Abstract
Evidence for a CO docking site near the FeMo-cofactor in nitrogenase has been obtained by FT-IR monitored low temperature photolysis. We investigated the possible migration paths for CO from this docking site using molecular dynamics calculations. The simulations support the notion of a gas channel with multiple internal pockets from the active site to the protein exterior. Travel between pockets is gated by motion of protein residues. Implications for the mechanism of nitrogenase reactions with CO and N2 are discussed.
Keywords: nitrogenase, FT-IR, molecular dynamics, myoglobin, carbon monoxide
Introduction
Nitrogenase (N2ase) is the enzyme responsible for biological nitrogen fixation.1–3 For molybdenum-containing N2ase from Azotobacter vinelandii, which this work will focus on, X-ray diffraction has revealed a unique [Mo-7Fe-9S-C]-homocitrate cluster at the active site of the MoFe protein of N2ase,4, 5 with an interstitial carbide at the center of a prismatic 6 Fe cage.5–7 This cluster, known as the FeMo-cofactor (FeMo-co), is capable of reducing a wide variety of triply bonded substrates, such as N2, C2H2, HCN, as well as protons.8 It has recently been shown that this enzyme can also produce CxHy hydrocarbons from CO9–11 and even CH4 from CO2.12, 13 The migration of substrates, such as: N2 and CO and protons, to the active site, as well as the exit of products such as NH3, various hydrocarbons CxHy, and H2, is clearly a critical part of N2ase reactivity.
Several different channels have been proposed for access to or from the FeMo-cofactor. 14–21 In 2003, Igarashi and Seefeldt pointed out a candidate substrate channel starting near surface residues α-K209 and α-W205. This mostly hydrophobic pathway (now called the ‘IS channel’20) ultimately passes the α-V70, α-H195, and α-R96 region and terminates at the Fe2,3,6,7 face of the FeMo-cofactor.14 At the opposite end of the FeMo-cofactor, a hydrophilic channel extends from a ‘water pool’15 proximal to the homocitrate ligand, through the interface between α and β subunits, and finally to the surface. Durrant offered this ‘interstitial channel’ as an efficient path for diffusion of both dinitrogen and ammonia,16 and his proposition was later tested through site-directed mutagenesis work by Barney and coworkers.17 Dance has noted a similar channel for the egress of ammonia, starting at α-Q191.18 Smith and coworkers have recently proposed a very different dynamic channel that opens and closes on the tens of nanoseconds time scale, starting near surface residues α-R281 and α-H383 and leading to the Fe2,3,6,7 face,19 which may have a role in proton transport.22, 23 Additional substrate/product pathways have most recently been proposed by Morrison and coworkers, based on Caver calculations combined with analysis of binding sites for Xe and small molecules.20 Apart from these small molecule channels, multiple proton relay chains (‘proton bays’ and ‘proton wires’), leading to S2B, S3B, and S5A were noted by Durrant 16 and more recently analyzed by Dance.21
Pockets and channels for small molecules like CO and O2 have proven important in the study of myoglobin (Mb) dynamics,24 and they have also been found important for hydrogenases,25 cytochrome oxidase,26 carbon monoxide dehydrogenase/acetyl-CoA synthase,27 and many other ‘gas-processing’ enzymes.28 There is a flourishing literature on how to deduce and evaluate these conduits by computational methods.29–34 On the experimental side, the migration of CO following MbCO photolysis has been followed by infrared spectroscopy, including static,35 picosecond time-resolved,36 and temperature derivative37 methods. Two photolysis product IR bands at 2131 and 2119 cm−1, respectively labeled B1 and B2, are observed for wild-type MbCO.35 These have been taken as evidence for two different CO orientations at the Mb ‘docking site’,38 and the magnitude of the splitting has allowed calculation of the interior electric field.37 Extensive time-resolved x-ray diffraction studies have allowed observation of CO migration between different pockets within Mb39 and the correlation of CO pockets with those observed under high-pressure Xe.40
Pockets and channels should certainly be relevant for understanding N2ase. Analogous to Mb, Xe pockets have been identified by x-ray crystallography in Klebsiella pneumoniae41 and Azotobacter vinelandii (Av).42 Recently, a detailed comparison of Xe sites in Av and Clostridium pasteurianum has been made, along with binding sites for CO and other small molecules.43 In this paper, we present FT-IR cryophotolysis data that supports a docking site for CO near the FeMo-cofactor. We then use molecular dynamics calculations to identify a location for the docking site in Av Mo N2ase as well as a channel allowing for escape of CO and for entry of N2.
