accepted that this S phase arrest, which is independent of checkpoint signaling factors and HIF, is due to the depletion or imbalance of cellular deoxyribonucleotides since certain nucleotide biosynthesis enzymes, including dihydroorotate dehydrogenase and ribonucleotide reductase, require oxygen to function [29, 30]. The hypoxic modulation of DNA repair pathways by PTMs serves to coordinate stabilization of replication forks, though it may also induce cell cycle arrest, initiate apoptosis, generate chromatin changes, and affect DNA repair itself (Figure 1).
Figure 1. DNA damage response signaling pathways activated by acute hypoxia and mediated by post-translational modifications.
ATR, activated by replication stress, and ATM, activated by the combination of replication stress and chromatin alterations, signal to downstream transducer and effector proteins. This signaling cascade leads to replication fork stabilization, chromatin relaxation, regulation of DNA repair pathway choice, cell cycle arrest, and potentially apoptosis. Several DNA damage response factors also contribute to stabilization and activation of HIF-1α.
The ataxia telangiectasia and Rad3-related (ATR) checkpoint kinase responds to DNA damage that impedes replication fork progression and generates single-stranded DNA [31]. Under hypoxia-induced replication stress, ATR forms nuclear foci and is required for phosphorylation of downstream targets, including CHK1 (S317/S345), H2AX (S139), RAD17 (S645), and NBS1 (S343) [24–26]. Activated CHK1 phosphorylates and inactivates TLK1, a serine/threonine kinase involved in cell cycle progression and processing newly replicated DNA [32]. Phosphorylated H2AX (γH2AX) exhibits pan-nuclear staining under hypoxia but also forms foci that colocalize with RPA foci, likely marking sites of single-stranded DNA [25, 27]. In addition, ATR is required for hypoxia-induced monoubiquitination of FANCD2 and FANCI [33]. These PTMs appear critical for stabilizing replication forks, as loss of ATR activity or FANCD2 monoubiquitination leads to DNA damage during hypoxia [28, 33]. In addition to promoting stabilization of replication forks, hypoxia-induced ATR also regulates p53. Hypoxia induces ATR-dependent phosphorylation of p53 (S15) and CHK1-dependent MDMX (S367) phosphorylation, which together lead to p53 stabilization, gene transrepression, G1 cell cycle arrest, and promotion of apoptosis [24, 34, 35].
The ataxia telangiectasia mutated (ATM) checkpoint kinase primarily senses and responds to DNA double-strand breaks (DSBs), which lead to ATM autophosphorylation at serine 1981, foci formation, and phosphorylation of downstream targets [31]. Although hypoxia does not generate DSBs, ATM does undergo autophosphorylation within six hours of severe hypoxic stress [27, 36, 37]. However, unlike activation of ATM by DSBs, hypoxic activation of ATM is independent of the MRN (Mre11-Rad50-Nbs1) complex and leads to diffuse localization throughout the nucleus rather than the formation of distinct nuclear foci [27]. Hypoxia-induced ATM is required for phosphorylation of downstream targets, including CHK2 (T68), KAP1 (S824), 53BP1 (S25), and DNA-PKcs (T2609) [27, 36–38]. CHK2 in turn is required for phosphorylation of BRCA1 (S988), which is postulated to regulate the choice between error-free and error-prone DSB repair, and p53 (S20) which is required for its stabilization [38, 39]. BRCA1, as well as 53BP1, do not form distinct foci under hypoxia, further suggesting that their phosphorylation is not a response to DNA DSBs [27]. Recent evidence by Olcina et al. instead indicates that ATM signaling activation under hypoxia is due to replication stress in the setting of specific chromatin alterations [40]. Hypoxia induces an increase in histone H3 lysine9 trimethylation (H3K9me3), preferentially in the vicinity of replication forks. Increased H3K9me3 is dependent upon the histone methyltransferases Suv39h1 and Suv39h2 whose protein levels increase under hypoxia. Olcina et al. demonstrated that the combination of replication stress and increased H3K9me3 together activate ATM, though neither is independently sufficient for ATM activation. Moreover, they suggest that hypoxia-induced ATM signaling is required for promoting replication progression by protecting forks in heterochromatin-like regions of DNA as inhibition of ATM leads to a decreased rate of DNA replication and an accumulation of DNA damage in S phase cells under hypoxic, but not normoxic conditions. ATM-dependent KAP1 phosphorylation may be important for chromatin relaxation to allow replication progression, but this hypothesis has not been formally tested. Of note, loss of CHK2 also leads to increased apoptosis under hypoxia [38], suggesting that hypoxia-induced ATM signaling may serve multiple distinct functions.
Following transient hypoxia, reoxygenation leads to accelerated and detectable DNA damage, primarily due to the production of reactive oxygen species. In response to this oxidative DNA damage, the ATM-CHK2 pathway is activated in a classical manner, resulting in 53BP1 foci formation and maintenance of p53 phosphorylation [25, 27]. Upon reoxygenation, ATM and CHK2 are required for cell cycle arrest in G2 phase via Cdc25C and Cdc2 phosphorylation and for protection from apoptosis [36, 37, 41]. If reoxygenation occurs within 8 to 12 hours, replication forks can reinitiate DNA synthesis; otherwise, the replisome is disassembled, preventing replication restart [29]. Both the ATM and ATR pathways appear to regulate firing of new origins of replication as inhibition of either CHK1 or ATM leads to an increase in new origin firing [29, 40]. Loss of ATR, ATM, CHK1, or CHK2 also sensitizes cells to reoxygenation [28, 36, 37], likely reflecting their importance in protecting cells from both hypoxia-induced replication stress and reoxygenation-induced DNA damage.
Hypoxia-induced activation of ATR and ATM signaling is independent of HIF, yet recent studies have revealed interesting crosstalk between the DNA damage response and HIF pathways. ATR kinase activity is required for accumulation of HIF-1α in the early hours of hypoxic exposure via an uncharacterized translational mechanism [42]. Under hypoxic stress, ATM also directly phosphorylates HIF-1α (S696), resulting in its stabilization [43]. DNA-PK similarly regulates HIF-1 protein stability, potentially via phosphorylation of HSF1 and upregulation of Hsp70/90 [44, 45]. BRCA1 also interacts with HIF-1, enhances its stability under hypoxia, and increases HIF-mediated gene transactivation [46]. Finally, hypoxia-induced phosphorylation of H2AX is partially HIF-dependent and is important for endothelial cell proliferation and neovascularization [47, 48]. Further studies to fully understand all the functions of hypoxia-modified DNA repair proteins will be critical, particularly as they are now being investigated as potential therapeutic targets (see below).
