Abstract
Over the past half-century, we have become increasingly aware of the ubiquity of DNA damage. Under the constant exposure to exogenous and endogenous genomic stress, cells must attempt to replicate damaged DNA. The encounter of replication forks with DNA lesions triggers several cellular responses, including the activation of translesion DNA synthesis (TLS), which largely depends upon specialized DNA polymerases with flexible active sites capable of accommodating bulky DNA lesions. A detrimental aspect of TLS is its intrinsic mutagenic nature, and thus the activity of the TLS polymerases must ideally be restricted to synthesis on damaged DNA templates. Despite their potential clinical importance in chemotherapy, TLS inhibitors have been difficult to identify since a direct assay designed to quantify genomic TLS events is still unavailable. Herein we discuss the methods that have been used to validate TLS inhibitors such as USP1, p21 and Spartan, highlighting research that has revealed their contribution to the control of DNA synthesis on damaged and undamaged templates.
Keywords: USP1, p21, Spartan, PCNA, DNA replication, mutagenesis, UV irradiation
The basics of translesion DNA synthesis
To promote damaged-DNA replication, TLS relies on the Y-family of DNA polymerases (Polη, Polι, Polκ and Rev1) and on the B-family member, Polζ. Either one polymerase, or two TLS polymerases in concert, operate to achieve the bypass of most types of DNA lesions. As depicted in Figure 1, while TLS across moderate distortions such as UV-induced cyclobutane pyrimidine dimers (CPDs) depends exclusively on Polη, TLS across bulkier adducts including UV-induced 6-4 photoproducts (6-4PPs) comprises at least two specialized polymerases, in which Polζ carries out an extension step that follows the lesion bypass step driven by Y-pols [1].
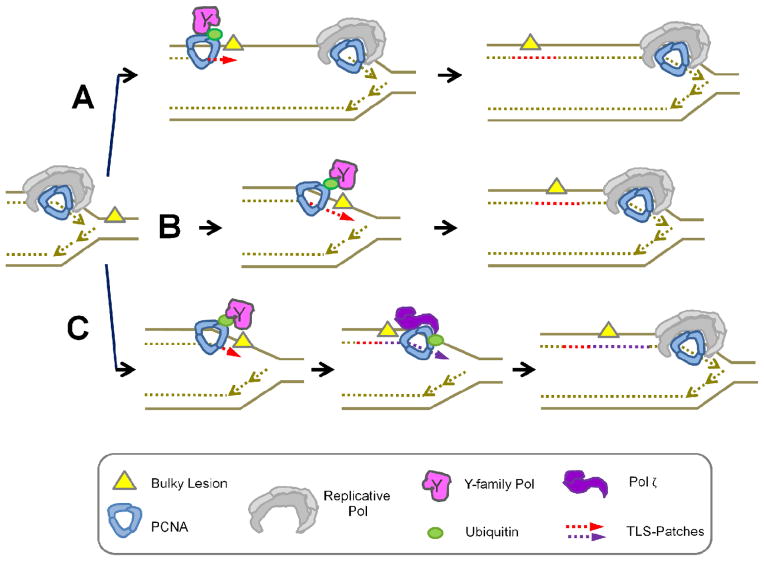
Figure 1.
The models for TLS activation. A) TLS is a post-replicative event: when replication forks encounter DNA lesions a gap is left behind the fork. PCNA-ubi marks the gap in front of the DNA lesions, which is filled by Y-polymerases at a later time. B) TLS is a replication-coupled event: when replication forks encounter DNA lesions, the replisome is modified by e.g. PCNA ubiquitination and Y-polymerases are loaded to elongate DNA across the DNA lesions. Replicative pols are re-loaded after lesion bypass. C) TLS is a two-steps process: while few lesions require only one TLS pol, many require two specialized pols. The first one inserts the first nucleotide in front of the DNA lesion while the second fills the gap.
Specialized DNA polymerases have no proofreading activity, their processivity is low and they are highly mutagenic, with a few exceptions as in the case of the Polη when it bypasses CPDs. Polη deficiency in humans causes the xeroderma pigmentosum variant (XPV), with clinical features that resemble those of defective nucleotide excision repair (NER) [2]. Loss of TLS capability also jeopardizes the survival of whole organisms as demonstrated by the embryonic lethality of Polζ deficiency in mouse models [3]. In addition, the overexpression of some Y-family polymerases has been detected in cancer cells, suggesting that dysregulated TLS may contribute to the genesis of human diseases including cancer and to the resistance to chemotherapy [4]. In general, the extent of DNA synthesis by TLS must be tightly regulated to achieve the best balance between cell survival and mutagenesis. In Escherichia coli the DNA stretches synthesized by TLS were shown to be no longer than ~60 nucleotides [5], suggesting an exquisite control of both loading and removal of specialized polymerases at replication forks.