Experimental
Photolysis of CO-Nitrogenase
The N2ase enzyme was prepared from Av and reacted with CO using the same protocol from our previous studies.23, 44 The infrared spectra were collected with a Bruker Vertex 70v FT-IR under cryogenic temperatures. Photolysis was induced by a broadband Sutter Instruments xenon-arc lamp. Samples were held in custom built cells with Teflon spacers to give a pathlength of 70 microns.
Molecular Dynamics
Simulations were performed using a customized forcefield in the Gromacs molecular dynamics package.45 The MoFe N2ase-CO α-subunit was first energy minimized, then followed by dynamics at multiple temperatures.
More detailed information about all experimental methods is available in the supporting information.
Results
Infrared Spectroscopy
As shown in Figure 1, positive product bands appear upon low temperature photolysis of CO-inhibited wild-type N2ase as well as the α-H195Q variant. The 37 cm−1 downshifts with 13CO substitution confirm that these are indeed CO bands. By analogy with the MbCO literature, the absorption bands at 2135 and 2123 cm−1 are consistent with ‘free CO’ species adopting two orientations at a docking site.38 We label the N2ase free CO bands respectively NB1 and NB2 by analogy to the myoglobin (Mb) CO labels.35 For Mb mutants, additional bands have been observed to range from 2108 to 2152 cm−1, and we cannot rule out the existence of such minor species in N2ase.
Figure 1.
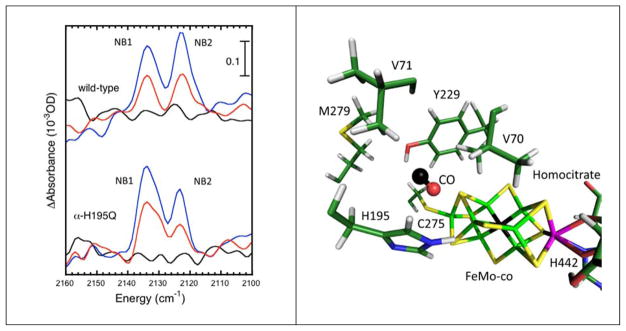
Left: ‘free CO’ IR bands in wild-type N2ase44
vs. H195Q N2ase. Curves represent photolysis difference (subtracting with a no photolysis background) spectra: baseline noise before photolysis (—), halfway through photolysis (
 ), and endpoint of photolysis (
), and endpoint of photolysis (
 ). Right: a model for CO in the docking site in one of the two orientations that could give rise to the observed splitting.
). Right: a model for CO in the docking site in one of the two orientations that could give rise to the observed splitting.
There are differences between wild-type and α-H195Q spectra that suggest that altering the amino acid environment around the FeMo-cofactor has an effect on the photolyzed CO. In the wild type enzyme, the NB2 band at 2123 cm−1 is more intense than the 2135 cm−1 NB1 band. In α-H195Q, the intensity pattern is reversed, and the asymmetry favors the NB1 band, suggestive of two or more unresolved sub-species. Since a small change in side-chain at the 195 position affects the photolysis spectra, these results are at least consistent with locating the ‘free CO’ close to that position.
Molecular Dynamics
Inspired by the experimental evidence for a CO docking site in N2ase, we began a molecular dynamics study of CO migration from that site–analogous to those used for myoglobin and related proteins.15, 46 In this work we took advantage of a force field for the FeMo-cofactor that had been previously validated by comparison with results from nuclear resonance vibrational spectroscopy (NRVS), as well as structural models for CO binding to the FeMo-cofactor from DFT calculations that have been tested against NRVS and EXAFS data.23 We chose to initially place the CO near Fe2. Our previous work has shown the photolabile “Hi-1” CO form has a terminal CO and a formyl-like species.47 Likewise, Fe6 has been implicated as the most reactive site on the FeMo-cofactor,48 therefore we presume Fe6 binds the more reduced formyl-like species and Fe2 binds the terminal CO species that photolyzes to free CO. The entire protein structure was then relaxed to minimize any forces resulting from CO insertion. This yielded an initial Fe2-CO distance of 4.2 Å. The CO was then allowed to migrate through the protein at various temperatures, including 10, 80, 150, 200, 250, and 300 K. The motion of the CO and the protein was followed out to 10 ns (Figure 3).
Figure 3.