3. Transcriptional Downregulation of DNA Repair
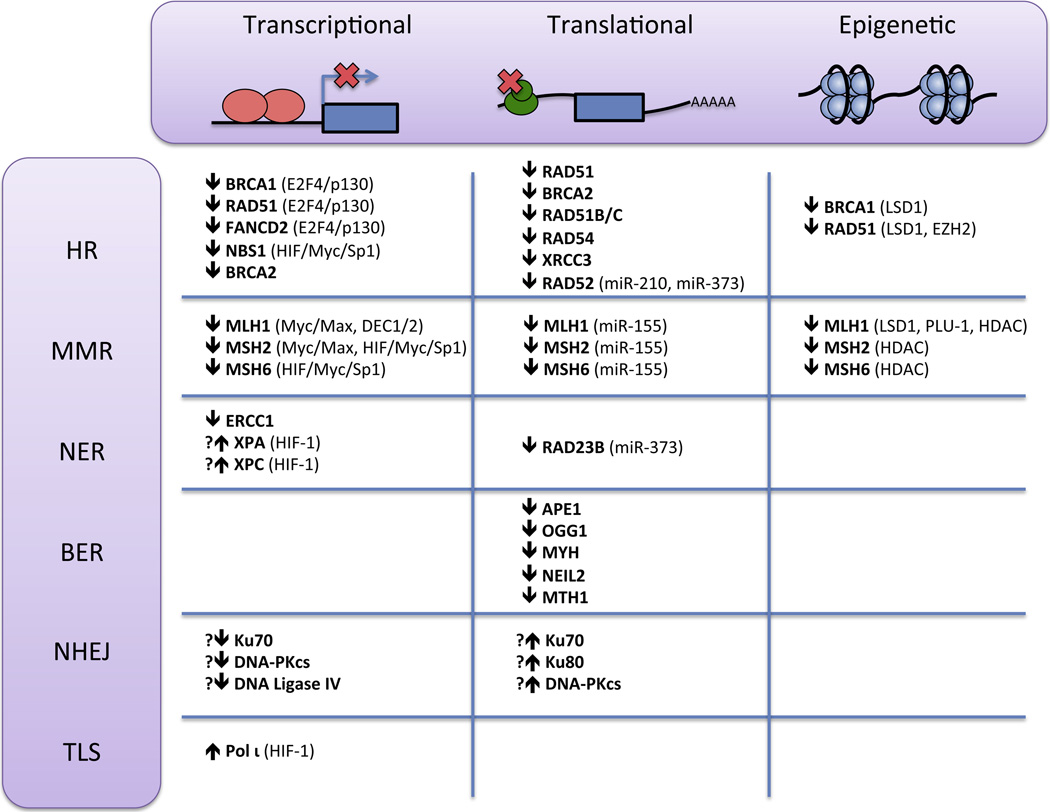
Transcriptional and translational control of gene expression enables cells to exert more sustained changes in protein function. Accordingly, more prolonged exposure, generally 12 to 72 hours, to either severe or moderate hypoxia results in transcriptional regulation of many DNA repair genes in various DNA repair pathways (Figure 2). Certain genes involved in mismatch repair (MMR), homologous recombination (HR), and potentially nucleotide excision repair (NER) undergo downregulation, while some reports suggest that non-homologous end joining (NHEJ) and translesion synthesis (TLS) genes may undergo upregulation. These changes in gene expression can persist upon initial return to normoxia but eventually reverse over the course of a few days.
Figure 2. Mechanisms involved in hypoxia-induced regulation of DNA repair gene expression.
The HR, MMR, NER, BER, NHEJ, and TLS DNA repair pathways undergo hypoxic regulation via transcriptional, translational, or epigenetic modulation of multiple DNA repair proteins. Transcription factors, microRNAs, and chromatin modifying enzymes implicated in regulating these proteins are indicated in parentheses.
MMR capacity is reduced under hypoxia as hypoxic cells display microsatellite instability, instability of dinucleotide-repeat sequences, and increased frameshift mutations in reporter genes [49–51]. Hypoxia has been shown to lead to transcriptional downregulation of the MMR genes, MLH1, MSH2, and MSH6, by a variety of different mechanisms. Under severe hypoxia (0.01% oxygen), MLH1 and MSH2 are downregulated at the protein and mRNA levels in a HIF-independent manner [49]. Mechanistically, severe hypoxia causes a decrease in Myc expression leading to a shift in occupancy at E-box motifs in proximal promoter regions of MLH1 and MSH2 from c-Myc/Max transcriptional activators to Mad1/Max and Mnt/Max transcriptional repressors [52]. Under more moderate hypoxia (1% oxygen), MSH2 and MSH6 are repressed in a HIF- and p53-dependent manner [50, 53]. In this case, HIF-1α displaces Myc from the MSH2 and MSH6 promoters to interact with constitutively bound Sp1 and repress expression. Moderate hypoxia also decreases MLH1 transcription in a HIF-dependent manner via the DEC1/2 transcriptional repressors, which are induced by hypoxia and bind to the E-box motif in the MLH1 promoter [54]. Finally, in stem cells, moderate hypoxia can induce decreased Sp1 binding at the MLH1, MSH2, and MSH6 promoters leading to their transcriptional repression in a HIF-independent manner [51]. Most of these mechanisms of hypoxia-induced MMR downregulation also require histone deacetylase (HDAC) activity, suggesting that coordination between histone modifications and transcription factor activity plays an important, though incompletely understood, role [49, 51, 54]. Altogether, these studies suggest that moderate hypoxia downregulates MMR genes in a HIF-dependent manner, while severe hypoxia can lead to transcriptional repression independent of HIF. The specific predominant pathway is also likely influenced by the cell type, genetic background, and experimental conditions.
Cellular HR capacity is diminished 3 to 8-fold in hypoxic cells as measured by recombination of a shuttle vector plasmid with donor DNA or by an intrachromosomal DSB repair assay [55–58]. The major mechanism underlying this repression of HR appears to be coordinated transcriptional downregulation of key HR mediators, BRCA1, RAD51, and FANCD2, by the E2F transcription factor network [33, 56, 59]. BRCA1, RAD51, and FANCD2 have conserved E2F consensus sites in their proximal promoter regions. In normoxic log phase cells, the E2F1 transcriptional activator and E2F4 and p130 transcriptional repressors bind simultaneously to the BRCA1 promoter and mediate a basal level of expression [60]. Upon severe hypoxic stress, independently of HIF and cell cycle phase, p130 undergoes dephosphorylation, nuclear accumulation, and increased binding to E2F4, leading to a shift in binding from the activating E2F1 factor to the repressive E2F4/p130 factor at the BRCA1 and RAD51 promoters [56, 59]. Hypoxia-induced FANCD2 mRNA and protein downregulation can be blocked by inhibition of p130, suggesting that its repression is also likely mediated by the E2F pathway in a manner analogous to BRCA1 and RAD51 [33]. Additional studies have suggested that hypoxia may lead to decreases in the mRNA and protein levels of other HR factors, including RAD51C, RAD51D, and XRCC3 [61]. BRCA2 was also identified in a microarray screen as being repressed by moderate hypoxia and confirmed to be downregulated at the protein and mRNA levels [62]. However, the mechanisms underlying these observations are unknown. Finally, moderate hypoxia (1% oxygen) leads to repression of another HR gene, NBS1, in manner similar to the repression of MSH2 under moderate hypoxia [63]. HIF-1α binds to Sp1 at the NBS1 promoter, displacing Myc and resulting in transcriptional repression. Whether NBS1 downregulation contributes to a decrease in HR function remains to be determined.
Unlike the clear inhibitory effect of hypoxia on MMR and HR, the impact of hypoxia on NER and NHEJ is less certain. Severe hypoxia reduces cellular NER capacity, as cells exposed simultaneously to hypoxia and low pH have decreased ability to repair a UV-damaged plasmid [64]. Cells also demonstrate hypermutability to UV irradiation on a chromosomal reporter gene when cultured under hypoxic and acidic conditions immediately after irradiation [64]. However, a recent study reported increased NER efficiency under moderate hypoxia (1.8% oxygen) [65]. The role of transcriptional regulation of NER genes is similarly not definitive. Many NER genes contain hypoxia response elements (HREs) in their promoter regions, and HIF has been shown to compete with the Sp1 transcriptional activator at the XPC promoter and to bind directly to the XPA promoter [66, 67]. It has not been determined whether these events could lead to downregulation under hypoxia, although HIF appears to upregulate rather than downregulate XPA and XPC gene expression under the conditions studied, cobalt chloride treatment and UV irradiation, respectively [66, 67]. Moreover, the protein levels of XPA, XPB, XPD, and XPG have been reported to be unchanged under hypoxic conditions [49, 64]. Recently, another gene involved in NER, ERRC1, was found to be downregulated at the mRNA and protein levels after moderate hypoxic exposure [68]. The underlying mechanism was not investigated, but these results suggest a promising explanation for the hypoxia-induced repression of NER. In addition, as ERCC1 also contributes to DNA crosslink repair, its hypoxic downregulation may have implications for tumor sensitivity to chemotherapeutic agents.