How and when
While Y-family DNA polη, κ and ι are recruited to Proliferating Cell Nuclear Antigen (PCNA) through a PCNA interacting protein (PIP) box, Rev1 utilizes its BRCT domain and/or its PAD domain for localization. All Y-family pols have one or two ubiquitin binding domains (UBD), which consolidates their interaction with PCNA at sites for translesion DNA synthesis, as several genotoxic treatments prompt Rad6/Rad18-dependent PCNA mono-ubiquitination at Lys164. Another mechanism that facilitates specialized pol localization to damaged DNA is the direct recruitment to Rev1, which can act as a scaffold protein [1, 6]. Conversely, it has been postulated that the removal of the ubiquitin moiety from PCNA facilitates the reverse exchange to replicative pols after lesion bypass [7]. PCNA can also be polyubiquitinated to promote non-TLS events but the biological relevance of such modification is not within the scope of this review [1, 6].
TLS events can take place at or behind the replication fork [8] (Figure 1). The initial characterization of polη indicated a post-replicative mode of action [9]. Following the discovery of PCNA ubiquitination, the replication-coupled mode of TLS dominated the field until experiments performed in S. cerevisiae demonstrated that TLS events can be postponed to the G2-phase without affecting cell viability [10, 11]. Currently, it is accepted that both TLS modes aid DNA replication although it is unclear whether this is an arbitrary choice or if signals arising from the DNA lesion or its surroundings are variables that affect such a decision. The post-replicative mode is particularly supported by a paradigm-breaking model that proposes discontinuity of DNA replication in both DNA strands following replication stress [8, 12]. Interestingly, a novel specialized polymerase with primase activity, PrimPol, could be essential for the onset of such discontinuous DNA synthesis events [13, 14]. It is therefore possible that discontinuous DNA synthesis in both strands and post-replicative TLS are frequent events.
Methods for assessing TLS
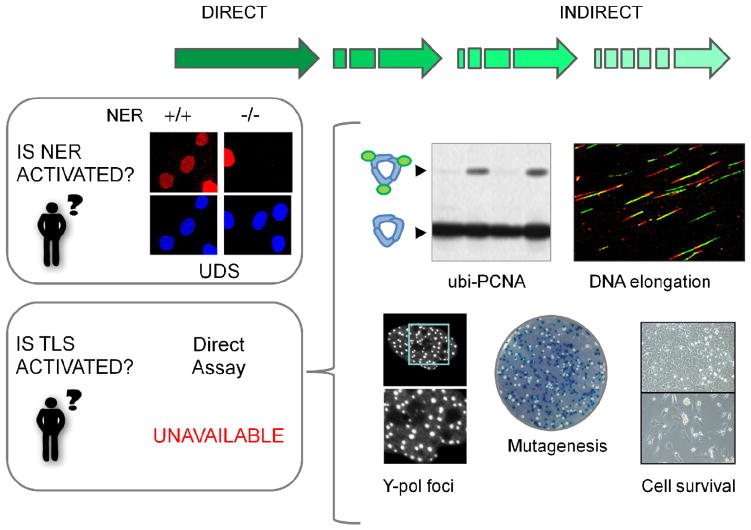
While the precise quantification of restricted DNA synthesis events is possible (e.g. unscheduled DNA synthesis (UDS) reveals NER), so far, it is impossible to identify TLS stretches of only a few nucleotides within the background of bulk DNA replication of normal DNA. Nevertheless, TLS efficiency may be inferred indirectly by monitoring various accepted TLS markers (Figure 2).
Figure 2.
The battery of assays used to study TLS. While specific assays such as unscheduled DNA synthesis (UDS) selectively reveal other DNA synthesis processes such as NER, there is no direct way to quantify TLS-triggered DNA synthesis. Biochemical markers of TLS and biological processes affected by TLS onset are used instead to indirectly infer modulations in TLS activation.
The recruitment of Y-pols to replication factories and their interaction with PCNA in the chromatin fraction
DNA replication takes place in defined subnuclear replication factories, in which a cluster of replication forks is initiated and elongates nascent DNA [1]. Y-pols are recruited to replication factories in response to replication stress (triggered by UV, MMS, BPDE, but not DSB-inducing agents such as ionizing irradiation-IR) in a manner that depends upon their PIP-box and/or UBD domains [6]. The interaction between chromatin-bound PCNA and specialized pols is also enriched following DNA damage induction. However, the upregulation of these markers is not sufficient proof of TLS occurrence. First, nuclear foci of specialized pols have been visualized outside of S-phase, c.f. [15] and have been associated in some cases with DNA repair, c.f. [16]. Second, increased UV sensitivity was reported using Polη mutants defective in PCNA binding, and are therefore unable to organize into detectable nuclear foci, e.g. [17]. Hence, the organization of specialized pols in foci must be interpreted in the context of other assays to infer the extent of TLS activation.