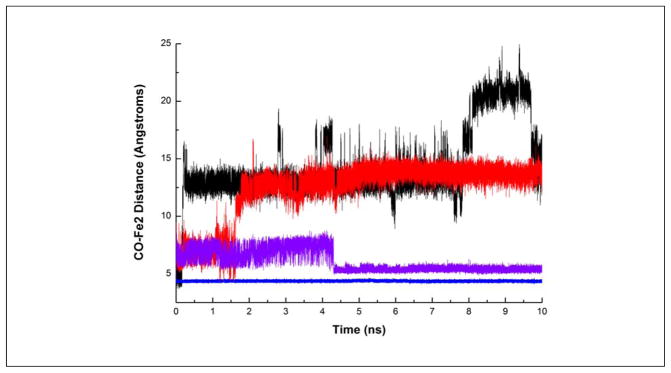
Typical time-dependent traces for CO migration at different temperatures (black 300K, red 250K, purple 150K, blue 10K).
The molecular dynamics calculations find a candidate ‘docking site’ with the CO about 5 Å from Fe2 (Figure 1b and Figure 3). In this location, CO is within H-bonding distance to α-His-195 Nε (3.1 Å). Another residue within H-bonding distance is α-Y229 (O-O distance 4.1 Å). Finally, the critical α-V70 side-chain is in position to constrain the CO motion. This bears some resemblance to the docking site in myoglobin, where photolyzed CO is within H-bonding distance to H64 and is further trapped by aliphatic side-chains of L29 and I107 after photolysis.38, 39, 49 As illustrated by the time courses plotted for different temperatures (Figure 2), at 10K the CO remains in the docking site, but at higher temperatures it rapidly migrates to 3 distinct regions at progressively larger distances from the FeMo-cofactor.
Figure 2.
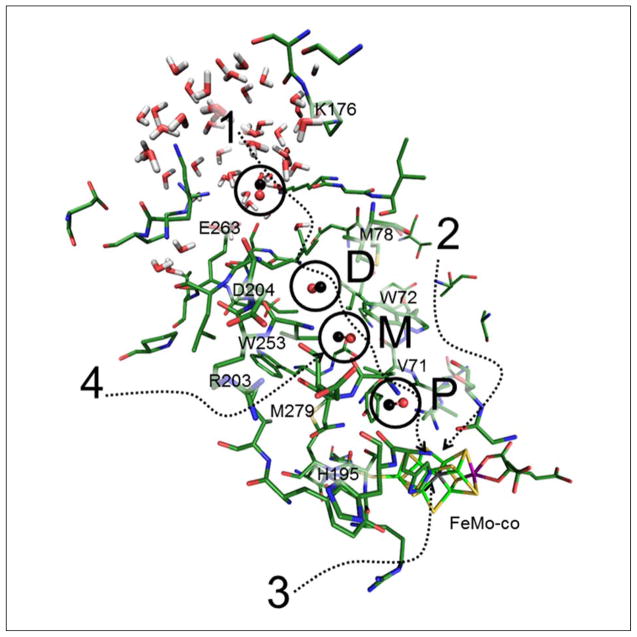
Some of the many pathways that have been proposed 1: Igarashi & Seefeldt,14 2: Barney,17 3: Smith,19 and Morrison,20 Also, the pockets observed in this work P: Proximal, M: Medial, and D: Distal pockets. Residues labels are given to provide a frame of reference and outline the channel path. The CO in the dynamics calculations follows the channel predicted by Igarashi (labeled 1 in the figure).
Upon leaving the docking site, the overall motion was stochastic and bidirectional. However, the path that was most frequently taken was strikingly similar to the IS channel.14 Our result is compared with other proposed channels and pockets in Figure 2. The channel begins near key residues α-H195 and α-V70, and it ends near surface residues α-M78, α-V179, and α-V206 and α-I259 (Figure 2). Although this might only seem a confirmation of the Igarashi channel, from the dynamics calculation we see that the migration of CO is not uniform over time (Figure 2). Rather, just as in myoglobin and other proteins,50 the CO was relatively stable in several distinct sites–the initial ‘docking site’ and 3 distinct ‘pockets’. Passage from the docking site and between pockets is governed by occasional gate openings by key residues along the channel through thermodynamic fluctuations. We now discuss the sites where the CO spent most of the time.
The first pocket encountered (after leaving the docking site) has the CO center-of-mass (COM) 9.5 Å from the FeMo-cofactor central carbide, with a closest approach to Fe2 of 8.5 Å (Figure 4a). Key side-chains in this pocket include α-M279, α-V71, and α-S278. Unlike the later pockets, this first pocket also includes a tyrosine side-chain, α-Y229, a good candidate for hydrogen-bonding to CO. As the closest pocket to the docking site, we label this the proximal or P-pocket.