NHEJ and TLS are error-prone repair pathways, implying that their upregulation could lead to increased mutagenesis under hypoxia. Pol ι, an extremely low fidelity polymerase involved in TLS, undergoes transcriptional upregulation under hypoxia mediated by HIF-1 binding to a consensus HRE site located in intron 1 of the POLI gene [69]. By bypassing 8-oxo-dG lesions, Pol ι may contribute to oxidative point mutagenesis during hypoxia or reoxygenation, though this possibility has not been fully investigated [69]. Various studies have reported NHEJ gene expression and function to be unchanged, increased, or decreased. First, a luciferase plasmid religation assay showed no statistically significant change in end joining activity in hypoxic cells, though there did appear to be a trend towards increased activity under hypoxia [56]. This study found no change in the protein levels of Ku70 or Ku80. Another study found decreases in the mRNA levels of Ku70, DNA-PKcs, and DNA Ligase IV, but similarly found no change in the protein level of Ku70 after hypoxia [61]. In contrast, a recent proteomic study identified several NHEJ proteins to be upregulated under severe hypoxia [70]. Increased protein expression of Ku70, Ku80, and DNA-PKcs was confirmed in epithelial carcinoma cells, but changes in mRNA expression were not investigated. Another recent study found that moderate hypoxia (1.8% oxygen) increased NHEJ repair efficiency with a corresponding increase in NHEJ protein expression [65]. It has been suggested that BRCA1 S988 phosphorylation may promote precise NHEJ over error-prone alternative NHEJ, but also that downregulation of BRCA1 may favor the use of NHEJ over HR to repair DNA DSBs [39, 56]. Further studies, for example utilizing assays to measure competition between HR and NHEJ or to detect precise versus error-prone NHEJ, will be necessary to clarify the activity of NHEJ and whether it ultimately has a stabilizing or destabilizing effect on genomic integrity under hypoxia.
4. Translational Downregulation of DNA Repair
Under hypoxic stress, control of gene expression at the translational level occurs through at least two major mechanisms: regulation of translational efficiency via changes in translation initiation and induction of microRNA activity (Figure 2). Anoxia and hypoxia both lead to a HIF-independent global decrease in translation initiation mediated acutely by PERK-mediated phosphorylation of the eIF2α translation initiation factor and chronically by disruption of the eIF4F cap-binding complex [71]. Under anoxic conditions, translation efficiency decreases about 60% within 1–2 hours, and then recovers slightly and stabilizes at about 50% of control levels [72]. Less stringent hypoxia (0.2% oxygen) leads to a rapid, but less severe, decrease in translation efficiency to about 85% of control levels, which remains stable over 72 hours [73]. In addition to global repression of translation efficiency, hypoxia also leads to gene-specific regulation of translation, with different mRNA transcripts showing divergent patterns of translational repression or upregulation under acute and prolonged hypoxia [72, 74].
Recent studies have revealed that hypoxia can control expression of DNA repair genes via specific changes in translational efficiency of their mRNA transcripts [58, 75]. Chronic exposure to 0.2% oxygen, which produced little cellular toxicity, no impact on cell cycle distribution, and a mild repression of global translation, led to decreased protein expression of genes involved in HR, including RAD51, BRCA2, RAD51B/C, RAD54, and XRRC3, without changes in their mRNA expression levels [58]. Using polyribosome analysis, the authors demonstrated a decrease in translational efficiency of the RAD51 and BRCA2 mRNA, as evidenced by a decrease in the number of attached ribosomes per transcript. These conditions also induced a corresponding 3-fold reduction in HR activity. Under the same conditions, multiple genes involved in base excision repair (BER) are also translationally repressed, including APE1, OGG1, MYH, NEIL2, and MTH1 [75]. Importantly, this study demonstrated a functional deficiency in BER and increased cellular sensitivity to DNA base damaging agents under hypoxia, adding to the list of DNA repair pathways that are functionally downregulated by chronic hypoxia. These studies suggest that translational control of gene expression may play a significant role in repressing HR and BER under hypoxic conditions that do not induce transcriptional downregulation, and it will be interesting to determine whether additional DNA repair pathways are regulated in a similar manner.
A second major mechanism of translational adaptation induced by hypoxic stress is the regulation of microRNA activity. Hypoxic stress has been shown to regulate the expression of more than 90 microRNAs, which play a role in modulating cellular metabolism, angiogenesis, apoptosis, proliferation, and invasion, as well as DNA repair [76]. To date, three individual hypoxia-induced microRNAs, miR-210, miR-373, and miR-155, have been implicated in targeting DNA repair factors [77–79]. MiR-210, whose levels increase several-fold under both moderate and severe hypoxia in a HIF-dependent manner, correlates with tumor hypoxia in vivo and is associated with poor clinical outcomes in breast and head and neck cancer [80–83]. In addition to impacting many cellular pathways of hypoxic adaptation, it targets the DNA DSB repair gene RAD52 likely via a predicted binding site in the 3’ untranslated region of RAD52 mRNA [77]. RAD52 protein levels are reduced under hypoxia and can be partially restored with anti-miR-210, indicating yet another way in which hypoxia may downregulate HR activity. MiR-373, which is similarly induced by hypoxia, targets both RAD52 and RAD23B mRNA, mediating the repression of their protein products [77]. As RAD23B plays a role in DNA damage recognition for NER and potentially BER [84], hypoxia may also repress these repair pathways via miR-373. Finally, miR-155 targets several MMR factors, and its overexpression leads to downregulation the MLH1, MSH2, and MSH6 proteins without affecting their mRNA levels [78, 79]. Overexpression of miR-155 generates microsatellite instability, and in vivo, elevated expression of miR-155 is found in microsatellite unstable tumors with unknown MMR defects [78]. MicroRNAs thus appear to suppress multiple DNA repair pathways, and given the vast number of microRNAs regulated by hypoxia, additional effects of microRNAs on DNA repair are likely to exist.
5. Epigenetic Regulation of DNA Repair
Hypoxia-induced transcriptional and translational changes, though important mechanisms of gene regulation, are eventually reversible upon return to normoxia. Epigenetic changes, including histone modifications and DNA methylation, can cooperate with and modulate the transcriptional control activity of transactivators and transrepressors. Furthermore, these chromatin modifications can additionally permit long-term stable changes in gene expression that persist in the absence of continued hypoxic stress (Figure 2).
Both severe and moderate hypoxic conditions induce global epigenetic alterations, generating a hypoxic signature of chromatin modifications [85–87]. A recent quantitative proteomic study of the chromatin-associated proteome identified a large number of proteins that changed their chromatin association under hypoxia, including over 100 proteins involved in chromatin and transcription modulation, which likely represent factors mediating epigenetic changes as well as those responding to the changes [88]. The most commonly reported global histone changes under hypoxia involve histone H3 lysine 4 and lysine 9 di- and tri-methylation (H3K4me2, H3K4me3, H3K9me2, and HeK9me3). Hypoxia increases the total levels of H3K9me2 and H3K9me3, which generally promote gene repression [85, 89, 90]. The increase in H3K9me2 is partially dependent upon the methyltransferase G9a, which itself is upregulated under hypoxia [89]. In addition, decreased activity of oxygen-dependent Jumonji homology domain (JHDM) demethylases may contribute to increased H3K9me2 and H3K9me3 [90]. Global levels of H3K4me2 and H3K4me3, which facilitate an open chromatin conformation and transcriptional activation, are also increased under hypoxia, potentially via decreased activity of the JHDM demethylase JARID1A [85, 91]. Additional global hypoxia-induced histone alterations include the activating changes of increased H3K14 acetylation, H3K79me2, and H4R3me2, and the repressive changes of decreased H3K9 and H4 acetylation and increased H3K36me3 and H3K27me2/3 [85, 90, 92]. Finally, hypoxia alters DNA methylation patterns, with acute anoxia leading to a reduction in DNA methylation and prolonged exposure to moderate hypoxia resulting in increased DNA methylation that correlates with increased DNMT3b activity [86, 93].