PCNA mono-ubiquitination
DNA damaging agents that initiate accumulation of bulky adducts and/or cause replication stalling increase the mono-ubiquitination of PCNA (PCNA-ubi) [2]. While the ubiquitination of PCNA is undoubtedly biologically relevant e.g. [18, 19], a number of results suggest that PCNA-ubi is not an unequivocal marker of TLS activation. First, PCNA-ubi in vertebrates is not always epistatic with Polη, Polκ, Polζ and Rev1, e.g. [20]. Second, some TLS events occur in the absence of PCNA-ubi, e.g. [20], and Polη recruitment to damaged-DNA can be independent of its UBD, e.g. [21]. Third, the function of PCNA-ubi might not overlap completely with TLS since: a) it can be upregulated when there is no damage to bypass (e.g. after hydroxyurea (HU)/aphidicolin (Aph) treatments), e.g. [22]; b) it precedes PCNA polyubiquitination which can trigger TLS-independent events, e.g. [23]; c) it can take place in cells transiting or arrested in G1, e.g. [24]. Thus, changes in PCNA-ubi must be also studied in combination with other TLS markers.
DNA elongation assays
Defects in the expression of TLS polymerases or in the extent of PCNA ubiquitination have been shown to modulate at least one of the following DNA replication assays: a) fiber assay, b) Alkaline Unwinding Assay (ADU), c) Alkaline Sucrose Gradient sedimentation assay (ASG). The fiber assay can measure the average replication speed before and after DNA damage within the same replication fork [25]. This approach relies on the direct visualization of denatured nascent DNA via immunodetection of thymidine analogs added before and after the DNA damage insult. The length of each DNA track is then utilized to infer the average rate of nascent DNA elongation within the time frame of the pulse. While a reduction in the length of the DNA track synthesized upon DNA damage is interpreted as a delay in continuous DNA elongation, it is yet uncertain if re-priming downstream from the DNA lesions might reduce the replication speed as well. To distinguish between the replication-coupled and the re-priming TLS models, the fiber assay must be combined with the ADU or the ASG assay. The ADU consists on a partial unwinding from the ssDNA at the tip of each fork [26, 27]. The protocol involves pulse-labelling with titrated thymidine; followed by the immediate exposure of the samples to DNA-damaging agents and incubation with a medium containing unlabeled thymidine. Sample collection at different times after chase are subjected to partial unwinding, sonication and separation of dsDNA from ssDNA fragments with hydroxyapatite columns. The ratio between [H3]-labelled ssDNA and the total [H3]-labelled DNA at each chase time is then used to infer the progression of the replication fork from the labelled area. Both stalled and discontinuous replication is expected to result in the formation of persistent [H3]-labelled ssDNA ends. The ASG is the “oldest” assay [9, 28] to study the growth of molecules replicated shortly after DNA insults. Similarly to the ADU assays, cells are labelled with titrated thymidine, but in this case the [H3]-thymidine pulse is delivered after exposure to the DNA-damaging agent. Samples are then chased for different times and incubated with a strong alkaline solution to achieve full denaturation before resolution in a sucrose gradient. A reduction in the size of [H3]-thymidine labelled DNA is interpreted as evidence for DNA replication stalling and/or re-priming.
While the utilization of ASG, ADU and fiber assays in isolation might not suffice to reveal whether TLS events are occurring at or behind the fork, they have been used in combination to seek an answer for such a challenging question (e.g. [29]). As detailed in Supplementary Table 1, these assays revealed a contribution to nascent DNA elongation of all specialized pols or PCNA-ubi. Hence, it is expected that every TLS regulator must affect at least one or more of these assays.
Mutagenic Signature
A number of assays have been designed to assess TLS-triggered mutagenesis. 1) The earliest and easiest-to-set-up assay is the supF assay which utilizes a UV-irradiated shuttle DNA plasmid to infer mutagenesis, using β-galactosidase activity as a read-out [30]. 2) The more sophisticated duplex vectors assay combines β-galactosidase activity and antibiotic resistance to distinguish between TLS and other replication-associated events [31]. 3) The gap-filling plasmid assay specifically focuses on post-replicative TLS, by employing a plasmid that cannot replicate in mammalian cells [32]. This assay has been adapted to compare TLS with other replication-associated events [33]. 4) The chromatinic HPRT assay focuses on the ability of HPRT mutant cells to survive the treatment with an otherwise toxic purine analogue (6-thioguanine) [34]. DNA sequencing is then required to link a mutation in the hprt gene with a TLS defect. 5) The recently described “genomic lesion tolerance assay” uses the integrase of phage ϕC31 to “chromatinize” two staggered closely-opposed lesions permitting a distinction between homologous recombination and accurate or mutagenic TLS [35].