Figure 4.
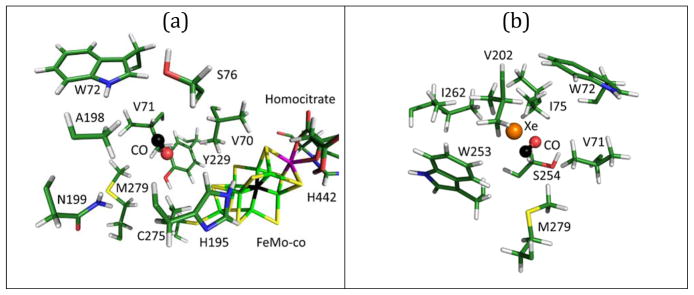
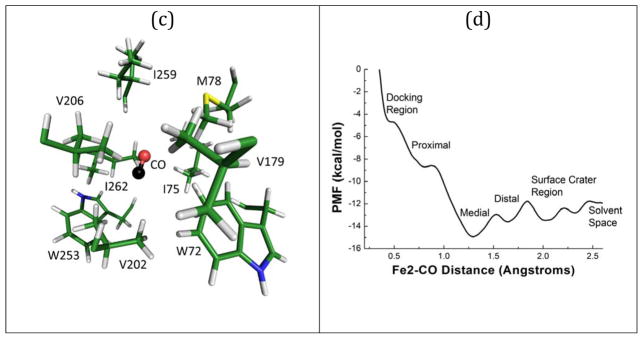
(a) Illustration of the proximal pocket, (b) the medial pocket with xenon overlayed from Morrison et al.,20 (c) the distal pocket, and (d) a profile of the interaction energy between CO and the system vs. the Fe2-CO distance along the proposed channel from the FeMo-cofactor to the solvent space.
The second (‘medial’ or ‘M’) pocket seen in the MD simulations contains α-W253, α-S254, α-I282 (Figure 4b). It is gated from the previous pocket by α-N199, α-M279, α-V71 and α-A198. The four residues sterically prevent CO from returning to the previous pocket, but they can open during thermal fluctuations. When CO is trapped within the M-pocket, the Fe2 to CO distance is about 13.0 Å. We gain confidence in this predicted location for CO because it coincides with the Xe1 pocket seen by x-ray diffraction.20 As shown in Figure 4b, our typical CO position overlaps nicely with the Xe position observed by Rees and coworkers.20,42
The third (‘distal’ or ‘D’) pocket observed in our calculations involves α-I259, α-V206, α-M78, α-V179 (Figure 4c). Access to the D-pocket from the M-pocket is gated by residues α-V202, α-I75, and α-W72. All of the aforementioned side-chains are part of the IS channel proposed by Igarashi and Seefeldt.14 The average Fe2-CO distance from within the D-pocket was 17 Å. We note that a final region for CO occupation was located beyond the D-pocket and within a solvent interaction crater on the surface of the protein. This region had an average Fe2-CO distance of ~21 Å. Penetration into the solvent occurred at a distance of about 25 Å.
Since N2 is the natural substrate for N2ase, we also performed a simulation at 300K using N2 as the diatomic in motion. As expected, N2 behaved similarly to CO and bidirectionally traversed through the three pockets and into the solvent space (Figure S1). In this simulation, the N2 spent most of its time in the M pocket–just as we observed with CO.
An Energy Surface
The motion of CO within N2ase appears to involve capture within pockets interlaced with occasional gate openings. We decided to map the potential energy surface for this motion of CO within the protein by performing a potential of mean force (PMF) calculation using GROMACS.51 This involved sampling multiple Fe2-CO distances and evolving them individually in time. The results from these calculations are shown in Figure 4d. The first minimum is a shallow well at 5 Å that corresponds to the proposed docking site. At longer distances there are three distinct potential minima corresponding to each identified pocket in the channel. Beyond these wells we see the solvent interaction crater at 20 Å and solvent space at about 25 Å. An important result from these calculations is that the location identified as the medial pocket is the lowest energy location where CO can reside.
The overall scale of the PMF is similar to what is seen in previous work for a ligand in the different channel proposed recently,19 however the PMF defined in this work has potential wells with depths greater than what is seen for the that channel. The M pocket has the largest barriers in both direction: 1.9 kcal/mol towards the D-pocket and 7.4 kcal/mol for CO moving towards the P-pocket.