In addition to global changes in histone modifications and DNA methylation, hypoxia can induce distinctive changes at specific promoters, leading to gene-specific epigenetic regulation. The promoters of DNA repair genes involved in MMR and HR are among those specifically modified under hypoxia. The importance of histone modifications at the promoters of MLH1, MSH2, and MSH6 was initially inferred from the finding that the pathways mediating their downregulation by transcription factors could be blocked by the HDAC inhibitor trichostatin A (TSA) [49, 51, 54]. Indeed, hypoxia leads to early repressive histone modification changes at the MLH1 promoter, including decreased H3K4 methylation via the LSD1/CoREST and PLU-1 demethylases, increased H3K9me2/3, and decreased H3K9 acetylation [89, 94]. Similar changes occur at the BRCA1 and RAD51 promoters, except that the decrease in H3K4 methylation is dependent on only LSD1 [95]. In addition, increased H3K27me3 at the RAD51 promoter, mediated by hypoxia-induced histone methyltransferase EZH2, leads to decreased RAD51 expression in breast tumor initiating cells [96]. Exactly how these histone modifications cooperate with transcription factors mediating MLH1, BRCA1, and RAD51 downregulation is not fully understood, but a likely possibility is that the histone modifications may impair or enhance binding of transcription factors to the promoters.
Beyond the early epigenetic changes induced by hypoxia, recent evidence suggests that prolonged moderate hypoxia (0.5–1% oxygen for several weeks) can induce stable silencing of the MLH1 and BRCA1 promoters [94, 95]. Reporter constructs containing the MLH1 or BRCA1 promoter underwent silencing at significantly higher frequency in hypoxic compared to normoxic cells, and the silencing persisted long after the cells were returned to normoxia. MLH1 promoter silencing was partially reversed with the DNA methylation inhibitor 5-aza-dC, while BRCA1 promoter silencing was reversed with TSA but not 5-aza-dC. Though similar, there may be subtle differences in the hypoxia-induced silencing of MLH1 and BRCA1. Given the array of chromatin changes induced by hypoxia, it will be interesting to determine whether additional DNA repair genes are epigenetically regulated.
6. Clinical Implications & Future Directions
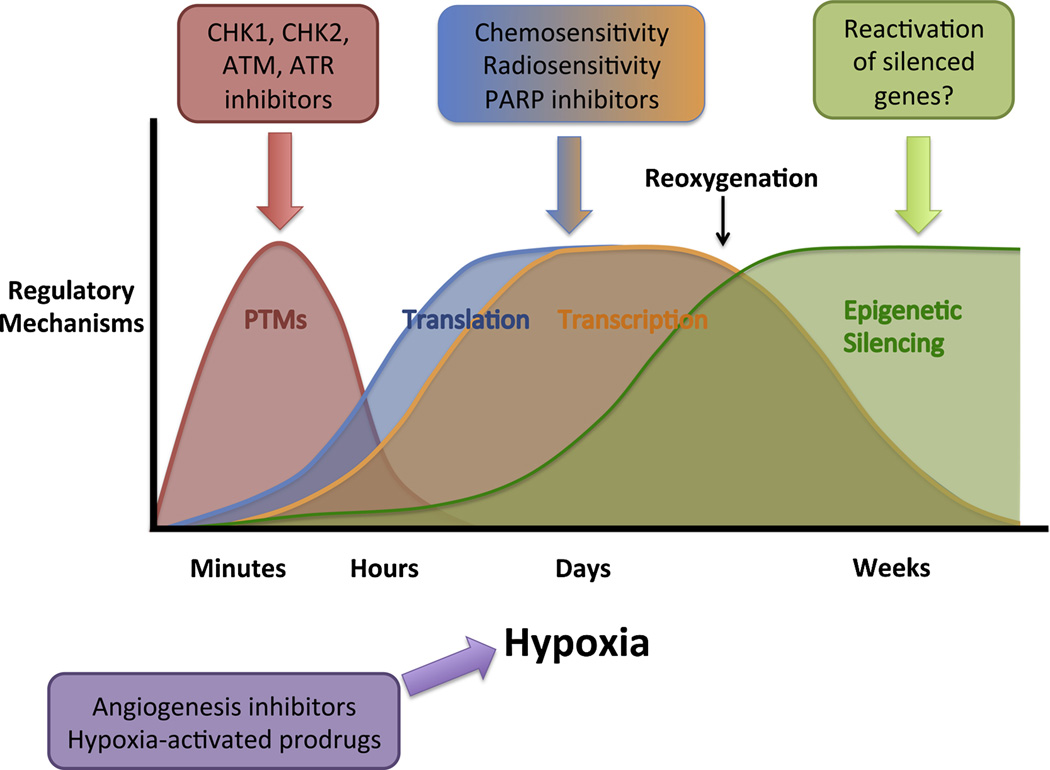
The knowledge accumulated over the past several decades about the influence of hypoxia on DNA repair has yielded suggestions for potential therapies targeting susceptibilities of hypoxic tumor cells, some of which are now in various stages of clinical development. As detailed in the previous sections, hypoxia modulates DNA repair by diverse mechanisms, many of which offer independent therapeutic promise (Figure 3). Acutely, PTMs of DNA repair proteins are required for responding to replication stress induced by severe hypoxia, and targeting these signaling pathways has been shown to sensitize cells to hypoxia-induced apoptosis. Under chronic moderate or severe hypoxia, transcriptional and translational downregulation of DNA repair introduces susceptibilities in hypoxic cells to DNA damaging agents or synthetic lethal approaches. Finally, prolonged hypoxia induces epigenetic changes that can lead to long-lasting changes in DNA repair gene expression, raising the possibility of trying to reactivate silenced tumor suppressor genes to mitigate hypoxia-induced genomic instability. An important step in the process of translating these findings to clinical medicine is the validation of the many mechanisms of DNA repair pathway modulation by hypoxia in in vivo tumor models. In this concluding section, we will describe the known in vivo correlations, discuss current investigations targeting DNA repair in hypoxic tumors, and end with focuses for future research.
Figure 3. General time course over which hypoxia induces different regulatory mechanisms controlling DNA repair and potential associated clinical implications.
Acute severe hypoxia activates PTM-mediated DNA damage signaling pathways that can be abrogated via inhibition of DNA damage signaling kinases. More chronic hypoxia leads to translational and transcriptional downregulation of DNA repair capacity, which generates cellular sensitivity to certain DNA damaging agents, radiotherapy, and PARP inhibitors. Prolonged moderate hypoxia induces stable silencing of specific DNA repair genes, which may be candidate tumor suppressor genes for reactivation. Drugs targeting hypoxia-induced DNA repair alterations may benefit from combination with agents that interact with hypoxia, such as angiogenesis inhibitors or hypoxia-activated prodrugs.