While these approaches have certain limitations [e.g. utilization of episomal substrates (SupF, duplex vectors and gap filling assay), lack of a site specific lesion (SupF and HRPT assay), incapacity to assess accurate TLS events (SupF, HRPT assay), and refractory response to stress conditions such as checkpoint activation [32])], they have nonetheless been fundamental for the disclosure of important mechanistic aspects of TLS as detailed in Supplementary Table 2.
Survival rates
While the preponderant role of TLS pols in cell survival has been described at the beginning of this review, it should be noted that conclusions regarding a causative role of TLS dysfunction on survival rates should be approached with caution since specialized pols might contribute to cell survival independently of TLS. For example, the significant sensitivity to UV irradiation of Polκ deficient cells has been attributed to its repair replication role in NER and not in TLS [16].
Negative regulators of TLS
Our current understanding envisions TLS as a locally constrained event targeted only to locations in damaged DNA. TLS inhibitors are in turn strongly regulated both by the cell cycle and by TLS activating signals. The implication of such tight regulation for the appropriate onset of TLS will be discussed below.
USP1
The identification of the deubiquitinase of PCNA, USP1/UAF1, led to the suggestion of a potential negative regulator of TLS [36]. USP1 reverts both basal and DNA damage-induced mono-ubiquitination of PCNA at K164 [36, 37]. The treatment of cells with UV irradiation triggers enhanced, yet mechanistically controversial, USP1 proteolysis [36]. However, it is intriguing that other stimuli that upregulate TLS such as MMS, MMC or HU do not upregulate USP1 proteolysis [36, 37]. A non-degradable USP1 reduced UV-initiated Polη focal organization and PCNA interactions [36]. The supF assay revealed spontaneous and UV-induced mutagenesis in USP1-depleted cells [36], while the downregulation or inactivation of USP1/UAF1 triggered a Polη-dependent mild increase in UV sensitivity [38]. Surprisingly, the effect of USP1 modulation in DNA elongation after UV irradiation has not yet been reported. Instead, the role of USP1 in undamaged cells has been revealed in a pioneering study from the group of Tony T. Huang: USP1 prevents accumulation of micronuclei during unstressed replication by restraining excessive recruitment of Polκ to undamaged DNA synthesis [39]. Since USP1 expression is restricted to S, G2/M-phases by the E3 ligase APC/C(Cdh1) [40], high USP1 levels in S-phase might prevent Polκ loading at undamaged DNA replication forks. Interestingly, USP1 also de-ubiquitinates FANCD2, a key member of the Fanconi Anemia (FA) pathway, required for the repair of DNA interstrand crosslinks (ICLs). The loss of the FA pathway causes multiple abnormalities leading to cancer, which correlate with USP1 overexpression in several tumour types [41]. Given the utmost importance of the FA pathway during DDR, the inability to separate the contribution of USP1 to FANCD2- and PCNA-dependent signalling complicates the identification of the direct contribution of USP1 to TLS signalling.
p21 waf1/cip1
The cyclin kinase (CDK) inhibitor, p21, is well known because of its role in the maintenance of cell cycle arrest outside S-phase [42]. Its ability to consolidate G1 and G2 arrest depends upon its CDK binding domain and on its major upregulation following several different genotoxic stimuli. Thereafter, the low levels of p21 in S-phase, for a long time, have been considered residual. During the last decade, overwhelming evidence from many groups has demonstrated that genotoxic stimuli such as UV irradiation upregulate p21 proteolysis to the extent of eliminating such “residual” levels. e.g.[24]. Since no cellular process is simply “ornamental”, this indicates that so-called residual levels of p21 might impair at least one aspect of the cellular response to UV irradiation [42]. To date, there is good evidence that low levels of p21 are sufficient to prevent TLS onset. Mechanistically, this has been linked to the control of PCNA ubiquitination by the CDK binding domain of p21 [24] and later on, to the p21 PIP-box, which binds PCNA with strong affinity, displacing weaker PIP-boxes in vitro [43]. In cells, sustained p21/PCNA binding precludes Y-pol focal organization and the interaction of PCNA with Polη, Polκ, Pol ι and Rev1 in chromatin after UV irradiation [42, 44, 45]. Interestingly, this happens without compromising the interaction of PCNA with the replicative pols, which have more than one PCNA binding domain [42, 44]. Remarkably, endogenous p21 recapitulates the effect of stable p21/PCNA binding in a manner that inversely correlates with p21 degradation, since both stable and endogenous p21 constrain DNA elongation at replication forks after UV irradiation [44, 45]. These observations suggest that p21 is a global inhibitor of Y-pols, and they are consistent with the defective DNA elongation observed after depletion of two or more Y-pols following UV irradiation [46]. When assessing the role of p21 on TLS-driven mutagenesis, Z. Livneh and colleagues showed that the PCNA binding domain of p21 reduces the efficiency but increases the accuracy of TLS events [32]. We therefore propose that the timely degradation of p21 slows down the onset of TLS events by promoting the selection of the less mutagenic Y-pol. In support of this model, the CRL4Cdt2 E3 ligase has been shown to trigger local degradation of chromatin-bound p21 within PCNA complexes [47, 48], and this depends upon a specific PIP-degron sequence in p21 [49]. Such choreographic control of TLS might extend to other PIP-degron proteins such as CDT1, which interferes with the recruitment of Polη and Polκ to replication factories [50]. While the timely removal of p21 from PCNA might promote more accurate TLS events, a failure to eliminate p21 from the clamp loader could permanently block TLS thereafter, leading to the cessation of DNA replication. Consistent with this hypothesis, the expression of a p21 mutant that resists UV-induced degradation triggers 53BP1 focal organization, micronuclei formation and cell death [45]. Moreover, when other PIP-degron proteins are not removed from PCNA, the UV sensitivity of cells increases as well [50]. Taken together, these findings indicate that p21, through its CDK and PIP-box can affect all parameters of TLS discussed in this review. It might control TLS at ongoing replication forks through PCNA-binding while it might modulate gap-filling by relying on its CDK binding domain. Independently of such speculations, the data discussed herein robustly demonstrate that p21 levels, which might be considered residual from the perspective of cell cycle arrest, are sufficient to control TLS, thus revealing an unexpected and important role for low p21 levels during S-phase.