Discussion
Our calculations are the first use of molecular dynamics to include an experimentally constrained force field for the FeMo-cofactor. By combining these calculations with IR-monitored cryogenic photolysis, we have found the first evidence for a CO docking site in N2ase. We favor this docking site as the most likely position for free CO following cryogenic photolysis. The docking site is adjacent to the FeMo-cofactor and has two possible hydrogen bonding partners for CO–α-H195 and α-Y229. In myoglobin, H-bonding to H64 is considered important for the 12 cm−1 splitting of product bands B1 and B2 at 2131 and 2119 respectively. Substitution of histidine by leucine in the H64L variant in myoglobin causes the loss of that splitting and yields a single product band at 2126 cm−1.38 H-bonding to α-H195 or α-Y229 may play a similar role in splitting the ‘free CO’ bands of photolyzed N2ase.
Using the MD calculations, by following the path of CO or N2 starting near this docking site, we discovered a channel for these molecules from the FeMo-cofactor to the protein exterior, in a location consistent with a previous proposal from static CAVER calculations.14 A benefit from the dynamics simulations is that they reveal a novel gating mechanism for CO or N2 migration through this hydrophobic channel–a feature not observable from static calculations. We find that CO spends most of its time in distinct pockets, and travel between pockets is enabled by gating motions of neighboring amino acids.
Although the exact mechanism of N2ase catalysis is far from understood, all proposed reaction pathways require multiple electron and proton transfers to the FeMo-cofactor region before any ligand binding can occur. In particular, the popular Lowe-Thornley (LT) model for the N2ase reaction pathway52 posits 3 or 4 electron/proton transfers to the FeMo-cofactor and/or its associated ligands before N2 binding. In the absence of substrate binding, the LT model proposes that the FeMo-cofactor will oxidize through H2 evolution.8
It has been argued that because N2ase is a slow enzyme, it has no need for a hydrophobic tunnel that would allow rapid gas access to the FeMo-cofactor.28 However, the pockets and gates that we observe may play a role in optimizing N2ase catalytic efficiency. By trapping an N2 molecule in the M-pocket, the enzyme would have substrate available for binding within a few nanoseconds of reaching the E3 or E4 levels. This might help minimize ‘futile’ H2 production that might occur if N2 were not immediately available. These gated “storage” pockets would allow for N2 availability whenever the FeMo-cofactor reaches the appropriate level of reduction. In a similar vein, it has been proposed that large sections of tunnels serve as H2 ‘gas reservoirs’ in hydrogenases. 53, 54
Weyman and coworkers studied the α-V75I and α-V76I variants in a similar N2ase from Anabaena variabilis (which would correspond to α-V7OI and α-V71I in Av).55 Although as expected, their α-V75I variant showed reduced N2 fixation activity and α-V76I substitution had no effect. Since our proposal for gated access between pockets is dynamic, we can accommodate these findings by simply allowing for comparable gating by isoleucine or valine residues at the Av position 71.
Some of the other channels illustrated in Figure 2 presumably play a role in the egress of the mandatory H2 co-product as well as NH3 or NH4+. In this area we are agnostic, since Lautier has noted that hydrophobic molecules sometimes travel through hydrophilic channels and vice versa.56 However, it seems unlikely that evolved H2 would exit through our proposed substrate channel, since this might limit N2 influx to the FeMo-cofactor. It seems logical that N2ase would have a mechanism or ‘pressure relief’ for the active site region to release H2 from the active site without interfering with the catalytic process. Overall, N2ase has to handle the flow of electrons, protons, N2, H2, and NH3, and the nature of ‘traffic control’ in this remarkable enzyme remains to be completely understood.
Supplementary Material
Acknowledgments
Computational work was performed on the Hopper Supercomputing Cluster from the National Energy Research Scientific Computing center through ERCAP request #87354. The authors would also like to thank William Newton and Christie Dapper for scientific discussions and assistance with the manuscript.
Funding
This work was funded by the National Institutes of Health grant GM-65440.