Several of the specific effects of hypoxia on DNA repair pathways have been validated in tumor models, supporting their possibility as targets for therapeutic intervention. First, concerning PTMs of DNA damage signaling proteins, phosphorylated p53 and γH2AX have been shown to colocalize in hypoxic regions of tumors grown in mice [25]. Recently, phosphorylated ATM and the histone modification supporting ATM activation (H3K9me3) were found to associate with the hypoxia marker CAIX in xenograft tumors [40]. These findings support the conclusion that both the ATR and the ATM signaling pathways are activated in hypoxic tumor regions, though the activity of many of their downstream targets remain to be investigated in vivo. In terms of DNA repair repression, factors involved in the MMR and HR pathways have been found to undergo regulation associated with in vivo hypoxia. MLH1 and MSH2 expression has been shown to inversely correlate with markers of hypoxia (CAIX or pimonidazole) in colon cancer mouse xenografts [94, 97]. In addition, MSH2 expression was inversely correlated with HIF-1α expression in human sporadic colon cancers containing wild type p53 [50, 53]. The HR factor RAD51 has been found to be downregulated in hypoxic regions of tumors, both at the protein level via immunofluorescence staining in cervical and prostate cancer xenografts and at the mRNA level via laser-capture microdissection and mRNA expression analysis in a glioma tumor model [55, 98]. Finally, BRCA1 protein expression was also found to inversely correlate with CAIX staining in a human breast cancer cohort, and CAIX positively correlated with a BRCA1 mutant signature indicating loss of BRCA1 function [99]. Altogether, these findings provide strong evidence that many consequences of hypoxic modulation of DNA repair reported in vitro can be recapitulated in vivo.
The acute activation of the ATR and ATM signaling pathways by hypoxia-induced replication arrest represents one component of the hypoxic stress response amenable to therapeutic intervention. As previously described, cells in which ATR, CHK1, ATM, or CHK2 have been depleted or inhibited exhibit increased sensitivity to both hypoxia and reoxygenation [28, 36–38, 100]. These observations suggest that targeting these kinases may selectively kill cancer cells in the hypoxic tumor microenvironment. Currently, CHK1 and combined CHK1/CHK2 inhibitors are being evaluated in clinical trials, primarily for their ability to potentiate the effects of DNA-damaging chemotherapy and radiotherapy [101]. Several first-generation inhibitors entered Phase 1 or 2 clinical trials in combination with cytotoxic chemotherapy, but encountered problems with toxicity or pharmacokinetics. Two newer agents, which may show more favorable toxicity profiles, are currently in Phase 1 trials in combination with gemcitabine. In addition to these combination therapy studies, a dual CHK1/CHK2 inhibitor (LY2606368) has recently entered trials as a single agent in patients with advanced cancer or with BRCA1/2 mutant breast or ovarian cancer (NCT01115790 and NCT02203513). ATM and ATR inhibitors are still in preclinical development, but are generating promising results. The ATM inhibitor KU59403 was recently shown to sensitize cells to both chemotherapy and radiation and to enhance the antitumor activity of topoisomerase inhibitors in human colon cancer mouse xenografts without major toxicity [102]. A novel ATR inhibitor VE-821 also sensitizes cells to radiotherapy, both in normoxic and hypoxic conditions, and can inhibit the growth of cancer cells in 3D spheroid models containing regions of hypoxia [103]. As of yet, no clinical studies have directly investigated the ability of DNA damage response kinase inhibitors to target hypoxic tumors or to synergize with modulators of angiogenesis. However, given the efficacy of kinase inactivation under hypoxia in vitro, as well as the advances being made in in vivo hypoxia imaging, these should be important directions for future study.
Downregulation of DNA repair capacity under chronic hypoxia offers a second opportunity to exploit the hypoxic tumor microenvironment in a clinical setting. Although hypoxic cells are generally resistant to radiotherapy and chemotherapy requiring active cell proliferation, the repression of DNA repair induces sensitivity to certain genotoxic agents. For example, hypoxic cells demonstrate increased sensitivity to the DNA crosslinking agents mitomycin C and cisplatin, likely due to the functional decrease in HR capacity [58, 104]. The downregulation of HR also appears to confer radiosensitivity under chronic hypoxia and in the immediate post-hypoxic period, in contrast to the radioresistance observed during acute hypoxia [58, 105]. Fewer DSBs are generated during irradiation in chronically hypoxic cells, but they persist at higher levels, leading to increased cell death under continual hypoxia [105]. The hypoxia-induced downregulation of BER also sensitizes chronically hypoxic cells to oxidative or alkylating DNA damage with hydrogen peroxide or methyl methanesulfonate and results in an accumulation of residual base damage [75].
Hypoxia-induced downregulation of DNA repair can also be combined with inhibition of complementary repair mechanisms to generate a synthetic lethal interaction in hypoxic cells. As a prime example, inhibition of the single-strand break repair protein poly(ADP-ribose) polymerase-1 (PARP-1) leads to the formation of DSBs that fail to be repaired in the setting of hypoxia-induced HR repression [106]. Hypoxic cells are more sensitive to PARP inhibition, and this susceptibility can be partially reversed by RAD51 overexpression [106]. Additionally, in vivo treatment of colon cancer xenografts with a PARP inhibitor can induce DNA damage and increase cell death specifically in hypoxic cells [106]. Interestingly, PARP inhibitors themselves lead to downregulation of BRCA1 and RAD51, which may factor into their radiosensitizing effects [107, 108]. Several PARP inhibitors are currently undergoing clinical investigation in human trials, both as single agents and in combination with genotoxic chemotherapy, and have shown clinical benefit in BRCA1/2-mutant or BRCA-like cancers [109]. Recently, trials have begun to examine the possibility that hypoxia and PARP inhibition may have clinical synergistic effects by investigating the combined treatment of PARP inhibitors and angiogenesis inhibitors. Phase 1 studies have found this dual therapy to have promising activity with manageable toxicities, and a Phase 2 study is now in progress [110, 111]. If this pursuit continues to yield encouraging results, it will support the investigation of additional proposals to specifically target DNA repair susceptibilities under hypoxia, such as the combination of other DNA damage signaling inhibitors with angiogenesis modulators or the development of hypoxia-activated prodrugs whose active metabolites could exploit DNA repair dysfunction [112–114].
Through the research efforts directed at understanding the effects of hypoxia on DNA repair, much has been learned about the diverse mechanisms underlying hypoxia-induced genetic instability, and these findings are now beginning to be applied towards clinical therapies. Nevertheless, important questions remain regarding some of the specific pathways regulated by hypoxic stress. What additional DNA repair proteins are altered post-translationally by acute hypoxic stress? How does hypoxia ultimately regulate NHEJ and NER? Can hypoxia induce silencing of other genes involved in genomic maintenance and by what mechanisms? In addition to addressing these and other basic mechanistic questions, it will also be important to extend many of the results from artificial in vitro systems to more physiologic conditions. Several DNA repair factor PTMs and changes in DNA repair gene expression have been observed in tumor models, but many other findings have not yet been confirmed in vivo. The mechanisms of transcriptional or translational downregulation that are utilized in vivo and whether intra-tumor hypoxia induces gene silencing also remain unknown. In addition, the interaction between hypoxia and other microenvironmental factors, such as low pH, low glucose, and inflammation, and its impact on DNA repair require further study. Ultimately, a more complete understanding of DNA repair pathway regulation under hypoxia will aid in the development of cancer therapies that explicitly take advantage of DNA repair alterations in the hypoxic tumor microenvironment.
Acknowledgements
This work was supported by NIH grant R01ES005775 to PMG and NIH Medical Scientist Program Training Grant T32GM007205.