DVC1/Spartan
Spartan is an evolutionarily-conserved multidomain protein containing a SprT-like domain of unknown function, a SHP box that mediates its interaction with the VCP/p97 chaperone, a PIP-box, and a UBZ domain that binds mono- and polyubiquitinated substrates [51–53]. The E3 ligase APC/C(Cdh1) restricts Spartan expression to S phase, G2 and early M-phases [51]. Through its PCNA and UBZ domains Spartan localizes in nuclear S-phase foci in response to UV, MMS, HU, MMC and cisplatin but not after treatment with IR [51–55]. Moreover, Spartan depletion impairs cell survival after UV, cisplatin, MMS and camptothecin but not after IR [51–56]. Importantly, Spartan deficiency has been linked to genome instability, premature aging and cancer predisposition both in humans [56] and in mice [57].
There is tantalizing evidence that Spartan is a negative regulator of TLS [51, 54, 58]. First, Spartan is downregulated in a dose-dependent manner after UV irradiation [53]. Second, Spartan suppresses UV-induced mutagenesis [51, 54, 58, 59]. However, loss of Spartan diminishes DNA elongation after UV and Aph treatments, and that would not be expected from a global negative regulator of TLS [56, 57]. Lessel and coworkers have speculated that excessive Polη loading to replicating DNA could be the cause for such slower replication fork rates. However, the concomitant loss of Spartan and Polη could not rescue the short-fiber phenotype [56]. Since it is expected that the overexpression of a TLS inhibitor phenocopies the loss of one/multiple specialized pols (see Table 1), evaluating the effect of Spartan overexpression in DNA elongation assays might be informative for this matter.
Conflicting results were reported when analysing the effect of Spartan on biochemical markers of TLS activation. Some reports show that Spartan depletion after UV irradiation causes enhanced and persistent retention of Polη in the chromatin fraction which is accompanied by an increase in both the PCNA/Polη interaction and in the focal organization of Polη [51, 54, 57]. In concordance, overexpression of Spartan suppressed the interaction between Polη and PCNA-ubi after UV [51]. In contrast, others have reported that Spartan deficiency causes a reduction in UV-induced Polη focal organization [53] and that its overexpression enhances spontaneous Polη foci formation (in a manner that depends upon negative regulation of USP1 by Spartan) [55]. The role of Spartan in PCNA ubiquitination is also controversial. While some reports indicate that Spartan enhances PCNA ubiquitination [52, 53, 55] others suggest that PCNA ubiquitination is not significantly affected by Spartan depletion [51, 54, 57]. Such conflicting results lead to equally confusing models for the role of Spartan in TLS. The groups that postulate Spartan as a positive TLS regulator suggest that: a) Spartan establishes a self-perpetuating process involving its recruitment to PCNA-ubi, which in turn enhances Rad18 chromatin access to PCNA [53]; b) Spartan protects PCNA-ubi from USP1 triggered de-ubiquitination [55]; c) Spartan prevents PCNA-ubi and RAD18 removal from chromatin during TLS [52]. Those who suggest a negative role of Spartan in TLS propose that: a) Spartan might directly interact with, and inhibit the extension step of Rev1/Polζ-dependent error-prone TLS [58]; b) Spartan prompts the removal of Polη from PCNA-ubi in a manner that facilitates the re-start of DNA synthesis by replicative polymerases [51, 54]. In conclusion, while Spartan has clearly a central role in TLS regulation, further work is needed to clarify whether it is a positive or a negative regulator of TLS (or both?).