Abbreviations
- Av
Azotobacter vinelandii
- N2
dinitrogen
- CO
carbon monoxide
- N2ase
nitrogenase
- FeMo-co
the iron-molybdenum cofactor of the MoFe protein
- MoFe protein
the larger molybdenum-iron containing protein of nitrogenase
- Fe protein
the smaller iron containing protein of nitrogenase
- IR
infrared
- FT-IR
fourier transform infrared spectroscopy
- MB
myoglobin
- MBCO
myoglobin inhibited with CO
- ns
nanoseconds
- ps
picoseconds
- PMF
potential of mean force
- LT
Lowe-Thorneley model of nitrogen fixation
Footnotes
Contains a detailed description of the methods and figures S1 and S2: the dynamics course for N2 and an image of the crater region respectively. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hoffman BM, Dean DR, Seefeldt LC. Climbing Nitrogenase: Toward a Mechanism of Enzymatic Nitrogen Fixation. Acc Chem Res. 2009;42:609–619. doi: 10.1021/ar8002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seefeldt LC, Hoffman BM, Dean DR. Mechanism of Mo-Dependent Nitrogenase. Ann Rev Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canfield DE, Glazer AN, Falkowski PG. The Evolution and Future of Earth’s Nitrogen Cycle. Science. 2010;330:192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 4.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Nitrogenase MoFe-Protein at 1.16 Å Resolution: A Central Ligand in the FeMo-Cofactor. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 5.Spatzal T, Aksoyoglu M, Zhang L, Andrade SLA, Schleicher E, Weber S, Rees DC, Einsle O. Evidence for Interstitial Carbon in Nitrogenase FeMo Cofactor. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancaster KM, Roemelt M, Ettenhuber P, Hu YL, Ribbe MW, Neese F, Bergmann U, DeBeer S. X-ray Emission Spectroscopy Evidences a Central Carbon in the Nitrogenase Iron-Molybdenum Cofactor. Science. 2011;334:974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiig JA, Hu Y, Lee CC, Ribbe MW. Radical SAM-Dependent Carbon Insertion into the Nitrogenase M-Cluster. Science. 2012;337:1672–1675. doi: 10.1126/science.1224603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess BK, Lowe DJ. Mechanism of molybdenum nitrogenase. Chem Rev. 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 9.Lee CC, Hu Y, Ribbe MW. Vanadium Nitrogenase Reduces CO. Science. 2010;329:642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CC, Hu YL, Ribbe MW. Tracing the Hydrogen Source of Hydrocarbons Formed by Vanadium Nitrogenase. Angew Chem. 2011;50:5545–5547. doi: 10.1002/anie.201100869. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZY, Dean DR, Seefeldt LC. Molybdenum Nitrogenase Catalyzes the Reduction and Coupling of CO to Form Hydrocarbons. J Biol Chem. 2011;286:19417–19421. doi: 10.1074/jbc.M111.229344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang ZY, Moure VR, Dean DR, Seefeldt LC. Carbon dioxide reduction to methane and coupling with acetylene to form propylene catalyzed by remodeled nitrogenase. Proc Nat Acad Sci. 2012;109:19644–19648. doi: 10.1073/pnas.1213159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seefeldt LC, Yang ZY, Duval S, Dean DR. Nitrogenase reduction of carbon-containing compounds. Biochim Biophys Acta. 2013;1827:1102–1111. doi: 10.1016/j.bbabio.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igarashi RY, Seefeldt LC. Nitrogen Fixation: The Mechanism of the Mo-Dependent Nitrogenase. Crit Rev Biochem Mol Biol. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 15.Elber R. Ligand diffusion in globins: simulations versus experiment. Curr Op Struc Biol. 2010;20:162–167. doi: 10.1016/j.sbi.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durrant MC. Controlled protonation of iron-molybdenum cofactor by nitrogenase: a structural and theoretical analysis. Biochem J. 2001;355:569–576. doi: 10.1042/bj3550569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barney BM, Yurth MG, Dos Santos PC, Dean DR, Seefeldt LC. A substrate channel in the nitrogenase MoFe protein. J Biol Inorg Chem. 2009;14:1015–1022. doi: 10.1007/s00775-009-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dance I. A molecular pathway for the egress of ammonia produced by nitrogenase. Sci Reports. 2013;3:1–9. doi: 10.1038/srep03237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith D, Danyal K, Raugei S, Seefeldt LC. Substrate channel in nitrogenase revealed by a molecular dynamics approach. Biochemistry. 2014;53:2278–2285. doi: 10.1021/bi401313j. [DOI] [PubMed] [Google Scholar]