Abbreviations
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related
- BER
base excision repair
- DSB
double-strand break
- H3K4me2
histone H3 lysine 4 dimethylation
- H3K4me3
histone H3 lysine 4 trimethylation
- H3K9me2
histone H3 lysine 9 dimethylation
- H3K9me3
histone H3 lysine 9 trimethylation
- HIF
hypoxia-inducible factor
- HDAC
histone deacetylase
- HR
homologous recombination
- HRE
hypoxia response element
- JHDM
Jumonji homology domain
- MMR
mismatch repair
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- PARP
poly(ADP-ribose) polymerase
- PTM
post-translational modification
- TLS
translesion synthesis
- TSA
trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors have no conflict of interest to declare.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Seminars in radiation oncology. 2004;14:198–206. doi: 10.1016/j.semradonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Multhoff G, Radons J, Vaupel P. Critical role of aberrant angiogenesis in the development of tumor hypoxia and associated radioresistance. Cancers. 2014;6:813–828. doi: 10.3390/cancers6020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer metastasis reviews. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 5.Jubb AM, Buffa FM, Harris AL. Assessment of tumour hypoxia for prediction of response to therapy and cancer prognosis. Journal of cellular and molecular medicine. 2010;14:18–29. doi: 10.1111/j.1582-4934.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaupel P, Mayer A, Hockel M. Tumor hypoxia and malignant progression. Methods in enzymology. 2004;381:335–354. doi: 10.1016/S0076-6879(04)81023-1. [DOI] [PubMed] [Google Scholar]
- 7.Vaupel P, Mayer A. Hypoxia in tumors: pathogenesis-related classification, characterization of hypoxia subtypes, and associated biological and clinical implications. Advances in experimental medicine and biology. 2014;812:19–24. doi: 10.1007/978-1-4939-0620-8_3. [DOI] [PubMed] [Google Scholar]
- 8.Chang J, Erler J. Hypoxia-mediated metastasis. Advances in experimental medicine and biology. 2014;772:55–81. doi: 10.1007/978-1-4614-5915-6_3. [DOI] [PubMed] [Google Scholar]
- 9.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nature reviews. Molecular cell biology. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 10.Webb JD, Coleman ML, Pugh CW. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cellular and molecular life sciences : CMLS. 2009;66:3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nature reviews. Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 12.Taylor CT, Cummins EP. The role of NF-kappaB in hypoxia-induced gene expression. Annals of the New York Academy of Sciences. 2009;1177:178–184. doi: 10.1111/j.1749-6632.2009.05024.x. [DOI] [PubMed] [Google Scholar]
- 13.Bindra RS, Glazer PM. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutation research. 2005;569:75–85. doi: 10.1016/j.mrfmmm.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Paquette B, Little JB. In vivo enhancement of genomic instability in minisatellite sequences of mouse C3H/10T1/2 cells transformed in vitro by X-rays. Cancer research. 1994;54:3173–3178. [PubMed] [Google Scholar]
- 15.Wilkinson D, Sandhu JK, Breneman JW, Tucker JD, Birnboim HC. Hprt mutants in a transplantable murine tumour arise more frequently in vivo than in vitro. British journal of cancer. 1995;72:1234–1240. doi: 10.1038/bjc.1995.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer research. 1996;56:5754–5757. [PubMed] [Google Scholar]
- 17.Li CY, Little JB, Hu K, Zhang W, Zhang L, Dewhirst MW, Huang Q. Persistent genetic instability in cancer cells induced by non-DNA-damaging stress exposures. Cancer research. 2001;61:428–432. [PubMed] [Google Scholar]
- 18.Papp-Szabo E, Josephy PD, Coomber BL. Microenvironmental influences on mutagenesis in mammary epithelial cells, International journal of cancer. Journal international du cancer. 2005;116:679–685. doi: 10.1002/ijc.21088. [DOI] [PubMed] [Google Scholar]
- 19.Rice GC, Hoy C, Schimke RT. Transient hypoxia enhances the frequency of dihydrofolate reductase gene amplification in Chinese hamster ovary cells. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:5978–5982. doi: 10.1073/pnas.83.16.5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young SD, Marshall RS, Hill RP. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9533–9537. doi: 10.1073/pnas.85.24.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Molecular cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- 22.Fischer U, Radermacher J, Mayer J, Mehraein Y, Meese E. Tumor hypoxia: Impact on gene amplification in glioblastoma. International journal of oncology. 2008;33:509–515. [PubMed] [Google Scholar]
- 23.Coquelle A, Rozier L, Dutrillaux B, Debatisse M. Induction of multiple double-strand breaks within an hsr by meganucleaseI-SceI expression or fragile site activation leads to formation of double minutes and other chromosomal rearrangements. Oncogene. 2002;21:7671–7679. doi: 10.1038/sj.onc.1205880. [DOI] [PubMed] [Google Scholar]
- 24.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia Links ATR and p53 through Replication Arrest. Molecular and cellular biology. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. The Journal of biological chemistry. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 26.Hammond EM, Green SL, Giaccia AJ. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutation research. 2003;532:205–213. doi: 10.1016/j.mrfmmm.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. ATM activation and signaling under hypoxic conditions. Molecular and cellular biology. 2009;29:526–537. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond EM, Dorie MJ, Giaccia AJ. Inhibition of ATR leads to increased sensitivity to hypoxia/reoxygenation. Cancer research. 2004;64:6556–6562. doi: 10.1158/0008-5472.CAN-04-1520. [DOI] [PubMed] [Google Scholar]
- 29.Pires IM, Bencokova Z, Milani M, Folkes LK, Li JL, Stratford MR, Harris AL, Hammond EM. Effects of acute versus chronic hypoxia on DNA damage responses and genomic instability. Cancer research. 2010;70:925–935. doi: 10.1158/0008-5472.CAN-09-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond EM, Giaccia AJ. The role of ATM and ATR in the cellular response to hypoxia and re-oxygenation. DNA repair. 2004;3:1117–1122. doi: 10.1016/j.dnarep.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 31.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Advances in cancer research. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 32.Pires IM, Bencokova Z, McGurk C, Hammond EM. Exposure to acute hypoxia induces a transient DNA damage response which includes Chk1 and TLK1. Cell cycle. 2010;9:2502–2507. doi: 10.4161/cc.9.13.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scanlon SE, Glazer PM. Hypoxic stress facilitates acute activation and chronic downregulation of fanconi anemia proteins. Molecular cancer research : MCR. 2014;12:1016–1028. doi: 10.1158/1541-7786.MCR-13-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, Giaccia A. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Molecular and cellular biology. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Jin Y, He G, Zeng SX, Wang YV, Wahl GM, Lu H. Hypoxia activates tumor suppressor p53 by inducing ATR-Chk1 kinase cascade-mediated phosphorylation and consequent 14-3-3gamma inactivation of MDMX protein. The Journal of biological chemistry. 2012;287:20898–20903. doi: 10.1074/jbc.M111.336875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freiberg RA, Hammond EM, Dorie MJ, Welford SM, Giaccia AJ. DNA damage during reoxygenation elicits a Chk2-dependent checkpoint response. Molecular and cellular biology. 2006;26:1598–1609. doi: 10.1128/MCB.26.5.1598-1609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freiberg RA, Krieg AJ, Giaccia AJ, Hammond EM. Checking in on hypoxia/reoxygenation. Cell cycle. 2006;5:1304–1307. doi: 10.4161/cc.5.12.2811. [DOI] [PubMed] [Google Scholar]
- 38.Gibson SL, Bindra RS, Glazer PM. Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer research. 2005;65:10734–10741. doi: 10.1158/0008-5472.CAN-05-1160. [DOI] [PubMed] [Google Scholar]