Concluding remarks and perspectives
While some aspects of the regulation of TLS by USP1, p21 and Spartan have been revealed, a number of issues require immediate attention. While it is accepted that the consequences of the inactivation of a single Y-pol must be different from those arising from the global block of all Y-pols, with the exception of p21 [45], the analysis of most inhibitors has been restricted to Polη [36, 51, 53–57, 60, 61]. Moreover, the overexpression/stabilization of TLS inhibitors should be exploited to support their negative role in TLS. In fact, the extensive use of gain-of-function-tools combined with the analysis of all Y-family pols served to define p21 as a global negative regulator of TLS in UV damage [45], while similar experiments with USP1 and Spartan are yet to be performed.
Application of the DNA fiber assay has shown that the functions of the TLS inhibitors do not totally overlap. After UV-irradiation, p21 degradation increases DNA elongation, thus supporting its role as a global TLS inhibitor [45], while Spartan dysfunction causes the opposite effect [56, 57]. Intriguingly, the role of USP1 in DNA elongation after UV irradiation has not been yet reported. Moreover, loss of either negative or positive TLS regulators cause hypersensitivity to DNA damage, which might indicate that an “appropriate” level of TLS events is required for cell viability, e.g. [53].
Another important issue that requires clarification is the contribution of TLS regulators to replication of undamaged DNA. TLS pols are certainly required for the synthesis across difficult-to-replicate DNA structures such as common fragile sites [4], but their participation in undamaged DNA replication must be restricted to minimize mutagenesis and other genomic instability parameters [39]. While USP1 has a well-documented role in the protection of undamaged DNA replication [39], diminished levels of Spartan during unperturbed replication affect the TLS parameter of DNA elongation [56]. This emphasizes the need for research to explore the contribution of TLS inhibition to the successful execution of the replication program in the absence of stress.
The information discussed in this review indicates that USP1 may have a more prominent role in the prevention of unleashed Y-pol loading on undamaged DNA than on the onset of TLS. On the other hand, p21 has been placed directly at the on-switch for TLS [42] and more conflicting evidence places Spartan at the off-switch for TLS [51, 54, 57] (Figure 3). In this regard, it is important to mention that recent reports bring the PCNA-interacting protein PAF15 and the ubiquitin-like protein ISG15 into play, being both factors potentially involved in the restoration of replicative DNA synthesis after TLS finalization [60, 61]. PAF15 may also prevent unleashed loading of Polη to undamaged DNA [60]. Additionally, emerging evidence highlights potential cross-regulation between TLS inhibitors, as USP1 and Spartan have been functionally linked [55]. Understanding the interconnections between TLS-regulators should foster the comprehension of the mechanisms that limit mutagenesis to optimal levels in cells.
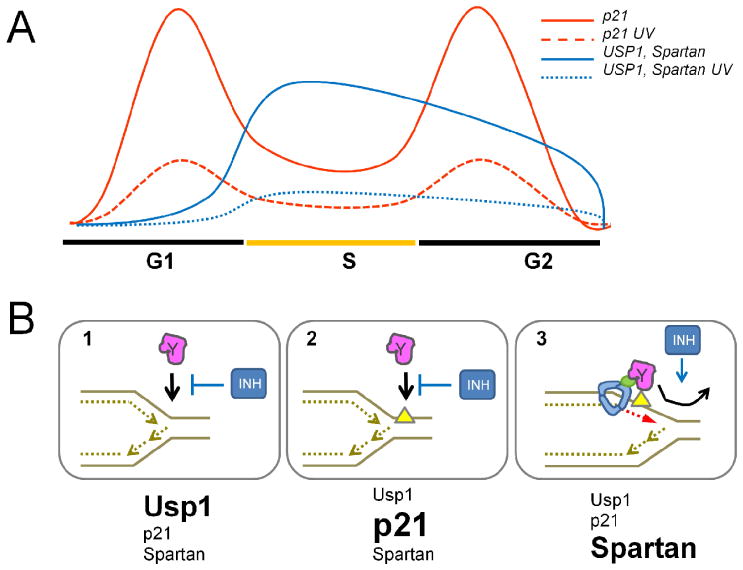
Figure 3.
The regulation and function of TLS inhibitors. A) In S-phase p21 is at its lowest levels while USP1 and Spartan are at their highest. Notably, they are all downregulated after UV irradiation. B) TLS inhibitors prevent the recruitment of Y-pols to non-TLS undamaged templates (1); choreograph the correct and timely activation of TLS at DNA lesions (2): and promote the switch-back to replicative synthesis (3). So far, the evidence indicates that each inhibitor may have prevalence at each one of these steps.