- 20.Morrison CN, Hoy JA, Zhang L, Einsle O, Rees DC. Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites. Biochemistry. 2015;54:2052–2060. doi: 10.1021/bi501313k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dance I. The controlled relay of multiple protons required at the active site of nitrogenase. Dalton T. 2012;41:7647–7659. doi: 10.1039/c2dt30518f. [DOI] [PubMed] [Google Scholar]
- 22.Fisher K, Hare ND, Newton WE. Another role for CO with nitrogenase? CO stimulates hydrogen evolution catalyzed by variant Azotobacter vinelandii Mo-nitrogenases. Biochemistry. 2014;53:6151–6160. doi: 10.1021/bi500546k. [DOI] [PubMed] [Google Scholar]
- 23.Scott A, Pelmenschikov V, Guo Y, Wang H, Yan L, George S, Dapper C, Newton W, Yoda Y, Tanaka Y, Cramer SP. Structural Characterization of CO-Inhibited Mo-Nitrogenase by Combined Application of NRVS, EXAFS, and DFT: New Insights into the Effects of CO Binding and the Role of the Interstitial Atom. J Am Chem Soc. 2014;136:15942–15954. doi: 10.1021/ja505720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meuwly M. Using small molecules to probe protein cavities: The myoglobin–XO (X = C, N) family of systems. Eur Phys J Special Topics. 2007;141:209–216. [Google Scholar]
- 25.Hong GY, Pachter R. Inhibition of Biocatalysis in Fe-Fe Hydrogenase by Oxygen: Molecular Dynamics and Density Functional Theory Calculations. ACS Chem Biol. 2012;7:1268–1275. doi: 10.1021/cb3001149. [DOI] [PubMed] [Google Scholar]
- 26.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the heme-Cu oxidases. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 27.Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontecilla-Camps JC, Amara P, Cavazza C, Nicolet Y, Volbeda A. Structure-function relationships of anaerobic gas-processing metalloenzymes. Nature. 2009;460:814–822. doi: 10.1038/nature08299. [DOI] [PubMed] [Google Scholar]
- 29.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaffe E, Fishelovitch D, Wolfson HJ, Halperin D, Nussinov R. MolAxis: Efficient and accurate identification of channels in macromolecules. Proteins - Struct Func Bioinform. 2008;73:72–86. doi: 10.1002/prot.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raunest M, Kandt C. dxTuber: Detecting protein cavities, tunnels and clefts based on protein and solvent dynamics. J Mol Graphics Mod. 2011;29:895–905. doi: 10.1016/j.jmgm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Lin TL, Song G. Efficient mapping of ligand migration channel networks in dynamic proteins. Proteins - Struct Func Bioinform. 2011;79:2475–2490. doi: 10.1002/prot.23071. [DOI] [PubMed] [Google Scholar]
- 33.Schmidtke P, Bidon-Chanal A, Luque FJ, Barril X. MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics. 2011;27:3276–3285. doi: 10.1093/bioinformatics/btr550. [DOI] [PubMed] [Google Scholar]
- 34.Berka K, Hanak O, Sehnal D, Banas P, Navratilova V, Jaiswal D, Ionescu CM, Varekova RS, Koca J, Otyepka M. MOLEonline 2.0: interactive web-based analysis of biomacromolecular channels. Nuc Acids Res. 2012;40:W222–W227. doi: 10.1093/nar/gks363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alben JO, Beece D, Bowne SF, Doster W, Eisenstein L, Frauenfelder H, Good D, McDonald JD, Marden MC, Moh PP, Reinisch L, Reynolds AH, Shyamsunder E, Yue KT. Infrared spectroscopy of photodissociated carboxymyoglobin at low temperatures. Proc Nat Acad Sci. 1982;79:3744–3748. doi: 10.1073/pnas.79.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim MH, Jackson TA, Anfinrud PA. Midinfrared Vibrational-Spectrum of CO After Photodissociation From Heme - Evidence For A Ligand Docking Site in the Heme Pocket of Hemoglobin and Myoglobin. J Chem Phys. 1995;102:4355–4366. [Google Scholar]
- 37.Lehle H, Kriegl JM, Nienhaus K, Deng PC, Fengler S, Nienhaus GU. Probing electric fields in protein cavities by using the vibrational stark effect of carbon monoxide. Biophys J. 2005;88:1978–1990. doi: 10.1529/biophysj.104.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nienhaus K, Olson JS, Franzen S, Nienhaus GU. The origin of stark splitting in the initial photoproduct state of MbCO. J Am Chem Soc. 2005;127:40–41. doi: 10.1021/ja0466917. [DOI] [PubMed] [Google Scholar]