- 39.Gibson SL, Bindra RS, Glazer PM. CHK2-dependent phosphorylation of BRCA1 in hypoxia. Radiation research. 2006;166:646–651. doi: 10.1667/RR0660.1. [DOI] [PubMed] [Google Scholar]
- 40.Olcina MM, Foskolou IP, Anbalagan S, Senra JM, Pires IM, Jiang Y, Ryan AJ, Hammond EM. Replication stress and chromatin context link ATM activation to a role in DNA replication. Molecular cell. 2013;52:758–766. doi: 10.1016/j.molcel.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim BM, Choi JY, Kim YJ, Woo HD, Chung HW. Reoxygenation following hypoxia activates DNA-damage checkpoint signaling pathways that suppress cell-cycle progression in cultured human lymphocytes. FEBS letters. 2007;581:3005–3012. doi: 10.1016/j.febslet.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 42.Fallone F, Britton S, Nieto L, Salles B, Muller C. ATR controls cellular adaptation to hypoxia through positive regulation of hypoxia-inducible factor 1 (HIF-1) expression. Oncogene. 2013;32:4387–4396. doi: 10.1038/onc.2012.462. [DOI] [PubMed] [Google Scholar]
- 43.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Molecular cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang MJ, Jung SM, Kim MJ, Bae JH, Kim HB, Kim JY, Park SJ, Song HS, Kim DW, Kang CD, Kim SH. DNA-dependent protein kinase is involved in heat shock protein-mediated accumulation of hypoxia-inducible factor-1alpha in hypoxic preconditioned HepG2 cells. The FEBS journal. 2008;275:5969–5981. doi: 10.1111/j.1742-4658.2008.06725.x. [DOI] [PubMed] [Google Scholar]
- 45.Bouquet F, Ousset M, Biard D, Fallone F, Dauvillier S, Frit P, Salles B, Muller C. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. Journal of cell science. 2011;124:1943–1951. doi: 10.1242/jcs.078030. [DOI] [PubMed] [Google Scholar]
- 46.Kang HJ, Kim HJ, Rih JK, Mattson TL, Kim KW, Cho CH, Isaacs JS, Bae I. BRCA1 plays a role in the hypoxic response by regulating HIF-1alpha stability and by modulating vascular endothelial growth factor expression. The Journal of biological chemistry. 2006;281:13047–13056. doi: 10.1074/jbc.M513033200. [DOI] [PubMed] [Google Scholar]
- 47.Economopoulou M, Langer HF, Celeste A, Orlova VV, Choi EY, Ma M, Vassilopoulos A, Callen E, Deng C, Bassing CH, Boehm M, Nussenzweig A, Chavakis T. Histone H2AX is integral to hypoxia-driven neovascularization. Nature medicine. 2009;15:553–558. doi: 10.1038/nm.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrann S, Kaufmann MR, Wirthner R, Stiehl DP, Wenger RH. HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biological chemistry. 2013;394:519–528. doi: 10.1515/hsz-2012-0311. [DOI] [PubMed] [Google Scholar]
- 49.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Molecular and cellular biology. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Molecular cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Jimenez FJ, Moreno-Manzano V, Lucas-Dominguez R, Sanchez-Puelles JM. Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells. Stem cells. 2008;26:2052–2062. doi: 10.1634/stemcells.2007-1016. [DOI] [PubMed] [Google Scholar]
- 52.Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer letters. 2007;252:93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 53.To KK, Koshiji M, Hammer S, Huang LE. Genetic instability: the dark side of the hypoxic response. Cell cycle. 2005;4:881–882. doi: 10.4161/cc.4.7.1839. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E, Nishiyama M. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors, DEC1 and DEC2. Oncogene. 2008;27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- 55.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Molecular and cellular biology. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer research. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 57.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Alterations in DNA repair gene expression under hypoxia: elucidating the mechanisms of hypoxia-induced genetic instability. Annals of the New York Academy of Sciences. 2005;1059:184–195. doi: 10.1196/annals.1339.049. [DOI] [PubMed] [Google Scholar]
- 58.Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, Belmaaza A, Wouters B, Bristow RG. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer research. 2008;68:605–614. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- 59.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 60.Bindra RS, Glazer PM. Basal repression of BRCA1 by multiple E2Fs and pocket proteins at adjacent E2F sites. Cancer biology & therapy. 2006;5:1400–1407. doi: 10.4161/cbt.5.10.3454. [DOI] [PubMed] [Google Scholar]
- 61.Meng AX, Jalali F, Cuddihy A, Chan N, Bindra RS, Glazer PM, Bristow RG. Hypoxia down-regulates DNA double strand break repair gene expression in prostate cancer cells. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2005;76:168–176. doi: 10.1016/j.radonc.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 62.Fanale D, Bazan V, Caruso S, Castiglia M, Bronte G, Rolfo C, Cicero G, Russo A. Hypoxia and human genome stability: downregulation of BRCA2 expression in breast cancer cell lines. BioMed research international. 2013;2013 doi: 10.1155/2013/746858. 746858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. The EMBO journal. 2006;25:4784–4794. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan J, Narayanan L, Rockwell S, Glazer PM. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer research. 2000;60:4372–4376. [PubMed] [Google Scholar]
- 65.Madan E, Gogna R, Pati U. p53 Ser15 phosphorylation disrupts the p53-RPA70 complex and induces RPA70-mediated DNA repair in hypoxia. The Biochemical journal. 2012;443:811–820. doi: 10.1042/BJ20111627. [DOI] [PubMed] [Google Scholar]
- 66.Rezvani HR, Mahfouf W, Ali N, Chemin C, Ged C, Kim AL, de Verneuil H, Taieb A, Bickers DR, Mazurier F. Hypoxia-inducible factor-1alpha regulates the expression of nucleotide excision repair proteins in keratinocytes. Nucleic acids research. 2010;38:797–809. doi: 10.1093/nar/gkp1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Bernauer AM, Yingling CM, Belinsky SA. HIF1alpha regulated expression of XPA contributes to cisplatin resistance in lung cancer. Carcinogenesis. 2012;33:1187–1192. doi: 10.1093/carcin/bgs142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dudas J, Schartinger VH, Romani A, Schweigl G, Kordsmeyer K, Marta PI, Url C, Kral F, Riechelmann H. Cell cycle association and hypoxia regulation of excision repair cross complementation group 1 protein (ERCC1) in tumor cells of head and neck cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014 doi: 10.1007/s13277-014-2001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ito A, Koshikawa N, Mochizuki S, Omura K, Takenaga K. Hypoxia-inducible factor-1 mediates the expression of DNA polymerase iota in human tumor cells. Biochemical and biophysical research communications. 2006;351:306–311. doi: 10.1016/j.bbrc.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 70.Ren Y, Hao P, Dutta B, Cheow ES, Sim KH, Gan CS, Lim SK, Sze SK. Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Molecular & cellular proteomics : MCP. 2013;12:485–498. doi: 10.1074/mcp.M112.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koritzinsky M, Wouters BG. Hypoxia and regulation of messenger RNA translation. Methods in enzymology. 2007;435:247–273. doi: 10.1016/S0076-6879(07)35013-1. [DOI] [PubMed] [Google Scholar]
- 72.Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. The EMBO journal. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koritzinsky M, Rouschop KM, van den Beucken T, Magagnin MG, Savelkouls K, Lambin P, Wouters BG. Phosphorylation of eIF2alpha is required for mRNA translation inhibition and survival during moderate hypoxia. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2007;83:353–361. doi: 10.1016/j.radonc.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 74.Koritzinsky M, Seigneuric R, Magagnin MG, van den Beucken T, Lambin P, Wouters BG. The hypoxic proteome is influenced by gene-specific changes in mRNA translation. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2005;76:177–186. doi: 10.1016/j.radonc.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 75.Chan N, Ali M, McCallum GP, Kumareswaran R, Koritzinsky M, Wouters BG, Wells PG, Gallinger S, Bristow RG. Hypoxia Provokes Base Excision Repair Changes and a Repair-deficient, Mutator Phenotype in Colorectal Cancer Cells. Molecular cancer research : MCR. 2014 doi: 10.1158/1541-7786.MCR-14-0246. [DOI] [PubMed] [Google Scholar]