Supplementary Material
Acknowledgments
When citing an example please see the Supplementary Information for a more complete list of references. We apologize to colleagues whose work could not be cited due to space restrictions. We are indebted to Dr. Philip Hanawalt, Stanford University for insightful suggestions. We thank Dr. Gastón Soria, Universidad de Cordoba for very insightful comments. Research in the V.G. laboratory is supported by grants from NIH (R03 TW008924) and ANPCyT. A.P.B. is supported by a fellowship from ANPCyT and S.F:M. is supported by a fellowship from CONICET.
Abbreviations used in this paper
- TLS
translesion DNA synthesis
- UV
ultraviolet
- CPD
cyclobutane pyrimidine dimers
- 6-4PPs
6-4 photoproducts
- NER
nucleotide excision repair
- PCNA
Proliferating Cell Nuclear Antigen
- PIP
PCNA interacting protein
- UBD
ubiquitin binding domains
- UDS
unscheduled DNA synthesis
- MMS
methyl-methane-sulfonate
- HU
hydroxyurea
- Aph
aphidicolin
- MMC
mitomycin-C
- BPDE
benzo[a]pyrene-diol epoxide
- IR
ionizing irradiation
- DSB
double strand break
- FA
Fanconi anemia
- ICLs
interstrand crosslinks
- ADU
Alkaline Unwinding Assay
- ASG
alkaline Sucrose Gradient sedimentation assay
- CDK
cyclin-dependent kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature reviews Molecular cell biology. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell research. 2008;18:174–183. doi: 10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann JS, Cazaux C. Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Seminars in cancer biology. 2010;20:312–319. doi: 10.1016/j.semcancer.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA repair. 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 6.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and molecular biology reviews: MMBR. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA repair. 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betous R, Pillaire MJ, Pierini L, van der Laan S, Recolin B, Ohl-Seguy E, Guo C, Niimi N, Gruz P, Nohmi T, Friedberg E, Cazaux C, Maiorano D, Hoffmann JS. DNA polymerase kappa-dependent DNA synthesis at stalled replication forks is important for CHK1 activation. The EMBO journal. 2013;32:2172–2185. doi: 10.1038/emboj.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L, Mendez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nature structural & molecular biology. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, Terrados G, Powell C, Salido E, Mendez J, Holt IJ, Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Molecular cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soria G, Belluscio L, van Cappellen WA, Kanaar R, Essers J, Gottifredi V. DNA damage induced Pol eta recruitment takes place independently of the cell cycle phase. Cell Cycle. 2009;8:3340–3348. doi: 10.4161/cc.8.20.9836. [DOI] [PubMed] [Google Scholar]
- 16.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nature cell biology. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 17.Despras E, Delrieu N, Garandeau C, Ahmed-Seghir S, Kannouche PL. Regulation of the specialized DNA polymerase eta: revisiting the biological relevance of its PCNA- and ubiquitin-binding motifs. Environmental and molecular mutagenesis. 2012;53:752–765. doi: 10.1002/em.21741. [DOI] [PubMed] [Google Scholar]
- 18.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 19.Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee KY, Geacintov NE, Carell T, Myung K, Tateishi S, D’Andrea A, Jacobs H, Livneh Z. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS genetics. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Molecular cell. 2008;30:519–529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 21.Acharya N, Yoon JH, Hurwitz J, Prakash L, Prakash S. DNA polymerase eta lacking the ubiquitin-binding domain promotes replicative lesion bypass in humans cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10401–10405. doi: 10.1073/pnas.1005492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Molecular cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 23.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. The Journal of cell biology. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 25.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erixon K, Ahnstrom G. Single-strand breaks in DNA during repair of UV-induced damage in normal human and xeroderma pigmentosum cells as determined by alkaline DNA unwinding and hydroxylapatite chromatography: effects of hydroxyurea, 5-fluorodeoxyuridine and 1-beta-D-arabinofuranosylcytosine on the kinetics of repair. Mutation research. 1979;59:257–271. doi: 10.1016/0027-5107(79)90164-7. [DOI] [PubMed] [Google Scholar]
- 27.Johansson F, Lagerqvist A, Erixon K, Jenssen D. A method to monitor replication fork progression in mammalian cells: nucleotide excision repair enhances and homologous recombination delays elongation along damaged DNA. Nucleic acids research. 2004;32:e157. doi: 10.1093/nar/gnh154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Zeeland AA, Smith CA, Hanawalt PC. Sensitive determination of pyrimidine dimers in DNA of UV-irradiated mammalian cells. Introduction of T4 endonuclease V into frozen and thawed cells. Mutation research. 1981;82:173–189. doi: 10.1016/0027-5107(81)90148-2. [DOI] [PubMed] [Google Scholar]
- 29.Elvers I, Johansson F, Groth P, Erixon K, Helleday T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic acids research. 2011;39:7049–7057. doi: 10.1093/nar/gkr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidman MM, Dixon K, Razzaque A, Zagursky RJ, Berman ML. A shuttle vector plasmid for studying carcinogen-induced point mutations in mammalian cells. Gene. 1985;38:233–237. doi: 10.1016/0378-1119(85)90222-7. [DOI] [PubMed] [Google Scholar]