- 39.Srajer V, Ren Z, Teng TY, Schmidt M, Ursby T, Bourgeois D, Pradervand C, Schildkamp W, Wulff M, Moffat K. Protein conformational relaxation and ligand migration in myoglobin: A nanosecond to millisecond molecular movie from time-resolved Laue X-ray diffraction. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 40.Tetreau C, Blouquit Y, Novikov E, Quiniou E, Lavalette D. Competition with xenon elicits ligand migration and escape pathways in myoglobin. Biophys J. 2004;86:435–447. doi: 10.1016/S0006-3495(04)74120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer SM. Ph D. University of East Anglia; 2000. X-ray structure determination of Klebsiella pneumoniae nitrogenase component 1. [Google Scholar]
- 42.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Structural basis of biological nitrogen fixation. Phil Trans R Soc A. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 43.Daskalakis V, Varotsis C. Binding and Docking Interactions of NO, CO and O-2 in Heme Proteins as Probed by Density Functional Theory. Int J Mol Sci. 2009;10:4137–4156. doi: 10.3390/ijms10094137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan L, Dapper CH, George SJ, Wang HX, Mitra D, Dong WB, Newton WE, Cramer SP. Photolysis of ‘Hi-CO’ Nitrogenase–Observation of a Plethora of Distinct CO Species via Infrared Spectroscopy. Eur J Inorg Chem. 2011;2011:2064–2074. doi: 10.1002/ejic.201100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elber R, Karplus M. Enhanced Sampling in Molecular Dynamics - Use of the Time-Dependent Hartree Approximation for a Simulation of Carbon-Monoxide Diffusion through Myoglobin. Journal of the American Chemical Society. 1990;112:9161–9175. [Google Scholar]
- 47.Yan LF, Pelmenschikov V, Dapper CH, Scott AD, Newton WE, Cramer SP. IR-Monitored Photolysis of CO-Inhibited Nitrogenase: A Major EPR-Silent Species with Coupled Terminal CO Ligands. Chem-Eur J. 2012;18:16349–16357. doi: 10.1002/chem.201202072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarma R, Barney BM, Keable S, Dean DR, Seefeldt LC, Peters JW. Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70(Ile) MoFe protein variant. Journal of Inorganic Biochemistry. 2010;104:385–389. doi: 10.1016/j.jinorgbio.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nutt DR, Meuwly M. Theoretical investigation of infrared spectra and pocket dynamics of photodissociated carbonmonoxy myoglobin. Biophys J. 2003;85:3612–3623. doi: 10.1016/S0006-3495(03)74779-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abbruzzetti S, Spyrakis F, Bidon-Chanal A, Luque FJ, Viappiani C. Ligand migration through hemoprotein cavities: insights from laser flash photolysis and molecular dynamics simulations. Phys Chem Chem Phys. 2013;15:10686–10701. doi: 10.1039/c3cp51149a. [DOI] [PubMed] [Google Scholar]
- 51.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. Journal of Computational Chemistry. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 52.Burgess BK, Lowe DJ. Mechanism of Molybdenum Nitrogenase. Chem Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 53.Chandrayan SK, McTernan PM, Hopkins RC, Sun JS, Jenney FE, Adams MWW. Engineering Hyperthermophilic Archaeon Pyrococcus furiosus to Overproduce Its Cytoplasmic [NiFe]-Hydrogenase. Journal of Biological Chemistry. 2012;287:3257–3264. doi: 10.1074/jbc.M111.290916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montet Y, Amara P, Volbeda A, Vernede X, Hatchikian EC, Field MJ, Frey M, FontecillaCamps JC. Gas access to the active site of Ni-Fe hydrogenases probed by X-ray crystallography and molecular dynamics. Nature Struc Biol. 1997;4:523–526. doi: 10.1038/nsb0797-523. [DOI] [PubMed] [Google Scholar]
- 55.Weyman PD, Pratte B, Thiel T. Hydrogen production in nitrogenase mutants in Anabaena variabilis. FEMS Microbiology Letters. 2010;304:55–61. doi: 10.1111/j.1574-6968.2009.01883.x. [DOI] [PubMed] [Google Scholar]
- 56.Lautier T, Ezanno P, Baffert C, Fourmond V, Cournac L, Fontecilla-Camps JC, Soucaille P, Bertrand P, Meynial-Salles I, Leger C. The quest for a functional substrate access tunnel in FeFe hydrogenase. Faraday Discussions. 2011;148:385–407. doi: 10.1039/c004099c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.