- 76.Pocock R. Invited review: decoding the microRNA response to hypoxia. Pflugers Archiv : European journal of physiology. 2011;461:307–315. doi: 10.1007/s00424-010-0910-5. [DOI] [PubMed] [Google Scholar]
- 77.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer research. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, Costinean S, Sandhu SK, Nuovo GJ, Alder H, Gafa R, Calore F, Ferracin M, Lanza G, Volinia S, Negrini M, McIlhatton MA, Amadori D, Fishel R, Croce CM. Modulation of mismatch repair and genomic stability by miR-155. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Babar IA, Czochor J, Steinmetz A, Weidhaas JB, Glazer PM, Slack FJ. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer biology & therapy. 2011;12:908–914. doi: 10.4161/cbt.12.10.17681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM, Ragoussis J, Harris AL. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 81.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends in molecular medicine. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 84.Miao F, Bouziane M, Dammann R, Masutani C, Hanaoka F, Pfeifer G, O'Connor TR. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. The Journal of biological chemistry. 2000;275:28433–28438. doi: 10.1074/jbc.M001064200. [DOI] [PubMed] [Google Scholar]
- 85.Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutation research. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watson JA, Watson CJ, McCrohan AM, Woodfine K, Tosetto M, McDaid J, Gallagher E, Betts D, Baugh J, O'Sullivan J, Murrell A, Watson RW, McCann A. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Human molecular genetics. 2009;18:3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 87.Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics, the epicenter of the hypoxic response. Epigenetics : official journal of the DNA Methylation Society. 2010;5:293–296. doi: 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- 88.Dutta B, Ren Y, Lim SK, Tam JP, Sze SK. Quantitative profiling of chromatome dynamics reveals a novel role for HP1BP3 in hypoxia-induced oncogenesis. Molecular & cellular proteomics : MCP. 2014 doi: 10.1074/mcp.M114.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer research. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- 90.Tausendschon M, Dehne N, Brune B. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011;53:256–262. doi: 10.1016/j.cyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 91.Zhou X, Sun H, Chen H, Zavadil J, Kluz T, Arita A, Costa M. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer research. 2010;70:4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Q, Costa M. c-Myc mediates a hypoxia-induced decrease in acetylated histone H4. Biochimie. 2009;91:1307–1310. doi: 10.1016/j.biochi.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics : official journal of the DNA Methylation Society. 2007;2:119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 94.Lu Y, Wajapeyee N, Turker MS, Glazer PM. Silencing of the DNA Mismatch Repair Gene MLH1 Induced by Hypoxic Stress in a Pathway Dependent on the Histone Demethylase LSD1. Cell reports. 2014;8:501–513. doi: 10.1016/j.celrep.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Molecular and cellular biology. 2011;31:3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shahrzad S, Quayle L, Stone C, Plumb C, Shirasawa S, Rak JW, Coomber BL. Ischemia-induced K-ras mutations in human colorectal cancer cells: role of microenvironmental regulation of MSH2 expression. Cancer research. 2005;65:8134–8141. doi: 10.1158/0008-5472.CAN-05-0713. [DOI] [PubMed] [Google Scholar]
- 98.Marotta D, Karar J, Jenkins WT, Kumanova M, Jenkins KW, Tobias JW, Baldwin D, Hatzigeorgiou A, Alexiou P, Evans SM, Alarcon R, Maity A, Koch C, Koumenis C. In vivo profiling of hypoxic gene expression in gliomas using the hypoxia marker EF5 and laser-capture microdissection. Cancer research. 2011;71:779–789. doi: 10.1158/0008-5472.CAN-10-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neumeister VM, Sullivan CA, Lindner R, Lezon-Geyda K, Li J, Zavada J, Martel M, Glazer PM, Tuck DP, Rimm DL, Harris L. Hypoxia-induced protein CAIX is associated with somatic loss of BRCA1 protein and pathway activity in triple negative breast cancer. Breast cancer research and treatment. 2012;136:67–75. doi: 10.1007/s10549-012-2232-0. [DOI] [PubMed] [Google Scholar]
- 100.Hammond EM, Freiberg RA, Giaccia AJ. The roles of Chk 1 and Chk 2 in hypoxia and reoxygenation. Cancer letters. 2006;238:161–167. doi: 10.1016/j.canlet.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 101.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacology & therapeutics. 2014;142:1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 102.Batey MA, Zhao Y, Kyle S, Richardson C, Slade A, Martin NM, Lau A, Newell DR, Curtin NJ. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Molecular cancer therapeutics. 2013;12:959–967. doi: 10.1158/1535-7163.MCT-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pires IM, Olcina MM, Anbalagan S, Pollard JR, Reaper PM, Charlton PA, McKenna WG, Hammond EM. Targeting radiation-resistant hypoxic tumour cells through ATR inhibition. British journal of cancer. 2012;107:291–299. doi: 10.1038/bjc.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strese S, Fryknas M, Larsson R, Gullbo J. Effects of hypoxia on human cancer cell line chemosensitivity. BMC cancer. 2013;13:331. doi: 10.1186/1471-2407-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumareswaran R, Ludkovski O, Meng A, Sykes J, Pintilie M, Bristow RG. Chronic hypoxia compromises repair of DNA double-strand breaks to drive genetic instability. Journal of cell science. 2012;125:189–199. doi: 10.1242/jcs.092262. [DOI] [PubMed] [Google Scholar]
- 106.Chan N, Pires IM, Bencokova Z, Coackley C, Luoto KR, Bhogal N, Lakshman M, Gottipati P, Oliver FJ, Helleday T, Hammond EM, Bristow RG. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer research. 2010;70:8045–8054. doi: 10.1158/0008-5472.CAN-10-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2008;88:258–268. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 108.Hegan DC, Lu Y, Stachelek GC, Crosby ME, Bindra RS, Glazer PM. Inhibition of poly(ADP-ribose) polymerase down-regulates BRCA1 and RAD51 in a pathway mediated by E2F4 and p130. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2201–2206. doi: 10.1073/pnas.0904783107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25:32–40. doi: 10.1093/annonc/mdt384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dean E, Middleton MR, Pwint T, Swaisland H, Carmichael J, Goodege-Kunwar P, Ranson M. Phase I study to assess the safety and tolerability of olaparib in combination with bevacizumab in patients with advanced solid tumours. British journal of cancer. 2012;106:468–474. doi: 10.1038/bjc.2011.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, Lee H, Whalen C, Tyburski K, Winer E, Ivy P, Matulonis UA. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. European journal of cancer. 2013;49:2972–2978. doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hunter FW, Hsu HL, Su J, Pullen SM, Wilson WR, Wang J. Dual targeting of hypoxia and homologous recombination repair dysfunction in triple-negative breast cancer. Molecular cancer therapeutics. 2014 doi: 10.1158/1535-7163.MCT-14-0476. [DOI] [PubMed] [Google Scholar]
- 113.Olcina M, Lecane PS, Hammond EM. Targeting hypoxic cells through the DNA damage response. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:5624–5629. doi: 10.1158/1078-0432.CCR-10-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nature reviews. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]