- 31.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Molecular cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 33.Adar S, Izhar L, Hendel A, Geacintov N, Livneh Z. Repair of gaps opposite lesions by homologous recombination in mammalian cells. Nucleic acids research. 2009;37:5737–5748. doi: 10.1093/nar/gkp632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang JL, Maher VM, McCormick JJ. Amplification and direct nucleotide sequencing of cDNA from the lysate of low numbers of diploid human cells. Gene. 1989;83:347–354. doi: 10.1016/0378-1119(89)90121-2. [DOI] [PubMed] [Google Scholar]
- 35.Izhar L, Ziv O, Cohen IS, Geacintov NE, Livneh Z. Genomic assay reveals tolerance of DNA damage by both translesion DNA synthesis and homology-dependent repair in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E1462–1469. doi: 10.1073/pnas.1216894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D’Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature cell biology. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 37.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16125–16130. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nature chemical biology. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones MJ, Colnaghi L, Huang TT. Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. The EMBO journal. 2012;31:908–918. doi: 10.1038/emboj.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. The Journal of cell biology. 2011;194:177–186. doi: 10.1083/jcb.201101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Santisteban I, Peters GJ, Giovannetti E, Rodriguez JA. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Molecular cancer. 2013;12:91. doi: 10.1186/1476-4598-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soria G, Gottifredi V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA repair. 2010;9:358–364. doi: 10.1016/j.dnarep.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nature reviews Molecular cell biology. 2013;14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 44.Soria G, Speroni J, Podhajcer OL, Prives C, Gottifredi V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. Journal of cell science. 2008;121:3271–3282. doi: 10.1242/jcs.027730. [DOI] [PubMed] [Google Scholar]
- 45.Mansilla SF, Soria G, Vallerga MB, Habif M, Martinez-Lopez W, Prives C, Gottifredi V. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic acids research. 2013;41:6942–6951. doi: 10.1093/nar/gkt475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jansen JG, Temviriyanukul P, Wit N, Delbos F, Reynaud CA, Jacobs H, de Wind N. Redundancy of mammalian Y family DNA polymerases in cellular responses to genomic DNA lesions induced by ultraviolet light. Nucleic acids research. 2014;42:11071–11082. doi: 10.1093/nar/gku779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes & development. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. The Journal of biological chemistry. 2008;283:29045–29052. doi: 10.1074/jbc.M806045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Havens CG, Walter JC. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Molecular cell. 2009;35:93–104. doi: 10.1016/j.molcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsanov N, Kermi C, Coulombe P, Van der Laan S, Hodroj D, Maiorano D. PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase eta and kappa focus formation on UV damage. Nucleic acids research. 2014;42:3692–3706. doi: 10.1093/nar/gkt1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, Hartmann-Petersen R, Lukas J, Choudhary C, Pocock R, Bekker-Jensen S, Mailand N. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nature structural & molecular biology. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 52.Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. The Journal of biological chemistry. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Molecular cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nature structural & molecular biology. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 55.Juhasz S, Balogh D, Hajdu I, Burkovics P, Villamil MA, Zhuang Z, Haracska L. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic acids research. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lessel D, Vaz B, Halder S, Lockhart PJ, Marinovic-Terzic I, Lopez-Mosqueda J, Philipp M, Sim JC, Smith KR, Oehler J, Cabrera E, Freire R, Pope K, Nahid A, Norris F, Leventer RJ, Delatycki MB, Barbi G, von Ameln S, Hogel J, Degoricija M, Fertig R, Burkhalter MD, Hofmann K, Thiele H, Altmuller J, Nurnberg G, Nurnberg P, Bahlo M, Martin GM, Aalfs CM, Oshima J, Terzic J, Amor DJ, Dikic I, Ramadan K, Kubisch C. Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features. Nature genetics. 2014;46:1239–1244. doi: 10.1038/ng.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maskey RS, Kim MS, Baker DJ, Childs B, Malureanu LA, Jeganathan KB, Machida Y, van Deursen JM, Machida YJ. Spartan deficiency causes genomic instability and progeroid phenotypes. Nature communications. 2014;5:5744. doi: 10.1038/ncomms6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim MS, Machida Y, Vashisht AA, Wohlschlegel JA, Pang YP, Machida YJ. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic acids research. 2013;41:1661–1668. doi: 10.1093/nar/gks1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machida Y, Kim MS, Machida YJ. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 2012;11:3395–3402. doi: 10.4161/cc.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park JM, Yang SW, Yu KR, Ka SH, Lee SW, Seol JH, Jeon YJ, Chung CH. Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Molecular cell. 2014;54:626–638. doi: 10.1016/j.molcel.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 61.Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nature cell biology. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.